INTRODUCTION
Klebsiella pneumoniae is currently a life-threatening bacterial pathogen due to hypervirulence and antibiotic resistance and is considered a “critical priority” for research and development of new antibiotics (Tacconelli et al., 2019). Klebsiella pneumoniae causes an increasing number of severe infections with limited treatment options (Paczosa and Mecsas, 2016). The most common cause of microbial abscess is community-acquired and hypervirulent K. pneumoniae (Breurec et al., 2016; Chen et al., 2020; Siu et al., 2012).
The hypervirulence of K. pneumoniae is strongly influenced by its ability to produce a thick layer which consists of the cell envelope and capsule polysaccharide. The cell envelope and capsule polysaccharide protects the bacteria from the environment and antimicrobial compounds (Lee et al., 2014). The destruction of the integrity of the K. pneumoniae cell envelope and capsule is an avenue to overcome the pathogenic nature of K. pneumoniae (Hsieh et al., 2013; Lee et al., 2013; Nevermann et al., 2019).
The cell envelope in K. pneumoniae consists of a lipid bilayer membrane (Nevermann et al., 2019). The strength of membrane integrity is maintained by the intercalation between the murein layer (lpp), outer membrane protein (OMP), and peptidoglycan-associated lipoprotein (pal). OMP and pal interact non-covalently with the lpp (Hsieh et al., 2013). Meanwhile, pal plays a vital role in connecting the OM and the peptidoglycan layer (Graham et al., 2021). At the same time, lpp plays an essential role in the integrity and permeability of the membrane (Graham et al., 2021). Pal, lpp, and ompA are structural genes encoding OMPs that are highly conserved among enteric Gram-negative bacteria (Hellman et al., 2000). This indicates the importance of these three proteins in Gram-negative bacteria. Blocking the expression of the production of these proteins will damage the cell surface and weaken the bacteria. Murein layer- (lpp-) deficient bacteria exhibit increased OM permeability, periplasmic component leakage, and increased outer membrane vesicle (OMV) release (Nevermann et al., 2019; Schwechheimer et al., 2015). Bacteria lacking pal exhibit decreased outer membrane integrity, increased susceptibility to antibiotics and detergents, increased periplasmic protein leakage, and increased OMV release (Graham et al., 2021; Schwechheimer et al., 2013). The collapse of the cell envelope integrity will affect the structures dependent on it, such as capsular polysaccharides (CPSs), fimbriae type 1, and siderophores. Though these three structures are important virulence factors for the pathogenicity of extended-spectrum beta-lactamase-producing K. pneumoniae (Gharrah et al., 2017), the destruction of cell envelope integrity is an alternative measure in inhibiting the bacterial growth.
Aspergillus oryzae is known as a traditional fermented food fermenter in Japan. In addition, A. oryzae is a heterogeneous source of enzymes (Flipphi et al., 2009). Aspergillus oryzae received safety certification from the United States Food and Drug Administration and World Health Organization (Nacef et al., 2020). Many studies explored the enzymes produced by A. oryzae in food and fermentation fields. However, few studies have investigated the potential of A. oryzae against bacterial cell envelope integrity. Here, we looked at the effects of the secondary metabolites of A. oryzae (SMAO) on the integrity of the cell envelope of KPESBL .
MATERIALS AND METHODS
Microbial strains
The bacteria KPESBL (ID.100029) was obtained from the Microbiology Laboratory of Faculty of Medicine at Brawijaya University in Malang, Indonesia. The strain was cultured in Luria broth media and incubated at 37°C for 16–18 hours. The inoculum was diluted to 106 CFU/ml equivalent to McFarland standard using sterile saline. The fungi strain A. oryzae van Tieghem was obtained from the Indonesian Culture Collection.
Preparation of a fungal fermentation and protein production
In a 250 ml Erlenmeyer flask containing 2% glucose, 100 ml of potato dextrose broth media was inoculated with 8 mm (diameter) of fungal mycelium (Nacef et al., 2020; Rachma et al., 2022). Under static condition, the flask was incubated in a shaker incubator (Boxun BSD-TX270) at 27°C for 72 hours (OD600 = 1.2). Following incubation, the culture was filtered using 0.22-micron filter paper (Whatman, Sigma Aldrich). The supernatant was centrifuged at 12,000 rpm for 15 minutes at 4°C and used as an extracellular protein source. Extracellular proteins were precipitated at 80% saturation with ammonium sulfate (Wingfield, 1998). The precipitate was agitated in an ice bath for 1 hour. The protein was also centrifuged at 12,000 rpm for 15 minutes at 4°C. The protein precipitate was dialyzed with a dialysis bag (Sigma D0405). The dialysis lasted 12 hours in 0.01 M phosphate buffer pH 7 in a 100 ml sample volume (Duong-Ly and Gabelli, 2014). The protein was then used for the assay.
Qualitative real-time polymerase chain reaction (RT PCR)
Total RNA was extracted from K. pneumoniae bacteria treated with A. oryzae extracellular protein (AOEP) as well as without AOEP using the hot phenol technique as described previously by Jahn et al. (2008). The quality of RNA was evaluated using NanoDrop (ND-1000; Thermo Scientific) and an Agilent 2100 Bioanalyzer with a Pico chip (Agilent Technologies) after DNA was eliminated with TURBO DNA-free (Ambion, Inc.). The absence of contaminating DNA was controlled by the absence of amplification products after 35 cycles of quantitative PCR (qPCR). About 1 µg of RNA, random hexamer primers (0.2 µg/µl), and M-MulV-RT (20 U/µl, reverse transcriptase of Moloney Murine Leukemia Virus; Thermo Scientific) were used to produce cDNA. Primer3Plus software was used to designate certain primers (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/).
The pal protocol is GTCAACTGCTCTACCAACTG, the lpp protocol is CAACAGTCGGCTGATTGGG, and the ompA protocol is TTAAGCCGCCGGCTGAGTTAC. For LightCycler reactions, the following components were combined into a master mixture: 3.0 µl of PCR-grade water, 1.0 µl of forward primer (10 µM), 1.0 µl of reverse primer (10 µM), 10 µl of 2× SYBR Green I Master Mix, and 5.0 µl of cDNA (50–100 ng) are required. The LightCycler 480 instrument was filled with a multiwell plate that had been sealed with foil, centrifuged at 1,500 g for 2 minutes, and then loaded with the centrifuged samples (Roche). Each sample underwent amplification in triplicate. In addition to each reaction, reactions without template (water) and reverse transcriptase (RNA) served as controls. RT-PCR was performed under the following conditions: denaturation (95°C for 10 minutes); amplification and quantification repeated 45 times (95°C for 10 seconds, 57°C for 20 seconds, and 72°C for 30 seconds with a single presence measurement); melting curve (95°C for 10 seconds, 65°C for 1 minute with continuous fluorescence measurement at 97°C); finally, a cooling step of 10 seconds at 40°C. After each run, the melting curve was analyzed to ensure the specificity of the primers. 16S rRNA was utilized as a normalization reference gene, and relative gene expression was determined using the 2-1Ct technique (Livak and Schmittgen, 2001).
Production of CPS
The CPSs were isolated and measured according to the method of Lin et al. (2013), with some modifications. The culture grown overnight was injected into 9.5 ml of Luria-Bertani (LB) broth along with 0.5 ml of cell lysate and incubated for 24 hours at 30°C. The associated late-log phase cells were extracted by centrifuge at 8,500 rpm for 30 minutes at 2°C. The released CPS was precipitated by adding three volumes of cold ethanol to the filtered supernatant and incubation at 2°C overnight. After 30 minutes of centrifugation at 5,000 g, the precipitated CPS was recovered and dissolved in 1 µl of deionized water. Phosphate buffered saline (PBS) media culture containing no enzymes served as the control. The bacterial cells were extracted, resuspended in sterile PBS, and treated for 30 minutes at room temperature in the dark with 15 g/ml Congo red. The absorbance of the supernatant was measured at 490 nm (A490).
Siderophore quantification
Using a modified microtiter plate, siderophore manufacturing was accomplished. 0.5 ml of inoculated (5 µl inoculum containing 108 CFU/ml) broth yielded the supernatant. Each bacterial culture’s supernatant (100 µl) was placed in a separate well of the microplate (Sigma CLS3474), followed by 100 µl of chrome azurol sulphonate (CAS) reagent. Using a microplate reader, the optical density of each sample was measured at 630 nm following incubation (Spectra Max M5e). Four replicates of each strain were plated in 96-well plates, (the experiment was done four times) and siderophore was calculated using the following formula: siderophore = (Ar − As)/Ar × 100, where Ar = absorbance of reference (CAS solution and uninoculated broth) and As = absorbance of sample (CAS solution and cell-free supernatant of sample).
Scanning electron microscope (SEM)
According to the manufacturer’s instructions, biofilm production on the surface of plastic coverslips (13 mm in diameter) (Thermanox®) was measured. Each well of a 24-well plate containing 200 µl of K. pneumoniae in LB broth and 200 µl of SMAO was covered with a coverslip and incubated for 6 hours. The culture media was eliminated, and the wells were cleansed with water multiple times. Air-dried coverslips were affixed to larger (22 × 22 mm) glass coverslips for imaging with the FEI Verios 460 SEM (FEI, Oregon, USA).
Agglutination assay
Using yeast agglutination tests, the expression of type 1 fimbriae by the K. pneumoniae strains was evaluated. As mentioned earlier, the experiments were conducted on Kline concavity slides with minor modifications (Stærk et al., 2016). The bacterial strains were sub-cultured overnight before being inoculated (1:100) into either C8-HSL-enriched or unenriched LB broth. About 5% (w/v) concentration of bacteria was added to a PBS-prepared suspension of Saccharomyces cerevisiae cells. The level of the agglutination was captured using a digital camera. Type 1 fimbriae exclusively facilitate the agglutination of yeast cells due to their affinity for mannose, a relatively abundant residue on yeast cell surfaces. Experiments were also conducted with 5% D-(+)-Mannose (Sigma-Aldrich) to confirm that type 1 fimbriae were responsible for the agglutination. Three tests were performed on each sample. Each K. pneumoniae strain was tested three times.
Growth curve analysis
To confirm the antibacterial activity of SMAO at sub-minimum inhibitory concentration (MIC) (1/32, 1/64, 1/128, and 1/256 MIC or 9.375, 4.687, 2.343, and 1.171 µg/ml), K. pneumoniae producing extended-spectrum beta-lactamase (KPESBL) was incubated for 0–24 hours with or without various doses of SMAO. In a 250 ml Erlenmeyer flask containing 20 ml of LB broth supplemented with SMAO, 1% KPESBL (OD600 = 0.9) was incubated (sub-MICs = 9.375, 4.687, 2.343, and 1.171 µg/ml). In a rotatory shaker, the culture was shaken at 30°C and 150 revolutions per minute. From 0 to 72 hours, the growth of bacteria was measured by UV–Visible spectrophotometry (U-290, Hitachi Company, Tokyo, Japan) at OD600.
RESULTS
SMAO was able to reduce the growth curve velocity of KPESBL
The bacterial growth curve was measured with and without SMAO treatment to know the impact of SMAO on KPESBL. The concentrations of SMAO below the MIC test were used for 1/32, 1/64, 1/128, and 1/256 MIC or (9.375, 4.687, 2.343, and 1.171 μg/ml subsequently).To confirm the non-antibacterial activity of SMAO at sub-MIC concentrations (9.375, 4.687, 2.343, and 1.171 µg/ml), the 0–24 hours activity of KPESBL treated with or without various SMAO concentrations was measured (Table 1). The growth curve shows that KPESBL treated with SMAO in sub-MIC began to enter the logarithmic phase in the same time as the control. Nevertheless, the growth of treated KPESBL began to differ from the control group starting at 9 hours as it entered the stationary phase and after that. The SMAO treatment group at all concentrations demonstrated slower growth compared to the control group at the stationary phase (>14 hours) (Fig. 1). These results showed that SMAO at different concentrations efficiently inhibited growth under the conditions.
Determination of the siderophore activity
Determination of the siderophore activity A slight color change indicated signs of siderophore reduction by SMAO to orange (still bluish-black). The results of the CAS test showed that SMAO at 150 μg/ml had an R-value of 0.891 and decreased as the dose of SMAO decreased (75 μg/ml = 6%, 37.5 μg/ml = 1.5%, and 18.75 μg/ml = 0%) (Fig. 2). The linear regression analysis showed that SMAO could inhibit the siderophore KPESBL in a dose-dependent manner (R-value = 0.891).
Determination of capsular exopolysaccharide
The CPS was measured to determine the effect of the SMAO on the bacteria protective structure.
Centrifugation at high speed and precipitation using ethanol was used to determine the capsular exopolysaccharide production. KPESBL treated with 150 μg/ml SMAO can reduce matrix bonding with Congo red dye by as much as 50.14% (Fig. 3). The activity declined along with the decrease in SMAO dose by 17.27%, 33.06%, and 39.49% (75 μg/ml = 39.49%, 37.5 μg/ml = 33.06%, and 18.75 μg/ml = 17.27%). SMAO 150 μg/ml’s ability to reduce the amine bond of the aromatic capsular exopolysaccharide KPESBL was 25% higher than the antibiotic kanamycin (34%). From the linear regression analysis, SMAO could inhibit the growth of KPESBL biofilm in a dose-dependent manner (R-value = 0.959). This shows that SMAO might be able to kill microbes by breaking down cell-bound (capsular) polysaccharides.
SMAO enables the expression of cell envelope-related genes
The relative expression of the genes related to cell envelope formation resistance was examined in KPESBL isolates treated with or without SMAO using qRT-PCR. As shown in Figure 3, the relative expression of ompA, lpp, and pal KPESBL exposed to SMAO was significantly down-regulated, with decreased expression of the ompA gene showing a significant difference compared to the untreated control. Then, the cell envelope integrity was measured using qRT-PCR.
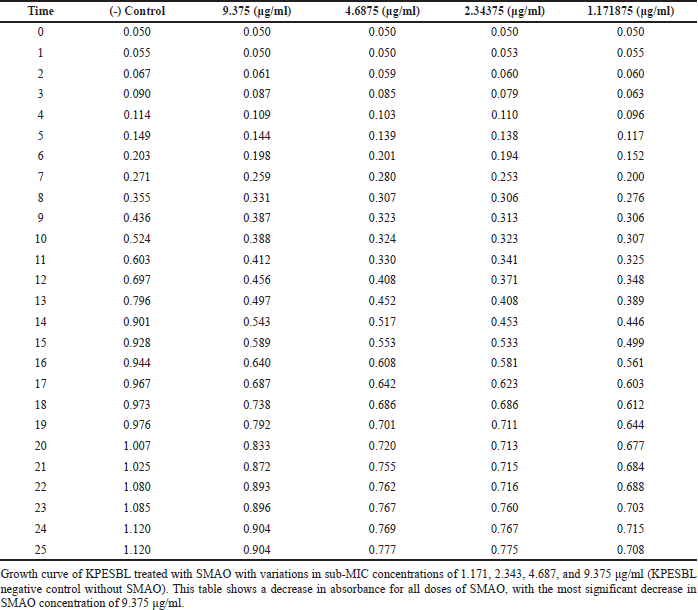 | Table 1. Effect of SMAO on the growth curve of KPESBL. [Click here to view] |
Using the delta-delta Ct method method (2 DDCt), quantitation of RT-qPCR results (Livak and Schmittgen, 2001) is represented as “Target/adh3 fold change.” The analysis showed that SMAO down-regulated the gene expression of ompA, lpp, and pal, compared to control cells that were not exposed to SMAO (Fig. 4). The ompA, lpp, and pal encoding porin outer membrane protein A; lipoprotein A and peptidoglycan-associated lipoprotein, respectively. As a result, this reflects the integrity of KPESBL’s cell envelope.
SEM analysis
To learn more about the effects of SMAO on cell envelope integrity, observations were made using SEM at 20,000× and 40,000× magnifications. KPESBL cells were observed without SMAO administration (Fig. 5).
The SEM confirmed the effect of SMAO on the cell envelope integrity of KPESBL. The group without SMAO treatment (Fig. 5A) showed intact cells that were thicker than the SMAO treatment group. Capsules in the SMAO treatment group appeared to be more intact with solid cell envelope integrity compared to the SMAO exposure group. Meanwhile, in the group without SMAO exposure, there was a decrease in cell size, thinning of the capsule, and deformation of cell shape, and the formation of pores was detected. This indicated the disruption of the integrity of the KPMDR cell envelope compared to cells not treated with AOEP (Fig. 5B). The SEM results show that SMAO effectively reduces the integrity of the cell envelope.
Agglutination assay
Because type 1 fimbriae have a high affinity for mannose-containing receptors on the surface of yeast cells, yeast agglutination is suggestive of type 1 fimbriae production. The yeast agglutination assay was utilized to demonstrate that SMAO therapy decreases the expression of type 1 fimbriae.
As seen in Figure 6, KPESBL that had not been exposed to SMAO could agglutinate yeast cells more intensely than that which had been exposed to it. Adding mannose apparently stopped KPESBL from agglutinating yeast cells, proving that type 1 fimbriae mediate agglutination. Mannose’s ability to prevent yeast cells from agglutinating proves that type 1 fimbriae alone are responsible for this process. Based on the yeast agglutination test results, SMAO treatment reduced yeast agglutination by KPESBL.
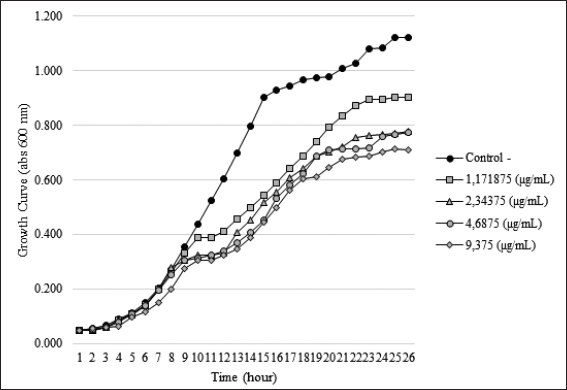 | Figure 1. Effect of SMAO on the growth of KPESBL. Growth curve of KPESBL treated with EO with variations in sub-MIC concentrations of 1.171, 2.343, 4.687, and 9.375 μg/ml (KPESBL negative control without SMAO). The figure shows a decrease in the growth curve for all SMAO doses, with the most significant decrease at the SMAO concentration of 9.375 μg/ml. KPESBL exposed to SMAO doses of 1.171, 2.343, 4.687, and 9.375 μg/ml entered the stationary phase at 9 hours, while in the negative control, KPESBL without SMAO exposure entered the stationary phase at 14 hours. [Click here to view] |
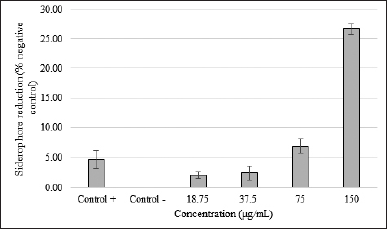 | Figure 2. SMAO activity against the siderophore KPESBL. After 24 hours of incubation with various concentrations of SMAO, the inhibitory effect of different concentration of SMAO on the siderophore KPESBL after incubation for 24 hours. Siderophore reduction by SMAO with a concentration of 18.75, 37.5, 75, and 150 μg/ml was 2.13%, 2.49%, 6.95%, and 26.65%, respectively. The kanamycin was only able to reduce 4.67% of the KPESBL siderophore. The siderophore inhibition value of KPESBL by SMAO was better than kanamycin (26.65% < 4.67%). The bars indicate the standard error. [Click here to view] |
DISCUSSION
Microbial resistance is a significant health problem and threat worldwide. The failure to treat bacterial infections has attracted the attention of researchers, as has been the case with diseases caused by KPESBL (World Health Organization, 2020). The severity of the KPESBL infection is due to the limited treatment options (Baker et al., 2019; Navan-Venezia et al., 2017; Virawan et al., 2020). Hence, it necessitates the need to develop new alternatives, including natural products. One strategy for finding new antibiotics is exploring limited studied ecosystems coupled with extreme factors (salinity, pH, etc.) (Alsharif et al., 2020). We used the isolate A. oryzae (Ahlb.) Cohn, which was found in the Teluk Kodek Pemenang, West Lombok, Indonesia, which is an area with a lot of toxic phytoplankton in water.
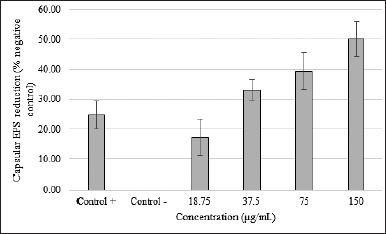 | Figure 3. SMAO effect on capsular exopolysaccharide KPESBL. SMAO on capsular exopolysaccharide KPESBL after incubation for 24 hours with different concentrations. The inhibition of capsular exopolysaccharide by SMAO at concentrations of 18.75, 37.5, 75, and 150 μg/ml was 17.27%, 33.06%, 39.49%, and 50.14%, respectively. The highest capsular exopolysaccharide inhibition by SMAO at a 150 μg/ml concentration was 50.14%, whereas antibiotic kanamycin was only able to reduce 24.81%. The value of CPS inhibition by SMAO was stronger than kanamycin (50.14% < 24.81%). The bars indicate the standard error. [Click here to view] |
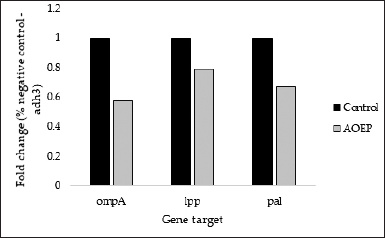 | Figure 4. Effect of 150 μg/ml SMAO on the expression of the gene responsible for the cell envelope stability of KPESBL. The genes whose expression was measured were ompA, lpp, and pal. The SMAO treatment concentration was 150 μg/ml, while the control group was KPESBL without exposure to SMAO. [Click here to view] |
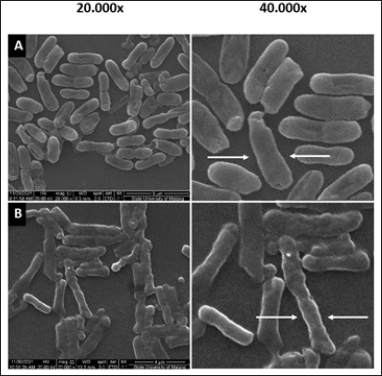 | Figure 5. The KPESBL group without SMAO treatment shows a thicker KPESBL cell size than the SMAO treatment group (A). The capsules in this group appeared to be intact with solid cell envelope integrity when compared to the KPESBL group treated with SMAO (B). In the SMAO group, decrease in cell size, thinning of the capsule, and deformation of the shape of the cells observed indicated the disruption of the integrity of the KPESBL cell envelope (see arrows in the figure). [Click here to view] |
To combat KPESBL infection, we offered a new strategy by inhibiting virulence factors. Currently, there are four virulence factors in K. pneumoniae that cause this bacterium to be more pathogenic, that is, capsule, lipopolysaccharides (LPS), fimbriae, and siderophores (Paczosa and Mecsas, 2016). The capsule is the extracellular polysaccharide matrix that protects the bacteria from antibacterial compounds (Cress et al., 2014), while LPS, also known as endotoxin, is a significant and essential component of the outer leaflet of the cell membrane (Willis and Whitfield, 2013). In contrast, fimbriae are membrane-bound structures that perform adherence to biotic and abiotic surfaces (Paczosa and Mecsas, 2016). In addition, K. pneumoniae makes and secretes siderophores to chelate iron from the environment (Chen et al., 2020). Therefore, the destruction of this structure can be an alternative to the retardation of bacterial growth. To the best of our knowledge, this study is the first to demonstrate the mechanism of inhibition of KPESBL by SMAO. This study demonstrated that SMAO can inhibit siderophore production. The decrease in siderophore levels mediated the inhibition of KPESBL growth (Chen et al., 2020; Nevermann et al., 2019; Wilson et al., 2016). Siderophores depletion results in hindrance in the bacteria’s ability to absorb iron from the media (Caza and Kronstad, 2013), leading to inhibition in bacterial (KPESBL) growth (Chen et al., 2020). Siderophore requirements occur in the early log phase in response to high iron requirements (Sinha et al., 2019). The growth curve showed that SMAO inhibits the exponential phase of KPESBL. Lack of iron due to loss of siderophore hampers the growth rate (Sinha et al., 2019). The decrease in siderophore in this phase will inhibit cell division.
Besides siderophore, SMAO was able to reduce the number of CPSs. CPS is one of the main virulence factors of K. pneumoniae (Lee et al., 2013). This structure protects the KPESBL from phagocytosis (Khatoon et al., 2018). Congo red used in this study can form complexes with polysaccharides in a helical conformation and cause a red color shift when measured at 490 nm (Jiao et al., 2021). However, if an enzyme breaks down the hydrogen polysaccharide, the three-stranded helical conformation will be destroyed. After that, the decrease in optical density is measured using a spectrophotometer. No data still describes the compound produced by Aspergillus spp. that can break down CPSs. Amylase is known to be able to hydrolyze glycosidic enzymes into simple sugars in polysaccharides (Houda et al., 2020). In addition, the decrease in CPSs can be caused by a low concentration in iron following siderophore depletion. CPS biosynthesis requires iron (Zubaidah et al., 2014). Iron is vital for the metabolism of L-tryptophan, 5’-MTA, spermidine, CMP, and L-leucine, which is essential for forming the extracellular matrix, that is, exopolysaccharide (Guo et al., 2021). The above results show that AOEP effectively reduces cell-free CPS in KPESBL. This damage will certainly weaken KPESBL.
SMAO also exhibited activity to down-regulate the expression of ompA, pal, and lpp genes, which leads to membrane instability. Disruption of the cell envelope by SMAO will destroy bacteria. This finding confirmed cell envelope integrity through SEM. This demonstrated that KPESBL exposed to SMAO decreased in cell size, thinning CPS, deformation of cell shape, and formation of pores. The cell envelope of Gram-negative bacteria is a multi-layered structure that is essential for the survival of bacteria (Misra et al., 2014). It serves as a barrier against harmful compounds in the external environment (Sutterlin et al., 2016). Maintaining its integrity is a matter of life and death for the bacteria. The interaction between ompA, pal, and lpp is essential in maintaining the integrity of the cellular envelope (Nevermann et al., 2019). These four factors lead to the disruption of the integrity of the KPESBL cell envelope. These results further strengthen SMAO’s ability to inhibit KPESBL growth through impaired cell envelope integrity.
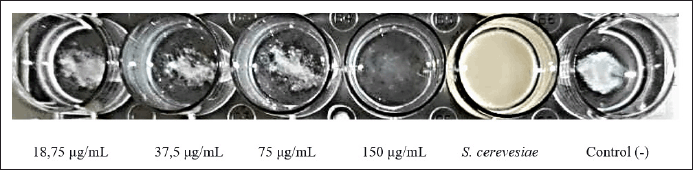 | Figure 6. The KPESBL group without SMAO treatment was able to agglutinate yeast cells with more intensity than the SMAO treatment group, while in the SMAO exposure group, the ability of KPESBL to agglutinate yeast cells decreased. The most significant decrease in agglutination was seen at 150 g/ml SMAO exposure. [Click here to view] |
Lastly SMAO also decreased the fimbrial adhesion ability of KPESBL. Fimbrial adhesions function in bacterial cells attachment to the host and protect them from being excreted from the urinary tract with urine (Gunther et al., 2001). A yeast agglutination assay represented the fimbrial adhesion ability of KPESBL after SMAO exposure (Stærk et al., 2016).
Showing inhibited yeast agglutination by the KPESBL strain indicates type 1 fimbriae dysfunction (Pacheco et al., 2021). The breakdown of the outer membrane might be responsible for AOEP that is inhibited on the fimbriae. This is due to the fact that the production of these fimbriae necessitates the presence of two specific proteins: a periplasmic chaperone and an outer membrane-located molecular usher (doorkeeper) (Mol and Oudega, 1996). The breakdown of the cell envelope’s integrity effectively inhibits fimbriae biosynthesis. Consequently, it reduces the ability of KPESBL to initiate host invasion (Culler et al., 2018). Our results show that SMAO interferes with KPESBL growth by disrupting cell envelope integrity. This change then affects the reduction of other virulence factors, like fimbria, CPS, and siderophore.
CONCLUSIONS
SMAO represents a bacteriostatic dose-dependent manner. SMAO bioactivity in KPESBL is mainly associated with decreased siderophore, inhibition of CPS and fimbriae, and loss of cell membrane integrity. Aspergillus oryzae can be used as a new source of anti-infectives to treat K. pneumoniae that has become resistant to drugs.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
We thank the Maulana Malik Ibrahim State Islamic University for financial support through the Bantuan Operasional Perguruan Tinggi Negeri (BOPTN) Grant.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Alsharif W, Saad MM, Hirt H. Desert microbes for boosting sustainable agriculture in extreme environments. Front Microbiol, 2020; 11:1666; doi: 10.3389/fmicb.2020.01666
Baker JL, Hendrickson EL, Tang X, Lux R, He X, Edlund A, McLean JS, Shi W. Klebsiella and Providencia emerge as lone survivors following long-term starvation of oral microbiota. Proc Natl Acad Sci, 2019; 116(17):8499–504; doi: 10.1073/pnas.1820594116
Breurec S, Melot B, Hoen B, Passet V, Schepers K, Bastian S, Brisse S. Liver abscess caused by infection with community-acquired Klebsiella quasipneumoniae subsp. Quasipneumoniae. Emerg Infect Dis, 2016; 22(3):529–31; doi: 10.3201/eid2203.151466
Caza M, Kronstad JW. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front Cell Infect Microbiol, 2013; 3; doi: 10.3389/fcimb.2013.00080
Chen T, Dong G, Zhang S, Zhang X, Zhao Y, Cao J, Zhou T, Wu Q. Effects of iron on the growth, biofilm formation and virulence of Klebsiella pneumoniae causing liver abscess. BMC Microbiol, 2020; 20(1):36; doi: 10.1186/s12866-020-01727-5
Cress BF, Englaender JA, He W, Kasper D, Linhardt RJ, Koffas MAG. Masquerading microbial pathogens: capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol Rev, 2014; 38(4):660–97; doi: 10.1111/1574-6976.12056
Culler H, Couto S, Higa J, Ruiz R, Yang M, Bueris V, Franzolin M, Sircili M. Role of SdiA on biofilm formation by atypical enteropathogenic Escherichia coli. Genes, 2018; 9(5):253; doi: 10.3390/genes9050253
Duong-Ly KC, Gabelli SB. Salting out of proteins using ammonium sulfate precipitation. Methods Enzymol, 2014; 541:85–94; doi: 10.1016/B978-0-12-420119-4.00007-0
Flipphi M, Sun J, Robellet X, Karaffa L, Fekete E, Zeng AP, Kubicek CP. Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp. Fungal Genet Biol, 2009; 46(1):S19–44; doi: 10.1016/j.fgb.2008.07.018
Gharrah MM, El-Mahdy AM, Barwa RF. Association between virulence factors and extended spectrum beta-lactamase producing Klebsiella pneumoniae compared to nonproducing isolates. Interdiscip Perspect Infect Dis, 2017:7279830; doi: 10.1155/2017/7279830.
Graham CLB, Newman H, Gillett FN, Smart K, Briggs N, Banzhaf M, Roper DI. A dynamic network of proteins facilitate cell envelope biogenesis in Gram-negative bacteria. Int J Mol Sci, 2021; 22(23):12831; doi: 10.3390/ijms222312831
Gunther NW, Lockatell V, Johnson DE, Mobley HLT. In vivo dynamics of type 1 fimbria regulation in uropathogenic Escherichia coli during experimental urinary tract infection. Infect Immun, 2001; 69(5):2838–46; doi: 10.1128/IAI.69.5.2838-2846.2001
Guo R, Luo X, Liu J, Lu H. Mass spectrometry based targeted metabolomics precisely characterized new functional metabolites that regulate biofilm formation in Escherichia coli. Anal Chim Acta, 2021; 1145:26–36; doi: 10.1016/j.aca.2020.12.021
Hellman J, Loiselle PM, Tehan MM, Allaire JE, Boyle LA, Kurnick JT, Andrews DM, Sik KK, Warren HS. Outer membrane protein a, peptidoglycan-associated lipoprotein, and murein lipoprotein are released by Escherichia coli bacteria into serum. Infect Immun, 2000; 68(5):2566–72; doi: 10.1128/IAI.68.5.2566-2572.2000
Houda SN, Rachid B, Julio G, Jesus RSM, Teresa V. In vitro antimicrobial, antiviral and cytotoxicity activities of Aspergillus oryzae isolated from El-Baida Marsh in Algeria. J Drug Deliv Ther, 2020; 10(4):191–5; doi: 10.22270/jddt.v10i4.4261
Hsieh PF, Liu JY, Pan YJ, Wu MC, Lin TL, Huang YT, Wang JT. Klebsiella pneumoniae peptidoglycan-associated lipoprotein and murein lipoprotein contribute to serum resistance, antiphagocytosis, and proinflammatory cytokine stimulation. J Infect Dis, 2013; 208(10):1580–9; doi: 10.1093/infdis/jit384
Jahn CE, Charkowski AO, Willis DK. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J Microbiol Methods, 2008; 75:318–24. doi: 10.1016/j.mimet.2008.07.004
Jiao L, Li J, Liu F, Wang J, Jiang P, Li B, Li H, Chen C, Wu W. Characterisation, chain conformation and antifatigue effect of steamed ginseng polysaccharides with different molecular weight. Front Pharmacol, 2021; 12:712836; doi: 10.3389/fphar.2021.712836
Khatoon Z, McTiernan CD, Suuronen EJ, Mah TF, Alarcon EI. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon, 2018; 4(12):e01067; doi: 10.1016/j.heliyon.2018.e01067
Lee KJ, Kim JA, Hwang W, Park SJ, Lee KH. Role of capsular polysaccharide (CPS) in biofilm formation and regulation of CPS production by quorum-sensing in Vibrio vulnificus: quorum-sensing represses CPS production and biofilm. Mol Microbiol, 2013; 90(4):841–57; doi: 10.1111/mmi.12401
Lee CH, Su LH, Liu JW, Chang CC, Chen RF, Yang KD. Aspirin enhances opsonophagocytosis and is associated to a lower risk for Klebsiella pneumoniaeinvasive syndrome. BMC Infect Dis, 2014; 14(1):47; doi: 10.1186/1471-2334-14-47
Lin TH, Huang SH, Wu CC, Liu HH, Jinn TR, Chen Y, Lin CT. Inhibition of Klebsiella pneumoniae growth and capsular polysaccharide biosynthesis by Fructus mume. Evid Based Complement Alternat Med, 2013:1–10; doi: 10.1155/2013/621701
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 2001:402–8; doi: 10.1006/meth.2001.1262.
Misra S, Sharma V, Srivastava AK. Bacterial polysaccharides: an overview. In: Ramawat KG, Mérillon JM (Eds.). Polysaccharides, Springer International Publishing, 2014; doi: 10.1007/978-3-319-03751-6_68-1
Mol O, Oudega B. Molecular and structural aspects of fimbriae biosynthesis and assembly in Escherichia coli. FEMS Microbiol Rev, 1996; 19(1):25–52; doi: 10.1111/j.1574-6976.1996.tb00252.x
Nacef HS, Belhattab R, Larous L. Chemical composition, antimicrobial study against human and plant pathogenic microorganisms and optimization of bioactive metabolites produced by the new strain Aspergillus oryzae 18HG80 isolated from saline soil (El-Baida Marsh, Algeria). J Microbiol Res, 2020; 10:11–21.
Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev, 2017; 41(3):252–75; doi: 10.1093/femsre/fux013
Nevermann J, Silva A, Otero C, Oyarzún DP, Barrera B, Gil F, Calderón IL, Fuentes JA. Identification of genes involved in biogenesis of outer membrane vesicles (OMVs) in Salmonella enterica Serovar Typhi. Front Microbiol, 2019; 10:104; doi: 10.3389/fmicb.2019.00104
Pacheco T, Gomes AÉI, Siqueira NMG, Assoni L, Darrieux M, Venter H, Ferraz LFC. SdiA, a quorum-sensing regulator, suppresses fimbriae expression, biofilm formation, and quorum-sensing signaling molecules production in Klebsiella pneumoniae. Front Microbiol, 2021; 12:597735; doi: 10.3389/fmicb.2021.597735
Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev, 2016; 80(3):629–61; doi: 10.1128/MMBR.00078-15
Rachma LN, Fitri LE, Prawiro SR, Raras TYM. Aspergillus oryzae attenuates quorum sensing-associated virulence factors and biofilm formation in Klebsiella pneumoniae extended-spectrum beta-lactamases. F1000Research, 2022; 11(1148):1148.
Schwechheimer C, Sullivan CJ, Kuehn MJ. Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry, 2013; 52(18):3031–40; doi: 10.1021/bi400164t
Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol, 2015; 13(10):605–19; doi: 10.1038/nrmicro3525
Sinha AK, Venkateswaran BP, Tripathy SC, Sarkar A, Prabhakaran S. Effects of growth conditions on siderophore producing bacteria and siderophore production from Indian Ocean sector of Southern Ocean. J Basic Microbiol, 2019; 59(4):412–24; doi: 10.1002/jobm.201800537
Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis, 2012; 12(11):881–7; doi: 10.1016/S1473-3099(12)70205-0
Stærk K, Khandige S, Kolmos HJ, Møller-Jensen J, Andersen TE. Uropathogenic Escherichia coli express type 1 fimbriae only in surface adherent populations under physiological growth conditions. J Infect Dis, 2016; 213(3):386–94; doi: 10.1093/infdis/jiv422
Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang, KC, Silhavy TJ. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci, 2016; 113(11); doi: 10.1073/pnas.1601375113
Tacconelli E, Mazzaferri F, de Smet AM, Bragantini D, Eggimann P, Huttner BD, Kuijper EJ, Lucet JC, Mutters NT, Sanguinetti M, Schwaber MJ, Souli M, Torre-Cisneros J, Price JR, Rodríguez-Baño J. ESCMID-EUCIC clinical guidelines on decolonization of multidrug-resistant Gram-negative bacteria carriers. Clin Microbiol Infect, 2019; 25(7):807–17; doi: 10.1016/j.cmi.2019.01.005
Virawan H, Nuryastuti T, Nirwati H. Multidrugresistant Klebsiella pneumoniae from clinical isolates at dr. Soeradji Tirtonegoro central hospital Klaten. J Kedokteran Kesehatan Indonesia, 2020; 11(2):109–20; doi: 10.20885/JKKI.Vol11.Iss2.art3
Willis LM, Whitfield C. 2013. Capsule and lipopolysaccharide. In: Donnenberg MS (Ed.). Escherichia coli, 2nd edition, Department of Oxford, Oxford, UK, pp 533–56; doi: 10.1016/B978-0-12-397048-0.00017-6
Wilson BR, Bogdan AR, Miyazawa M, Hashimoto K, Tsuji Y. Siderophores in iron metabolism: from mechanism to therapy potential. Trends Mol Med, 2016; 22(12):1077–90; doi: 10.1016/j.molmed.2016.10.005
Wingfield P. Protein precipitation using ammonium sulfate. In: Coligan JE, Dunn BM, Speicher DW, Wingfield PT (Eds.). Current protocols in protein science, John Wiley & Sons, 1998, pp A.3F.1–8; doi: 10.1002/0471140864.psa03fs13
World Health Organization. 2020. Core competencies for infection prevention and control professionals. World Health Organization. Available via https://apps.who.int/iris/handle/10665/335821
Zubaidah E, Suryawira YM, Saparianti E. Comparative study production of exopolysaccharide (EPS) by lactic acid bacteria (L. casei and L. plantarum) in different media (Dates and Mulberry juice). Agroindus J, 2014; 3(1):107.