INTRODUCTION
Dental caries is a common problem in oral health. Kassebaum et al. (2017) reported that 2.5 billion people globally suffered from untreated dental caries in permanent teeth. Furthermore, a study from Maharani et al. (2019) showed that 61% of 696 children aged 12 years old in Jakarta had experienced caries. Specifically, Amalia et al. (2019) conducted research on early childhood caries (ECC) in Indonesia and gained information on ton high prevalence of ECC in children aged 5 years old while who got treatment only 9.5%. The high prevalence of dental caries makes many people in the world search for a breakthrough to decrease this problem.
Dental caries is a disease that is due to the influence of many factors, especially bacteria, which involves chronic and progressive destruction of hard dental tissue (Niu et al., 2020). Some bacteria are involved in the stage development of the dental caries pathogen. However, Streptococcus mutans is a main dental caries-associated bacterium (Chinsembu, 2016; Gross et al., 2012; Hwang et al., 2018; Ribeiro et al., 2018). Streptococcus mutans commonly relates to the dental caries mechanism because of its important role in the biofilm formation known as dental plaque (da Silva et al., 2014). As the primary etiological agent, S. mutans produces a mechanism to develop glucosyl polymers using glucosyltransferases (Gtfs) to adhere to the tooth surface and build a bacterial colony that enhances caries progression (Nijampatnam et al., 2021). Furthermore, S. mutans also produces a glucan binding protein to promote adherence with another bacterium and antigen I/II as a major surface protein S. mutans regarding the development of dental caries (Matsumoto, 2018). In the presence of sucrose, Gtfs and the glucan-binding protein will digest sucrose to glucan for biofilm formation, and therefore these are called sucrose-dependent proteins. However, antigen I/II is different because it can conduct adhesive formation on the tooth surface without the presence of sucrose using the salivary pellicle as a sucrose-independent protein (Matsumoto, 2018). Emeka et al. (2020), Kong et al. (2021), and Rivera Quiroga et al. (2021) showed that inhibiting these proteins’ activity leads to disrupting biofilm formation and adherence mechanism of S. mutans in in vitro analysis.
A lot of therapies have been conducted to combat dental caries, such as using xylitol (Sharif et al., 2013), chlorhexidine, fluoride (Malhotra et al., 2013), and natural product derivations (Oh et al., 2020). However, in the mouth, many bacteria normally inhabit the oral cavity, such as lactobacilli and andomizedrial species related to the probiotic bacteria that have some useful benefits to human health (Haukioja , 2010). Therefore, ensuring the method is safe to maintain ecological oral cavity organisms is important and can be a different approach to combat dental caries. Previous research also showed that using a probiotic administration combination can reduce the plaque index in periodontitis patients (Alanzi et al., 2018; Grusovin et al., 2020; Shaik and Reddy, 2017; Vicario et al., 2013).
Propolis is a natural product that consists of resinous substances collected by bees from various plants. It has a lot of functional benefits for human health such as antiviral and immune boosters (Harisna et al., 2021) because propolis has abundant bioactive substances like phenolic acids, flavonoids, amino acids, minerals, and vitamins (Kurek Górecka et al., 2021). Interestingly, bioactive compounds from propolis are more potent with their antimicrobial activity to the Gram-positive bacteria than Gram-negative ones, which can be explored to inhibit S. mutans activity as a Gram-positive bacterium in biofilm formation (Kurek Górecka et al., 2021). However, research attempted to evaluate the potential inhibitor from propolis compounds to caries-related protein S. mutans through an in silico approach.
Computational research as an in silico method is important to examine the compound ability from the propolis content to specifically target the caries-related protein and ensure its safety for local treatment. As the main major proteins relating to biofilm formation, sucrose-dependent and sucrose-independent S. mutans are highly considered to be targeted by an anticaries drug. Therefore, this research aims to evaluate the compounds in propolis that have high activity to inhibit glucosyltransferases as sucrose-dependent and antigen I/II as sucrose-independent proteins through an in silico approach, mainly molecular docking and molecular dynamics simulation.
MATERIALS AND METHODS
Propolis screening
A propolis sample was provided by PT. Nano Herbaltama Internasional, Indonesia. Methanol 9 ml and propolis 1 ml were added to a 10 ml volumetric. The Minisart SRP15 Syringe Filter PTFE 0.2 μm (Sartorius, Germany) was used to filter the solution. LC-MS/MS analyses were done by adding a 2 ml vial of the filtered solution in the LC-MS/MS Xevo G2-XS Quadrupole Time-of-Flight Mass Spectrometer (Waters, New Zealand). The separation process was figured out using the ACQUITY UPLC HSS-T3 column (100 × 2.1 mm, 1.7 μm). The temperature was set at 40 and 25?C to maintain the column and sample, respectively. Formic acid of 0.1% in water (A) and formic acid of 0.1% in acetonitrile (B) were used as the mobile phase at a flow rate of 0.6 ml/minute. The preparation of the elution gradient was done using the following protocol: starts at 1%, at 0.5 minutes 1% A, at 16 minutes 35% A, at 18 minutes 100%, and 20 minutes 1% A. Flavonoids screening was done by injecting 2 μl of the test solution, and the chromatograph was recorded for 20 minutes. Electrospray Ionization positive ion mode was set with the range scan of mass at 50–1,200 Da. The source and desolvation temperatures were 120?C and 550?C, respectively. The low collision energy was 6 eV, and the high collision energy was 15–40 eV (ramp). The mass correction was done using a 1 ng/ml solution of leucine enkephalin as a lock spray during acquisition. Finally, a flow rate of 10 μl/minute was used to inject the lock spray, and the scan time was 0.1 seconds with an interval of 30 seconds.
Receptor and ligand preparation
A three-dimensional crystal protein structure of glucosyltransferase [Protein data bank (PDB) ID: 3AIC] and Ag I/II (PDB ID: 3IPK) from S. mutans were retrieved from Protein Data Bank PDB (www.rcsb.org). These proteins were analyzed for their similarity to the human protein, also Lactobacillus reuteri and Bifidobacterium lactis as oral probiotics using the Protein Basic Local Alignment Search Tool (BLAST-P) on the NCBI website (https://www.ncbi.nlm.nih.gov/). After conducting BLAST-P, all proteins were prepared by the AutoDockTools version 4.2 program (Scripps Research Institute, La Jolla, CA) to remove the cocrystallized ligand and water while missing polar hydrogen was added and merged with nonpolar hydrogen.
The structures of up to a total of 93 ligands from propolis screening compounds were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/) in the 3D structure data format form. Some ligands that did not have a 3D structure from PubChem were generated using Discovery Studio Visualizer 2021. All ligands were converted into the PDB, partial charge (Q), and atom type (T) form using PyRx 0.8 (Dallakyan and Olson, 2015). Thus, these were prepared to add the Gasteiger charges and merged with the nonpolar hydrogen molecules using AutoDockTools. Rotatable torsions of the ligands were activated. The grid box coordinate was determined using the center grid box option of the receptor-ligand complex.
Molecular docking validation and visualization
AutoDockTools was used to validate the molecular docking method. Grid coordination of the native ligand and protein was saved as a grid parameter file to create a grid log file (GLG) file using AutoGrid. The GLG file was used to build docking parameter file and docking log file (DLG) files while running AutoDock. The maximum number of evals while running was at a medium level using a Lamarckian genetic algorithm as the output function, and the rest of the parameters were set to default. The root mean square deviation (RMSD) value was analyzed from the DLG file and visualized using Discovery Studio Visualizer 2021.
Molecular docking and visualization
Molecular docking was carried out using the PyRx 0.9 version, which used AutoDock 4.2 as the docking software (Dallakyan and Olson, 2015) underperformed on Windows Pro 10, Intel Core® 7, and RAM 16 GB. All 93 ligands were loaded into PyRx and the grid box coordination that was obtained from AutoDockTools during preparation was set (193.74; 49.075; 195.987 and 1743.498; 44.166; 198.644 of x, y, z coordination 3AIC and 3IPK, respectively, with spacing in angstrom of 0.375). The docking process was done in PyRx using the Lamarckian genetic algorithm. All the other parameters were kept as default. Discovery Studio Visualizer 2021 was used to visualize the docking result.
ADMET analysis
Absorption, Distribution, Metabolism, and Excretion (ADME) analyses were done using the SwissADME web server (https://www.swissadme.ch/) (Daina et al., 2017). The top 10 ligands after conducting molecular docking were moved forward for ADMET analysis. The ligands of this study were conducted for dental oral local treatment; therefore, some parameters such as the maximum two violation of Lipinski’s rule, water-soluble drug, and both negatives of the blood-brain barrier and P-gp substrate were considered (Al-Khafaji et al., 2021; Sharif et al., 2013). Moreover, for toxicity for lethal dose 50 (LD50 mg/kg) prediction, the ProTox-II program was used (https://tox-new.charite.de) (Banerjee et al., 2018). A ligand that has been classified as having above class 3 toxicity was selected.
Molecular protein interaction
The best two ligands after molecular docking and ADMET screening were selected for molecular protein interaction analysis. Discovery Studio Visualizer 2021 was used to analyze and visualize some parameters such as pose conformation, amino acid residue, and chemical interaction between ligand and protein.
Conserved amino acid analysis
The ConSurf web server was used (https://consurf.tau.ac.il/) to analyze the conserved amino acid sequence. This server used the Bayesian method to identify the residues in the conserved region (Ashkenazy et al., 2016). PDB IDs of 3AIC and 3IPK were uploaded to the web server, and the identification region was limited to chain A only. Multiple sequence alignment (MSA) was automatically made from ConSurf, and the rest of the parameters were kept at default. Scaling conserved amino acid result was displayed in the gradation color starting from 1 as the variable until 9 as the conserved region.
Molecular dynamics
The molecular dynamics simulation studies were executed using GROMACS 2021.5 (Berendsen et al., 1995). A unit cell with a rhombic dodecahedral shape was utilized for the protein-ligand complex and positioned at its center at least 10 Å from the edge of the box. The system was neutralized by replacing some water molecules with Na+ and Cl−. The energy minimization was performed with the steepest descent algorithm until the maximum force reached less than 1,000 kJ mol−1 nm−1. Equilibration was conducted for 100 ps at 300 K and separated into two stages, the isothermal-isochoric ensemble and the isothermal-isobaric ensemble. The particle-mesh Ewald was used for electrostatic interaction calculation, and the short-range cut-off of Van der Walls was 12 Å. The 50 ns molecular dynamics production was carried out with a modified Berendsen thermostat as the temperature coupling and a Parrinello-Rahman barostat as the pressure coupling. RMSD and root mean square fluctuation (RMSF) were calculated from the molecular dynamics trajectories.
RESULTS AND DISCUSSION
Receptor and ligand preparation
To address the potential comprehensive inhibition of S. mutans as an important cariogenic bacterium, we used glucosyltransferase protein (Gtf) (PDB ID: 3AIC) as a sucrose-dependent agent and Ag I/II as a sucrose-independent agent (PDB ID: 3IPK). From the BLAST-P (NCBI) result, the 3IPK and 3AIC proteins were less identical to L. reuteri protein sequences. Moreover, 3AIC was also less identical to human protein sequences (Supplementary Figs. S1–S3).
As many as 93 compounds were identified in propolis screening by the LC/MS-MS method. Most of these compounds were determined as being in the flavonoids group. Flavonoids are built by a 15-carbon skeleton consisting of two rings of benzene that are linked by a heterocyclic pyrene ring (Khameneh et al., 2019; Wagh, 2013). Among 93 flavonoid ligands, the most prominent flavonoid subgroups were flavones, isoflavones, and flavonols. Most flavone groups have a hydroxyl group in the primary ring of position 5 while flavonols have the hydroxyl group in the 3 positions of ring C. Furthermore, the isoflavones subgroup has a substituent benzene ring at 3 positions of ring C (Mutha et al., 2021; Panche et al., 2016). Flavonoids are usually linked with health-promoting effects, and therefore they are common as components in nutraceutical, pharmaceutical, medicinal, and cosmetic products (Wagh, 2013). Their broad activities are structure dependent. For instance, the hydroxyl functional group in flavonoids induces an antioxidant effect by scavenging free radicals or chelating metal ions (Ko?evar et al., 2007). In addition, the antimicrobial effect of flavonoids was also recorded, which is dependent on the hydrophobic substituents’ structure such as the prenyl groups (Xie et al., 2014). There are several mechanisms in the flavonoid group to combat bacteria, such as inhibition of nucleic acid synthesis, inhibition of cytoplasmic membrane function by disrupting biofilm formation, and some crucial enzymes (Barbieri et al., 2017; Górniak et al., 2019; Khameneh et al., 2019). Furthermore, Yuan et al. (2021) revealed that flavonoids mainly disrupt the cell membrane of Gram-positive bacteria such as S. mutans as the major target. To examine the flavonoid content as a potential anticaries agent, an in silico method was conducted. All ligands’ details are shown in Supplementary Table S3.
Before conducting the molecular docking process between the ligand and protein, docking validation was carried out. Acarbose as a native ligand from the 3IAC protein and phenylmethanesulfonic acid (PMS) as a native ligand from the 3IPK protein were used for this validation. From validation of the molecular docking method, the lowest binding affinity of acarbose was −7.16 kcal/mol and −4.75 kcal/mol of PMS with the RMSD values of 1 and 1.24, respectively (Supplementary Tables S1 and S2). Supplementary Figure S4 showed the result from the docking validation and the binding site of the 3AIC and 3IPK proteins.
Molecular docking and ADMET analysis
The top 10 docking results of the 3AIC and 3IPK proteins were summarized in Tables 1 and 2, with the range of binding affinity from all ligands of 3AIC being −7.53 to −8.44 kcal/mol and the range of 3IPK from −6.5 to −7.32 kcal/mol. On the other side, acarbose and PMS as native ligands were −5.78 kcal/mol and −4.64 kcal/mol, respectively.
During the in silico process, molecular docking is important to conduct drug ability screening. The molecular docking process shows the list of interactions between two molecules to build a stable complex (Trezza et al., 2020). The lower binding affinity indicates the more stable and closer native state of protein-ligand efficiency. It exhibits a good potential drug (Fu et al., 2018). According to the docking results (Tables 1 and 2), it seemed that lower binding affinity results of some ligands after running molecular docking from the 3AIC and 3IPK proteins were affected by the more complex structure. This can be seen in the best ligand of both proteins, especially diosmetin, cosmosiin, genistin, and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside. The interactions among these proteins as the best ligand of the 3AIC and 3IPK proteins were visualized (Supplementary Fig. S5). Hydrogen, hydrophobic, Van der Waals, and electrostatic bonds are the main interactions between ligand-protein complexes, and these are very important to creating stable bonding with the receptor (Freitas and Schapira, 2017; Shawon et al., 2021). In contrast, acarbose and PMS as native ligands generally only had hydrogen and Van der Waals bonds without other interactions, such as a hydrophobic interaction, except for an electrostatic bond in the PMS-3IPK complex (Supplementary Fig. S10). It may result in the lower binding affinity of these native ligands than diosmetin, cosmosiin, genistin, and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside.
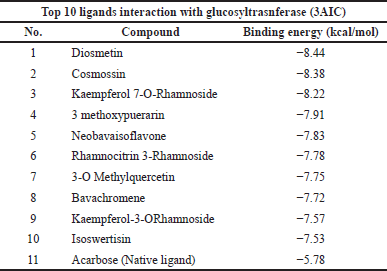 | Table 1. Top 10 binding affinity of 3AIC protein. [Click here to view] |
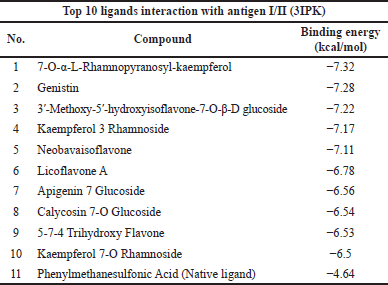 | Table 2. Top 10 binding affinity of 3IPK protein. [Click here to view] |
The top 10 best ligands from molecular docking were moved forward for ADMET analysis. Diosmetin and cosmosiin were the best ligand interactions with the 3AIC receptor that met all drug-like requirements besides their binding affinity score (Table 3). Furthermore, in the 3IPK receptor, genistin and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside were the best ADME (Table 4) and docking results. To assess toxicology, the ProTox-II web server showed the classification of toxic doses. Diosmetin and cosmosiin were classified as having class 5 toxicity, which is known as a safe compound (Supplementary Fig. S5). Moreover, genistin and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside were classified as having classes 5 and 6 toxicity (Supplementary Fig. S6), respectively.
Molecular protein-ligand interaction
According to the binding affinity and ADMET result, the two best ligands of each protein receptor were chosen to be visualized (Fig. 1). Diosmetin and cosmosiin were selected from the 3AIC protein (Fig. 1A–D) while genistin and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside were selected from the 3IPK protein (Fig. 1E–H). Diosmetin formed three classical hydrogen bonds with ASP 477, TRP 517, and GLU 515. Besides classical hydrogen, it also formed a weak hydrogen bond as a carbon-hydrogen bond with ASP 909 and ASN 481. There were four amino acids from diosmetin that built a hydrophobic interaction (TYR 916 as pi-pi interaction, HIS 587, ALA 478, and LEU 433 as pi-alkyl interaction). Another noncovalent bonding of a small ligand was the Van der Waals interaction. It was built by diosmetin to interact with the 3AIC protein with the residues ASN 914, ARG 475, ASN 862, and ASP 480. Furthermore, an additional interaction involved pi-anion bonding from ASP 588 also existing between diosmetin and the 3AIC protein. On the other side, cosmosiin did not build any electrostatic interactions like diosmetin. Cosmosiin built classical hydrogen bonds with ASP 477, ASP 480, ASP 588, GLU 515, and GLN 960. Weak hydrogen bonds only interacted with ASN 481 and ASP 909. Furthermore, hydrophobic interactions existed among TRP 517 as pi-pi interaction, LEU 382, LEU 433 as pi-sigma interaction, and ALA 478 as pi-alkyl interaction. The last interaction in cosmosiin with the 3AIC protein was the Van der Waals interaction that involved TYR 430, LEU 434, ARG 475, HIS 587, GLN 592, PHE 907, and TYR 916.
In the 3IPK protein, genistin built a hydrogen bonding interaction with ASP 512, ARG 824, ASP 760, TRP 816, and THR 586 as conventional hydrogen and SER 697 as a weak hydrogen bond. A hydrophobic interaction also existed in genistin interaction LYS 822 as pi-alkyl and TRP 816 as pi-pi stacked. The Van der Waals interaction involves LEU 653, ASN 820, SER 762, and VAL 587. Furthermore, 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside-built hydrogen bonds with THR 652, ASN 820, ARG 824, TRP 816, and THR 586 as conventional hydrogen bonds and ASP 909 and ASN 481 as weak hydrogen bonds. The hydrophobic interaction involved TRP 816 as pi-pi stacked, LEU 653 as alkyl, and LYS 822 as pi-alkyl. The Van der Waals interaction involved ASP 512, SER 762, and VAL 587. Detailed information on molecular interaction was displayed in Tables S4 and S5.
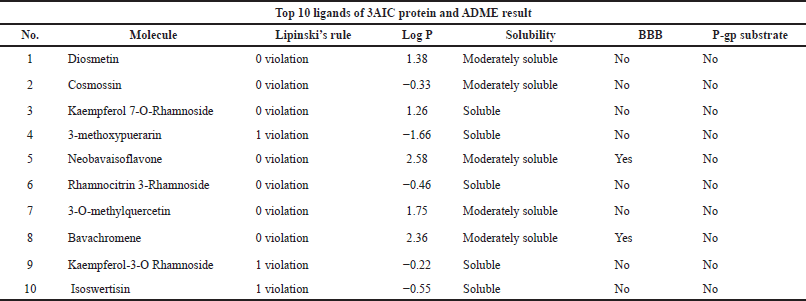 | Table 3. Ligand ADME analyses result in the 10 best potential ligands for 3AIC protein. [Click here to view] |
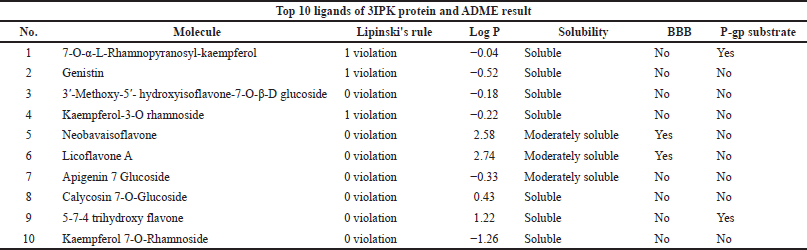 | Table 4. Ligand ADME analyses result in the 10 best potential ligands for 3IPK protein. [Click here to view] |
Freitas and Schapira (2017) showed that the hydrophobic interaction was the most prominent among the high-efficiency ligands. It is different from protein-fragment complexes that are more stable using hydrogen bond interaction. In the hydrophobic interaction, the aliphatic carbon of the receptor and aromatic carbon of the ligand are most prevalent. Regarding the aromatic carbon, a small ligand as an inhibitor generally contains aromatic carbon such as a benzene ring to build the interaction with its receptor, and it is owned by the flavonoids group, as seen in Supplementary Table S3. Moreover, leucine and valine are frequently involved in the hydrophobic interaction (Freitas and Schapira, 2017), and they also exist in the ligand-protein complex of both the 3AIC and 3IPK proteins, which can be seen in Supplementary Tables S4 and S5.
Analyzing the data from acarbose, diosmetin, and cosmosiin, amino acids Glu 515, Leu 433, ASN 481, and ASP 477 were the residues that were always involved in the interaction between the ligand and 3AIC receptor shown by their existence in the interaction (Supplementary Table S4). Moreover, ARG 824 and TRP 816 were amino acid residues that existed in PMS as the native ligand of 3IPK also in both genistin and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside as the best ligand compound (Supplementary Table S5). It can be known that these amino acids acted as key residues in the 3AIC and 3IPK interactions, respectively. The key residue that acts as a functional amino acid is only distributed in the active binding site of the protein (Chen et al., 2006). Amitai et al. (2004) and Ma et al. (2003) stated that the key residue is generally well conserved in the protein structure, which has an important role in protein interaction and drug development. It can be considered because of its highest binding energy related to the others. As displayed in Supplementary Figures S7 and S8, the key residues in the 3AIC and 3IPK proteins were well conserved compared to the other organisms that have high similarity to these proteins. Furthermore, a mutation in the key residue region will most affect the protein stability and substrate specificity (Yang et al., 2021).
Molecular dynamics
After conducting the molecular dynamics (MD) process within 50 ns, the RMSD and RMSF values from each protein-ligand complex were obtained (Fig. 2). From Figure 2B, it can be observed that the 3AIC protein with diosmetin reached stability after 10 ns with the structure overall remaining at 3.5 Å while cosmosiin kept stable at 4 Å (Fig. 2C) after 10 ns. Furthermore, the RMSF value from both ligands almost had a similar pattern (Fig. 2D).
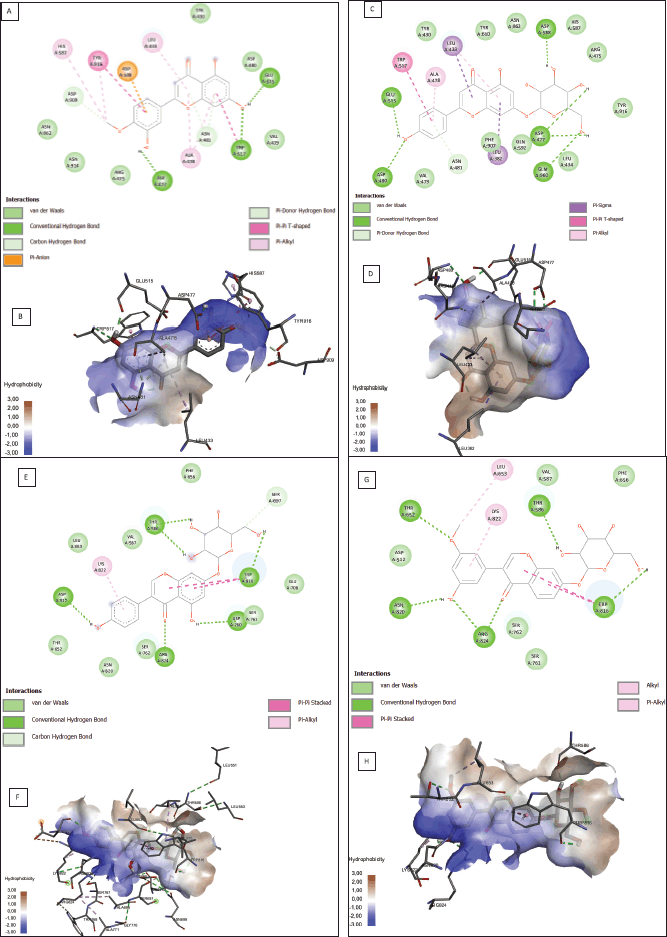 | Figure 1. Protein-ligand interaction results from potential ligands of the 3AIC (A–D) and 3IPK (E–H) proteins. Molecular interaction (A) and hydrophobicity area (B) of diosmetin. Molecular interaction (C) and hydrophobicity area (D) of cosmosiin. Molecular interaction(E) and hydrophobicity area (F) of genistin. Molecular interaction (G) and hydrophobicity area (H) of 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside in the 3IPK protein. [Click here to view] |
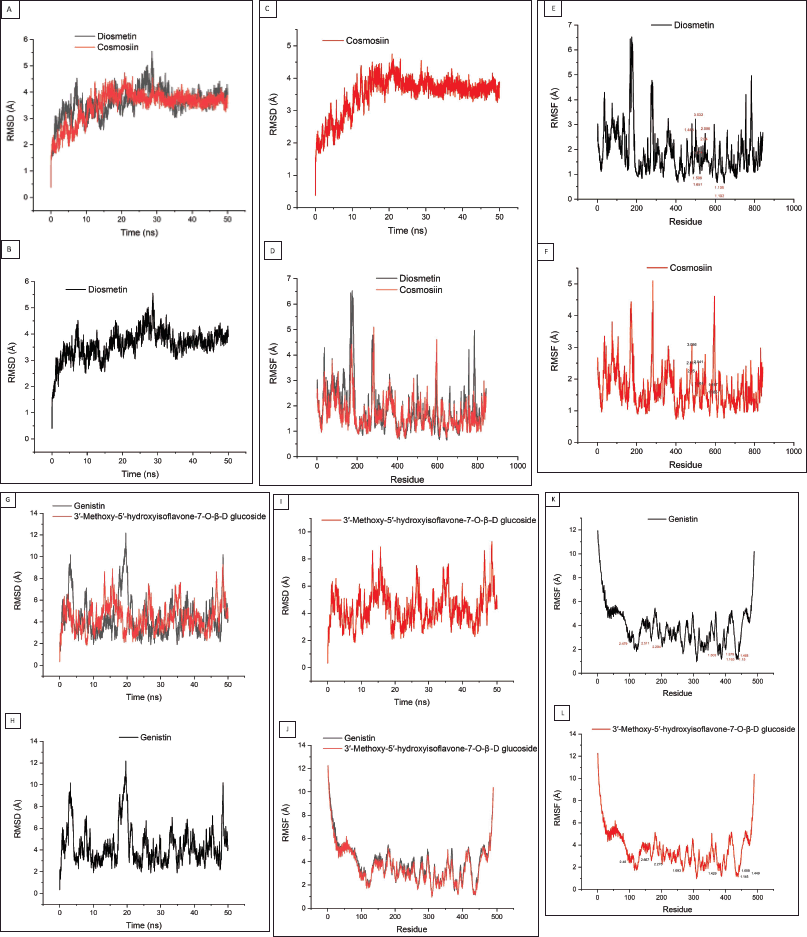 | Figure 2. Molecular dynamics results from potential ligands of the 3AIC (A–F) and 3IPK (G-L) proteins. RMSD (A–C) and RMSF (D–F) of diosmetin and cosmosiin, RMSD (G–I) and RMSF (J–L) of genistin and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside. The residue number displayed within the RMSF result showed the binding site residue from both ligands (7E, 7F, 7K, and 7L). [Click here to view] |
Similar to the ligand of the 3AIC protein, genistin and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside as the best candidate ligands of the 3IPK protein also were examined using MD. The RMSD value showed that genistin reached stabilization at around 3 Å (Fig. 2H) while 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside was stable at around 4 Å (Fig. 2I) with more fluctuations than the 3AIC complexes. On the other side, the RMSF value showed that both ligands had a similar pattern (Fig. 2J).
In contrast to molecular docking, molecular dynamics can show the flexibility of the ligand and receptor within a certain solvent, therefore allowing the receptor to change its conformation (Prasasty et al., 2020). The two most common methods to calculate the conformation change are RMSD and RMSF. RMSD shows the average atomic displacement from the entire protein against its reference structure in a time-dependent manner (Martínez, 2015; Ramírez and Caballero, 2018). Overall, both diosmetin and cosmosiin almost had a similar pattern of RMSD with a value maximum at around 5 Å (Fig. 2A). It showed that both ligands might have the same influence on the 3AIC protein. Conversely, the RMSD values from genistin and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside were slightly different from each other (Fig. 2G). Ligand 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside showed more fluctuation than genistin where the maximum fluctuation of both ligands was at around 12 Å. Some fluctuations in both complexes indicated that the protein searches for other conformations to stabilize (Prasasty et al., 2020).
Alongside the RMSD value, we also examined the RMSF value of the protein-ligand complex. RMSF is used to examine the fluctuation of each residue in the entire protein. Overall, RMSF from the 3AIC and 3IPK complexes almost showed a similar pattern (Fig. 2D and J). Yang et al. (2017) and Baweja et al. (2017) stated that the loop and surface region display higher RMSF values. Based on the RMSF value, the residues of 173 until 182 and 276 until 283 from diosmetin and cosmosiin (Fig. 2D) that exhibited high fluctuation were located in the loop and surface region of the 3AIC protein (Supplementary Fig. S9a). Similar to the 3AIC-ligand complexes, residues from 1 until 23 and 482 until 489 were highly fluctuating in both ligands of 3IPK (Fig. 2J) settled in the loop and surface region of the 3IPK protein (Supplementary Fig. S9b).
The binding site residues in both diosmetin and cosmosiin were LEU 433, ARG 475, ASP 477, ALA 478, ASN 481, TRP 517, GLU 515, ASP 588, and HIS 587 (Supplementary Table S4) while in the 3IPK-ligand complex with genistin and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside these were ARG 824, TRP 816, SER 762, ASP 512, THR 586, LYS 822, ASN 820, and VAL 587 (Table S5). Regarding the RMSF result (Fig. 2E, F, K, and L), all binding sites almost did not show any high fluctuations. Some research reported that binding residues within the structural protein have a lower RMSF value (Freitas and Schapira, 2017; Shawon et al., 2021). Moreover, De Vita et al. (2021) stated that a lower RMSF value in the binding site indicates strong interaction of the ligand to the protein. Therefore, these ligands from both proteins were able to stabilize strongly to the protein regarding the low fluctuation in the binding site residues.
CONCLUSION
To summarize, our in silico study showed that propolis as a natural source has good potential inhibition of S. mutans regarding caries tooth production. Furthermore, the glucosyltransferase (3AIC) protein and antigen I/II (3IPK) protein were safe to be targeted by local oral treatment drugs based on the protein homology sequence from human and normal oral bacteria. According to the molecular docking and dynamics result, diosmetin and cosmosiin exhibited the best potential inhibitors of the 3AIC protein while genistin and 3′-methoxy-5′-hydroxyisoflavone-7-O-β-D glucoside exhibited the best potential inhibitors of the 3IPK protein. Further research on these compounds is needed to elucidate deeper mechanisms and activity in the real biological system to develop caries drug treatment.
ACKNOWLEDGMENTS
The authors are grateful to PT Nano Herbaltama Internasional for providing the sample of propolis.
CONFLICTS OF INTEREST
The authors declared no conflicts of interest.
FUNDING
This study was funded by PT. Nanotech Indonesia Global Tbk under Contract No. 022/NCI-MOU/X/2021.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
SUPPLEMENTARY MATERIAL
Supplementary data can be downloaded from the journal’s website
REFERENCES
Alanzi A, Honkala S, Honkala E, Varghese A, Tolvanen M, Söderling E. Effect of Lactobacillus rhamnosus and Bifidobacterium lactis on gingival health, dental plaque, and periodontopathogens in adolescents: a randomized placebocontrolled clinical trial. Benef Microbes, 2018; 9(4):593–602.
Al-Khafaji ZM, Mahmood SB, Mahmood AB. In silico identification of potential inhibitors for Streptococcus mutans signal peptidase I using structure-based drug design. Lat Am J Pharm, 2021; 40:221–7. CrossRef
Amalia R, Chairunisa F, Alfian MF, Supartinah A. Indonesia: epidemiological profiles of early childhood caries. Front Public Heal, 2019; 7(Aug):210. CrossRef
Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S. Network analysis of protein structures identifies functional residues. J Mol Biol, 2004; 344(4):1135–46. CrossRef
Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res, 2016; 44(W1):W344–50. CrossRef
Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucl Acids Res, 2018; 46(W1):W257–63. CrossRef
Barbieri R, Coppo E, Marchese A, Daglia M, Sobarzo-Sánchez E, Nabavi SF, Nabavi S. Phytochemicals for human disease: an update on plant-derived compounds antibacterial activity. Microbiol Res, 2017; 196:44–68. CrossRef
Baweja M, Singh PK, Sadaf A, Tiwari R, Nain L, Khare SK, Shukla P. Cost effective characterization process and molecular dynamics simulation of detergent compatible alkaline protease from Bacillus pumilus strain MP27. Process Biochem, 2017; 58:199–203. CrossRef
Berendsen HJC, Van der Spoel D, Van Drunen R, GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun, 1995; 91(1–3):43–56. CrossRef
Chinsembu KC. Plants and other natural products are used in the management of oral infections and the improvement of oral health. Acta Trop, 2016; 154:6–18. CrossRef
Chen C, Li L, Xiao Y. Identification of key residues in proteins by using their physical characters. Phys Rev E Stat Nonlin Soft Matter Phys, 2006; 73(4):1–7. CrossRef
Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep, 2017; 7(October 2016):1–13. CrossRef
Dallakyan S, Olson A. Participation in global governance: coordinating “the voices of those most affected by food insecurity.” Glob Food Secur Gov, 2015; 1263:1–11.
Da Silva ACB, da Silva DR, Macêdo Ferreira SA de, Agripino GG, Albuquerque AR, Rêgo TG do. In silico approach for the identification of potential targets and specific antimicrobials for Streptococcus mutans. Adv Biosci Biotechnol, 2014; 05(04):373–85. CrossRef
De Vita S, Chini MG, Bifulco G, Lauro G. Insights into the ligand binding to bromodomain-containing protein 9 (BRD9): a guide to the selection of potential binders by computational methods. Molecules, 2021; 26(23):7192. CrossRef
Emeka PM, Badger-Emeka LI, Ibrahim HIM, Thirugnanasambantham K, Hussen J. Inhibitory potential of mangiferin on glucansucrase producing Streptococcus mutans biofilm in dental plaque. Appl Sci, 2020; 10(22):1–17. CrossRef
Ferreira De Freitas R, Schapira M. A systematic analysis of atomic protein-ligand interactions in the PDB. MedChemComm, 2017; 8(10):1970–81. CrossRef
Fu Y, Zhao J, Chen Z. Insights into the molecular mechanisms of protein-ligand interactions by molecular docking and molecular dynamics simulation: a case of oligopeptide binding protein. Comput Math Methods Med, 2018; 2018:3502514. CrossRef
Górniak I, Bartoszewski R, Króliczewski J. Comprehensive review of antimicrobial activities of plant flavonoids. Phytochem Rev, 2019; 18:241–72. CrossRef
Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One, 2012; 7(10):e47722. CrossRef
Grusovin MG, Bossini S, Calza S, Cappa V, Garzetti G, Scotti E, Gherlone EF, Mensi M. Clinical efficacy of Lactobacillus reuteri-containing lozenges in the supportive therapy of generalized periodontitis stage III and IV, grade C: 1-year results of a double-blind randomized placebo-controlled pilot study. Clin Oral Investig, 2020; 24(6):2015–24. CrossRef
Harisna AH, Nurdiansyah R, Syaifie PH, Nugroho DW, Saputro KE, Firdayani, Prakoso CD, Rochman NT, Maulana NN, Noviyanto A, Mardliyati E. In silico investigation of potential inhibitors to main protease and spike protein of SARS-CoV-2 in propolis. Biochem Biophys Reports, 2021; 26:100969. CrossRef
Haukioja A. Probiotics and oral health. Eur J Dent, 2010; 4(3):348–55. CrossRef
Hwang G, Klein MI, Koo H. Analysis of the mechanical stability and surface detachment of mature Streptococcus mutans biofilms by applying a range of external shear forces. Biofouling. 2014; 30(9):1079–91. CrossRef
Kassebaum NJ, Smith AGC, Bernabé E, Fleming TD, Reynolds AE, Vos T, Murray CJL, Marcenes W; GBD 2015 Oral Health Collaborators. Global, regional, and national prevalence, incidence, and disability-adjusted life years for oral conditions for 195 countries, 1990–2015: a systematic analysis for the global burden of diseases, injuries, and risk factors. J Dent Res, 2017; 96(4):380–7. CrossRef
Khameneh B, Iranshahy M, Soheili V, Sedigheh B, Bazzaz F. Review on plant antimicrobials: a mechanistic viewpoint. Antimicrob Resist Infect Control, 2019; 8:1–28. CrossRef
Ko?evar N, Glava? I, Kreft S. Flavonoidi. Farm Vestn, 2007; 58(4):145–8.
Kong J, Xia K, Su X, Zheng X, Diao C, Yang X, Zuo X, Xu J, Liang X. Mechanistic insights into the inhibitory effect of theaflavins on virulence factors production in Streptococcus mutans. AMB Express, 2021; 11(1):102. CrossRef
Kurek-Górecka A, Walczy?ska-Dragon K, Felitti R, Nitecka-Buchta A, Baron S, Olczyk P. The influence of propolis on dental plaque reduction and the correlation between dental plaque and severity of COVID-19 complications—a literature review. Molecules, 2021; 26(18):5516. CrossRef
Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci USA, 2003; 100(10):5772–7. CrossRef
Maharani DA, Zhang S, Gao SS, Chu CH, Rahardjo A. Dental caries and the erosive tooth wear status of 12-year-old children in Jakarta, Indonesia. Int J Environ Res Public Health, 2019; 16(16):2994. CrossRef
Malhotra A, Hegde M. Medical management of dental caries: a change in therapeutic approach. Int Res J Pharm, 2013; 4(June 2013):39–42.
Martínez L. Automatic identification of mobile and rigid substructures in molecular Dynamics simulations and fractional structural fluctuation analysis. PLoS One, 2015; 10(3):1–10. CrossRef
Matsumoto-Nakano M. Role of Streptococcus mutans surface proteins for biofilm formation. Jpn Dent Sci Rev, 2018; 54(1):22–9. CrossRef
Mutha RE, Tatiya AU, Surana SJ. Flavonoids as natural phenolic compounds and their role in therapeutics: an overview. Futur J Pharm Sci. 2021; 7(1):25. CrossRef
Nijampatnam B, Ahirwar P, Pukkanasut P, Womack H, Casals L, Zhang H, Cai X, Michalek SM, Wu H, Velu SE. Discovery of potent inhibitors of Streptococcus mutans biofilm with antivirulence activity. ACS Med Chem Lett, 2021; 12(1):48–55. CrossRef
Niu Y, Wang K, Zheng S, Wang Y, Ren Q, Li H, Ding L, Li W, Zhang L. Antibacterial effect of caffeic acid phenethyl ester on cariogenic bacteria and Streptococcus mutans biofilms. Antimicrob Agents Chemother, 2020; 64:e00251–20. CrossRef
Norsuzila Ya’acob M, Abdullah M, Ismail, et al. We are IntechOpen, the world’s leading publisher of Open Access books Built by scientists, for scientists TOP 1 %. Intech. 1989;32:137-144.
Oh DH, Chen X, Daliri EBM, Kim N, Kim JR, Yoo D. Microbial etiology and prevention of dental caries: exploiting natural products to inhibit cariogenic biofilms. Pathogens, 2020; 9(7):1–15. CrossRef
Panche AN, Diwan AD, Chandra SR. Flavonoids: an overview. J Nutr Sci, 2016; 5:e47. CrossRef
Prasasty VD, Cindana S, Ivan FX, Zahroh H, Sinaga E. Structure-based discovery of novel inhibitors of Mycobacterium tuberculosis CYP121 from Indonesian natural products. Comput Biol Chem, 2020; 85(December 2019):107205. CrossRef
Ramírez D, Caballero J. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules, 2018; 23(5):1–17. CrossRef
Ribeiro M, Malheiro J, Grenho L, Fernandes MH, Simões M. Cytotoxicity and antimicrobial action of selected phytochemicals against planktonic and sessile Streptococcus mutans. PeerJ, 2018; 2018(6):1–17. CrossRef
Rivera-Quiroga RE, Cardona N, Padilla L, Rivera W, Rocha-Roa C, Diaz De Rienzo MA, Morales SM, Martinez MC. In silico selection and in vitro evaluation of new molecules that inhibit the adhesion of streptococcus mutants through antigen i/ii. Int J Mol Sci, 2021; 22(1):1–17. CrossRef
Shaik JA, Reddy RK. Prevention and treatment of white spot lesions in orthodontic patients. Contemp Clin Dent, 2017; 8(September):11–9. CrossRef
Sharif MO, Ahmed F, Worthington H V. Xylitol-containing products for preventing dental caries in children and adolescents. Cochrane Database Syst Rev, 2013; 2013(9). CrossRef
Shawon J, Khan AM, Shahriar I, Halim MA. Improving the binding affinity and interaction of 5-Pentyl-2-Phenoxyphenol against Mycobacterium Enoyl ACP reductase by computational approach. Informatics Med Unlocked, 2021; 23:100528. CrossRef
Trezza A, Iovinelli D, Santucci A, Prischi F, Spiga O. An integrated drug repurposing strategy for the rapid identification of potential SARS-CoV-2 viral inhibitors. Sci Rep, 2020; 10(1):1–8. CrossRef
Vicario M, Santos A, Violant D, Nart J, Giner L. Clinical changes in periodontal subjects with the probiotic Lactobacillus reuteri Prodentis: a preliminary randomized clinical trial. Acta Odontol Scand, 2013; 71(3-4):813–9. CrossRef
Wagh VD. Propolis: a wonder bees product and its pharmacological potentials. Adv Pharmacol Sci, 2013; 2013. CrossRef
Xie Y, Yang W, Tang F, Chen X, Ren L. Antibacterial activities of flavonoids: structure-activity relationship and mechanism. Curr Med Chem, 2014; 22(1):132–49. CrossRef
Yang J, Zhu H, Zhang T, Ding J. Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations. Cell Discov, 2021; 7(1). CrossRef
Yang LQ, Sang P, Zhang RP, Liu SQ. Substrate-induced changes in Dynamics and molecular motions of cuticle-degrading serine protease PL646: a molecular dynamics study. RSC Adv, 2017; 7(67):42094–104. CrossRef
Yuan G, Guan Y, Yi H, Lai S, Sun Y, Cao S. Antibacterial activity and mechanism of plant flavonoids to gram-positive bacteria predicted from their lipophilicities. Sci Rep, 2021; 11(1):1–15. CrossRef