INTRODUCTION
A “nano-bio-interface” refers to the dynamic physicochemical interaction between nanoparticle (NP) surfaces and the biological components resulting in protein corona (PC) formation on the particle’s surface. PC gives the NP a new identity that differs from the pristine NP, which can change dynamically depending on the local environmental conditions, such as the biological fluid components, shear stress, interaction time, temperature, and pH (Ke et al., 2017; Tenzer et al., 2013). Another factor influencing PC formation is the NP surface charges and hydrophobicity. For instance, similarly charged NP and proteins have a repulsive effect, whereas opposite charges cause electrostatic attractions (Pfeiffer et al., 2014). PC formation also could alter the NP hydrodynamic diameters and colloidal stability while at the same time affecting the adsorbed proteins’ functionality via structural changes (Amin et al., 2012). Several changes happen to the NP characteristics after PC formation, leading to either NP aggregation or destabilization (Sund et al., 2011). It was assumed that the colloidal destabilization affects the coated NP’s ability to counter the van Der Waals forces, causing aggregation. The strength and nature of the NP-coated layers determine this destabilization process, further influencing the NP interaction with cells and the subsequent cellular uptakes (Ahsan et al., 2018).
Thus, PC compositions will vary depending on the type of biological fluid it interacts with, the NP materials, and testing benchmarks (in vitro or in vivo). This issue highlights the need for standardization in the PC research methodologies and online database system construction which will be essential for large-scale and translational PC research (Ban et al., 2020). Over the years, conventional liquid chromatography-mass spectrometry (LC-MS) has been the gold standard for PC profiling and quantification despite its complexities and time-consuming sample preparation. Recent advancements in proteomics analysis using the automated Proteograph workflow and its integrated software, for instance, have successfully improved proteomics analysis coverage, scalability, reproducibility, and accuracy. It simplifies the LC-MS preparation stage and significantly enhances the data analysis capability by incorporating genomics analysis enabling more comprehensive pathway mapping (Blume et al., 2020; Ferdosi et al., 2022).
Another important issue is the impact of PC on the NP biological mechanisms, including NP stability, toxicity, cellular uptakes, biodistribution, and elimination from the body (Ke et al., 2017). Therefore, it becomes of great importance to examine the major components and dynamic forces that play parts in the nano-bio-interface. However, most PC experimental techniques so far have been conducted in static conditions in vitro due to the challenging in vivo conditions.
A deeper understanding of the nano-bio-interface allows NP design modification based on the specific affinities between protein and NP materials, hence providing better NP behavior prediction in vivo while reducing undesirable nonspecific protein binding to the NP (Chetwynd and Lynch, 2020; Li and Lee, 2020). The modification usually involves the NP surface by either adding albumin coating, PEGylation, or attaching specific ligands to improve NP stability and reduce clearance (Mozar and Chowdhury, 2017). Furthermore, the specific interaction between the NP and blood plasma protein could be exploited for disease diagnostic purposes, depending on the NP capability to adsorb specific disease biomarkers from the patient’s plasma. Thus, improvement in proteome profiling methodologies by a more efficient system will be necessary for low-abundance protein detection and quantification (Blume et al., 2020).
The clinical relevance of the PC-based diagnostic will be discussed in this review, which consists of PC roles in disease screening, together with its respective methodology. The author will also explore several strategies to tackle issues surrounding PC analysis by using more innovative approaches such as machine learning algorithms, microfluidics models, the Proteograph workflow, and multiomics analysis. The focus on diagnostic and biomarker discoveries is the latest progress of PC research, while in the past more focus was put on its targeted therapeutics role (Kamaly et al., 2022). Therefore, personalized PC (PPC) is still a growing field that will shape the future of personalized medicine.
PERSONALIZED PC
So far, what has been widely investigated is the impact of NP physicochemical characteristics and experimental conditions on PC formation. However, in recent years, disease-specific PC characterization research for clinical purposes has gained more attention. Several studies have shown that PC components are also influenced by each patient’s specific health conditions. This experiment was done by incubating the NP in human plasma from patients suffering from different diseases, leading to varied PC components. It was known that some pathological conditions might alter the vascular system and its component affecting NP PC compositions (Corbo et al., 2017). Additionally, some changes in the plasma protein components can be used as a diagnostic tool to predict the severity of diseases. Since each disease is characterized by different plasma proteins, the resulting NP PC on those patients’ plasma would also be different. This theory eventually led to a PPC concept (Caputo et al., 2017).
Hajipour et al. (2015) utilized both polystyrene and silica NP to compare the PC components of patients suffering from several diseases and healthy individuals as the control. The human plasma was taken from breast cancer, diabetes, common cold, thalassemia, blood cancer, fauvism, and hypercholesterolemia patients, respectively. After SDS PAGE and silver staining, it was shown that PC components of rheumatism, thalassemia, and blood cancer patients were significantly different compared to healthy individuals. On the other hand, breast cancer patients’ PC patterns were slightly similar to the control. The NP size distribution and zeta potential (ZP) also varied among different disease samples. These results further proved that each disease affects PC differently depending on its specific pathophysiology and the patient demography.
Another study conducted by Colapicchioni et al. (2016) showed that different types of cancers created different PC components on liposomes. Blood plasma for PC analysis was taken from breast, gastric, and pancreatic cancer patients. Among this group of three patients, pancreatic cancer PC was found to be the most abundant and has the least negative charge, as confirmed by Zetasizer and SDS PAGE analysis. A specific band at 37 kDa associated with IgA and IgG might be contributed by autoantibodies production in cancer. This study opens up possibilities of using PC analysis for cancer diagnostic screening.
Gold NP (AuNP) PC’s potential to detect early stages of cancer was investigated by Zheng et al. (2015). Their team found that the amount of IgG in AuNP PC was higher after incubation in prostate cancer serum than in healthy men’s serum. This result supported the study by Colapicchioni et al. (2016), which explained that higher autoantibodies were produced in cancer patients as a part of the immunodefense against cancer. Further PC proteomic analysis was performed, and the result showed a difference in cancer and noncancer serum molecular profiles. Since high autoantibodies as a response to tumorigenesis were also found in many other cancer blood samples, this test can be used as a broad-spectrum cancer screening. This test has 90%–95% specificity and 50% sensitivity for early-stage prostate cancer detection according to pilot studies conducted in Florida hospitals. This result marked a significant improvement in the prostate-specific antigen (PSA) test, which is currently used as a gold standard for prostate cancer detection in the clinic (Zheng et al., 2015). Therefore, the emergence of NP-PC-based technology holds a promising future clinically, not only for targeted therapeutics but also for disease diagnostics.
PC CHARACTERIZATION METHODOLOGY
PC characterization is quite challenging due to the sheer number of proteins adsorbed to the NP surface. PC can be classified into two categories, namely, hard and soft PC, depending on the binding affinity and the protein exchange rate (Ahsan et al., 2018). Following the Vroman effect, large quantities and highly mobile proteins will bind first to the NP surface, which are later replaced by less mobile but higher-affinity proteins (Gupta and Roy, 2020).
Hard PC developed due to the strong binding affinity between the protein and NP surface. Conversely, soft PC has weak binding strength to the NP and mainly consists of protein–protein interaction (Docter et al., 2015). As a result, there is a greater protein exchange rate in soft PC causing it to be easily replaced and less stable. The competitive binding between the proteins is affected by many factors, including electrostatic forces, binding kinetics, protein size, and thermodynamic favorability (Cedervall et al., 2007).
Lundqvist et al. (2008) discovered that NP surface charge plays a key role in the PC formation, with positively charged NP tending to attract different types of proteins than both neutral and negatively charged NP. The protein adsorption kinetics is also influenced by the NP surface charge; for example, faster adsorption of bovine serum albumin (BSA) is observed in positively charged AuNP compared to neutral or negatively charged NP. This means that, in a mixture of proteins, some proteins will bind stronger and faster to the NP than the others, which occurs dynamically (Boulos et al., 2013).
Wang et al. (2016) developed a thermodynamic model to predict protein adsorption to NP in a complex biological fluid medium. They analyzed the competitive binding between GB3 and ubiquitin proteins toward AuNP using different pH mediums in time-dependent experiments. It was revealed that the protein’s pKa values, medium pH, and the order of protein addition heavily influence the PC formation and composition.
Most of the studies being performed are more effective at detecting hard PC since it has strong adsorption to the NP surface. The PC component could be easily characterized using SDS PAGE and MS techniques. This is called a direct method, where the PC component could be directly identified via a proteomics approach (Gupta and Roy, 2020). The other type of analysis is indirect methods, in which NP characteristics are observed before and after exposure to biological fluids, consisting of NP size, surface charges, absorbance, and mass. The differences between direct and indirect methods are summarized in Figure 1.
A technique such as dynamic light scattering (DLS) was used to measure NP size changes instigated by Brownian motion to obtain the NP hydrodynamic diameter and distribution of particle size (Z-average). Additionally, the same instrument also can measure ZP, which is an estimation of NP surface charges in the solution, indicating its colloidal stability. Previous studies using AuNPs showed that, after PC formation, AuNP size became larger and its surface charge remained negative. Some changes in the particle surface charges influence the NP electrostatic and steric stabilities (Piella et al., 2017). However, NP agglomeration often interferes with DLS analysis results; hence, additional imaging using transmission electron microscopy or atomic force microscopy is preferred to acquire more accurate NP size and shape measurement (Kokkinopoulou et al., 2017; Radauer-Preimi et al., 2016). Another useful analytical tool is isothermal titration calorimetry to monitor the NPs’ temperature changes due to protein adsorption into the surface of the particles (Winzen et al., 2015).
The direct method requires a purification step during sample preparation, mostly by centrifugation, to remove excess unbound protein before PC identification using MS. This method is highly specific, and the sample needs to be free from contaminants. Follow-up analysis for PC quantification also can be done by protein assay or targeted LC-MS after a specific protein of interest has been identified. On the other hand, indirect methods can be performed in situ without the purification step since it is not highly specific (Tenzer et al., 2013).
Many of the experiments were performed in vitro due to difficulties in the in vivo analysis. As a result, the shear forces of the bloodstream involvement in the PC formation have still not been explored much (Winzen et al., 2015). Hadjidemetriou et al. (2016) investigated the PC components in vivo for the first time by injecting PEGylated liposomal doxorubicin into mice intravenously and collecting the blood periodically, followed by PC purification and characterization. It was revealed that PC already formed 10 minutes after injection and the components varied widely between the in vitro and in vivo experiments. There were fluctuations of PC concentrations and compositions over time in 10 minutes, 1 hour, and 3 hours. Therefore, time-dependent PC characterizations and improved in vivo PC modeling are crucial for PPC translational research. A deeper investigation of the specific role of the PC component will be beneficial in identifying its relevance to the NP cellular interaction, internalization, and targeting efficiency.
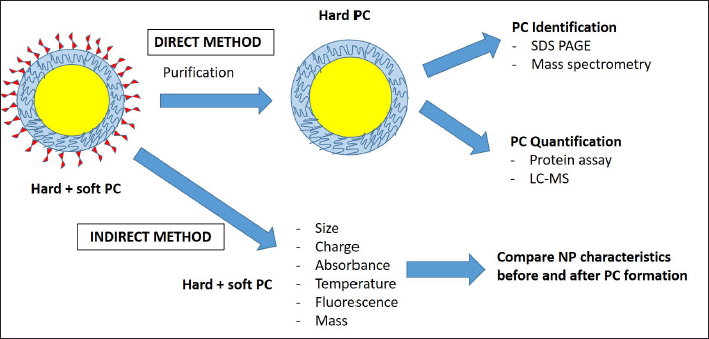 | Figure 1. NPs PC characterization methodologies. PC characterizations employed two methodologies, namely direct and indirect. The direct method requires a purification step followed by protein identification and quantification. Therefore, this method is more effective at detecting hard PC. Alternatively, the indirect method is less specific and works by comparing PC characteristics before and after PC formation on the NP surface. [Click here to view] |
PC ROLE IN NP BIOLOGICAL MECHANISM
PC will determine the fate of the NP inside of the body, starting from its first interaction with the blood components after administration until its elimination, as mentioned in Figure 2. NP stability is heavily affected by PC as the NP will immediately interact with opsonins in the bloodstream. Opsonin consists of many proteins, including immunoglobulin, laminin, C-reactive protein, C3, C4, and C5, that could trigger opsonin-mediated phagocytosis. After interaction with opsonins, there would be a massive and rapid clearance of NP from the body causing the treatment to be ineffective. Several strategies have been incorporated in NP design to hinder PC–opsonin interaction and reduce phagocytosis, including NP surface modification (Mozar and Chowdhury, 2018). Dysopsonin, such as albumin or apolipoprotein that was adsorbed to the NP surface, can prolong NP circulation, hence extending its half-life, making it an ideal surface precoating strategy for NP. Lu et al. (2019) modified gold nanospheres PC with ApoE, a member of the dysopsonin family, to extend the NP circulation time and prevent phagocytosis.
Further analysis using proteomics and fluorescence imaging showed that ApoE precoating before NP administration successfully increased NP tumor accumulation without cytotoxicity effect compared to IgE and human serum albumin (HSA) coated NP. Researchers also found that NP surface hydrophilicity significantly affects PC formation on HSA/IgE coated NP, whereas it does not affect ApoE coated NP (Papini et al., 2020). PC impacts on NP cellular internalization and elimination also have been broadly documented. Cheng et al. (2015) revealed that AuNP PC inhibits cellular uptake in a size- and cell-type-dependent manner. Stronger internalization inhibition was observed in large-sized AuNP compared to a smaller size by the phagocytic cell, which might be contributed by differences in endocytic pathways.
Other studies by Giulimondi et al. (2019) found that smaller-size precoated liposomes exhibited lower uptakes by immune cells, enabling longer circulation time in the blood. Artificial human plasma proteins were used as the liposome precoating method prior to static incubation in whole blood. They found that the plasma coating concentration determines the PC composition. Low plasma concentration created PC enriched with fetuin, which is an opsonin, whereas a higher plasma concentration contributed to higher antiopsonin adsorption, namely, clusterin. Therefore, small-size and negatively charged liposomes precoated with a 50% human plasma concentration were chosen for drug delivery in this study. Both internal and external factors that affect PC formation on the NP surface are summarized in Table 1.
Cationic charged liposomes NP tend to form aggregation through their interaction with anionic components in the blood, which hinder blood circulation leading to poor tumor accumulation. IgG also plays an important role in the cationic NP macrophage clearance by affecting its binding to Fc receptors, lamellipodia engulfment, cellular uptakes, and eventually delivery to the lysosomes for degradation (Moghimi and Hunter, 2001). Aside from opsonin-mediated phagocytosis, the NP will also be recognized by the adaptive immune system, either B or T lymphocytes triggering rapid clearance (Mozar and Chowdhury, 2018). A hydrophilic and neutral NP could suppress plasma protein adsorption, whereas a negatively charged NP induces phagocytosis.
Another important determinant is the NP shape. A spherical shape and larger NP size make the NP easier to be sequestered by the immune system and rapidly internalized by macrophages via a mannose receptor-mediated pathway. NP surface modification also plays a key role; for instance, PEGylation could hinder the opsonization process, hence reducing opsonin-mediated phagocytosis (Pozzi et al., 2014). As an alternative to PEGylation, Debayle et al. (2019) developed a coating with zwitterionic polymer ligands which eliminates both hard and soft PC formation on quantum dots NP. This offer improved protection against opsonization that could improve NP circulation time in the blood.
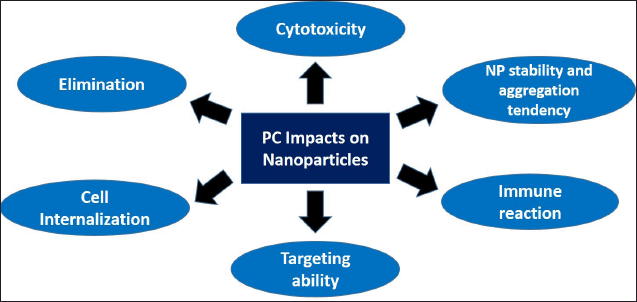 | Figure 2. PC impacts on NPs behavior in biological settings. PC formation determines NP characteristics in biological settings including its stability, targeting ability, cellular internalization, toxicity, and elimination. [Click here to view] |
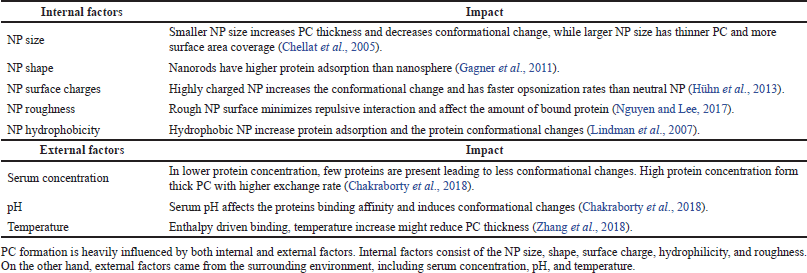 | Table 1. NP internal and external factors that affect PC formations. [Click here to view] |
PC impacts on the NP targeting ability were also investigated by Dai et al. (2015), which confirmed that PC formation did not significantly alter the targeting ability of antibody conjugated core-shell NP toward colon cancer cells. They found that different concentrations of human serum (between 10% and 100%) created a different composition of PC; however, it did not affect its targeting ability. Aside from its therapeutic advantage, PC impacts on cellular cytotoxicity are also important to explore. Shannahan et al. (2015) found that silver NPs form PC after interaction with BSA and human serum albumin which hinder its degradation. The team examined the PC toxicity toward both rat lung epithelial and rat aortic endothelial cells. After internalization, the PC component was lost enabling the NP to induce interleukin-6 expression that contributes to cellular toxicity. Besides, scavenger receptor BI facilitated the silver NP uptakes by the macrophage and its subsequent inflammatory responses and cytotoxicity. In another study, it was found that PC formation hinders radical oxygen species (ROS) formation, leading to lower NP cytotoxicity. NP with semiconductor materials such as ZnO is known to generate ROS, but PC formation can inhibit this phenomenon indicating its protective effect by reducing NP toxicity (Durán et al., 2015; Yin et al., 2015).
NP DIAGNOSTIC APPLICATION IN THE CLINIC
Despite its lack of availability in the clinic, there are already few patented PC-based NP technologies for disease diagnostics, as shown in Table 2. In the past decades, NP has been deployed for various health condition screening tests, by either imaging, blood, or urine testing. Gold and magnetic NP have been extensively studied and utilized for pregnancy testing and microalbuminuria detection via urine tests (Kuppusamy et al., 2014). Microalbuminuria is an indicator of severe kidney damage, which serves as an important marker for progressive cardiovascular and kidney diseases. NP application for pregnancy testing was based on a qualitative assay by detecting the human chorionic gonadotropin (HCG) hormone in the urine. It will lead to color changes, in which pink indicates pregnancy while gray points to the absence of pregnancy (Rojanathanes et al., 2008). A similar qualitative concept was applied to albumin detection in the urine (Wiwanitkit et al., 2007).
Another example of NP application in disease screening is the Nanosphere Verigene System for blood culture nucleic acid tests. The Verigene System is a patented AuNP technology used for various molecular diagnostic assays by identifying the specific target nucleic acid. The Verigene blood test could detect both gram-positive and gram-negative bacteria and their respective resistance markers from blood cultures (Farmer et al., 2017). The results were consistent with the matrix-assisted laser desorption ionization-time of flight- analysis, indicating its high sensitivity and precision. In the clinic, this test is employed to detect antimicrobial resistance markers among burn patients to improve targeted antibiotic therapy (Beal et al., 2013).
NP-enabled blood (NEB) test for cancer diagnostics
There is still a big gap between the investment toward plasma biomarker discovery and the actual biomarkers used in clinical settings. The low number of biomarkers used in the clinic indicates the need for more effective biomarker discovery techniques. Preventative measures and early diagnosis are better than the end-stage curative approach, further raising the need for finding a valid and reliable disease biomarker (Goossens et al., 2015; Selleck et al., 2017). The current proteomics approach is quite challenging due to the abundant components of blood proteins found in the plasma. Therefore, an NP-based approach for biomarker discovery was developed, taking advantage of the PC concept which involves NP interaction with the blood plasma to capture disease-specific proteins.
Some NP surface modifications by specific antibody coating enable specific interaction with the intended proteins. The differences in the PC protein profile pattern among different diseases could be analyzed via SDS PAGE. Assuming that the protein of interest is bound to the NP, it could potentially simplify blood proteomics profiling analysis. This approach is also called the NEB test (Hadjidemetriou et al., 2019). Digiacomo et al. (2021a) conducted the NEB test using AuNP on pancreatic ductal adenocarcinoma (PDAC) patient blood samples with healthy individual blood as the control. In this study, personalized PC characterization by SDS PAGE and densitometric analysis were performed to differentiate PDAC patients and nononcological protein patterns. This NEB test has high sensitivity (78.6%) and specificity (85.3%) rates as well as being affordable, rapid, and deliverable, making it an ideal tool for cancer screening and detection. In a separate study using liposomes, the Digiacomo et al. (2021b) successfully identified specific proteins in the PC, namely, ficolin 3 (FCN-3), that could serve as a potential PDAC biomarker in addition to other known markers such as complements and fibrinogen, which have been confirmed previously in other literature.
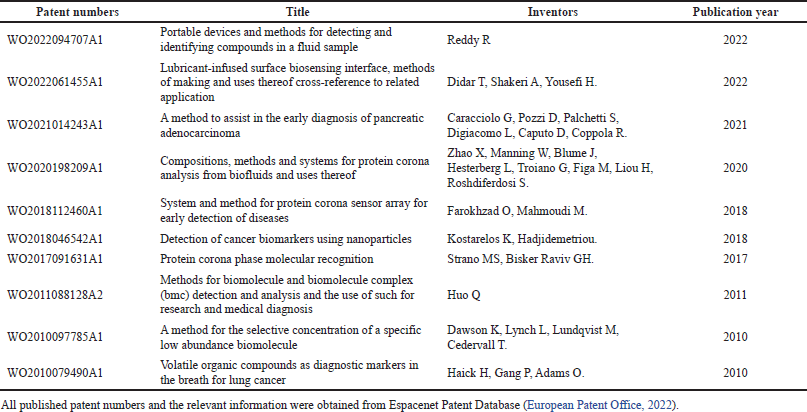 | Table 2. NP PC technology for diseases diagnostics patents list. [Click here to view] |
The complement system’s role in cancer pathogenesis is essential because it affects the elimination of apoptotic and necrotic cancer cells as well as other carcinogenic pathogens to prevent tumorigenesis (?wierzko et al., 2020). FCN itself is a part of the innate immune system that has a function of eliminating abnormal and non-self-antigens through either direct opsonization or the lectin complement pathway. The lectin complement pathway is activated via the binding of pathogen-associated molecular patterns to mannose-binding lectin and FCNs, including ficolin-3 (FCN3) which is mainly expressed in the liver (Lu et al., 2020).
Other research by Vidaurre-Agut et al. (2019) revealed that mesoporous silica NPs PC components were dominated by low molecular weight (MW) serum protein, which proved difficult to capture on standard MS analysis. This study aimed to find a more specific prostate cancer biomarker since elevated PSA levels are often not specific to prostate cancer and might occur due to other health conditions (Moyer et al., 2012). Through stochastic optical reconstruction microscopy analysis, it was shown that small proteins could rapidly fill the NP pores whereas large proteins could not fit into the pores. Additionally, quantitative coadsorption study results also exhibited that small protein presence hinders the adsorption of large MW proteins on the NP surface. Many of the small MW proteins usually are in low concentrations and are often masked by more abundant proteins such as albumin and immunoglobulin and are hence difficult to detect via MS analysis. Therefore, mesoporous silica NP’s ability to selectively capture low MW serum proteins will be useful in diagnosing certain disease states via the proteomics approach, which presents a simpler alternative to the standard MS procedure (Vidaurre-Agut et al., 2019).
NP immunoassay for cancer detection
NP application for clinical immunoassay has been widely studied through either NP-based colorimetric, electrochemical, or immunodipstick assays. The basic concept for NP immunoassay is that the NP-antibody-antigen formation can improve signal detection of disease-specific proteins or biomarkers. Future combination with next-generation sequencing (NGS) devices will potentially enable personalized genetic analysis and more targeted therapeutics for individual patients (Tang et al., 2013).
A previous study by Huo et al. (2011) initiated a similar approach to prostate cancer detection. They utilized a AuNP immunoassay to detect serum biomarkers using both human and mouse blood serum samples, in which they found differences in the NP PC composition of healthy, aggressive, and less aggressive prostate cancer samples. It was observed in mice studies that PC size and IgG levels also differ significantly. Additionally, in both mice and human prostate cancer serum samples, there was a lower level of vascular endothelial growth factor (VEGF) than a healthy individual. VEGF itself plays a key role in cancer angiogenesis. Based on this result, NP immunoassay techniques have the potential to be used as a tool for cancer screening and biomarker discoveries.
Recent research by Moyano et al. (2021) also implemented the NP immunoassay concept for colorectal cancer (CRC) biomarker detection, which has the potential to be a less invasive alternative to colonoscopy. It has been known that extracellular vesicles (EV) express CD147, one of the CRC biomarkers, to monitor treatment responses in CRC patients. The Moyano et al. (2021) developed a quantitative lateral flow immunoassay (LFIA) technique using magnetic NP-antibody conjugate to bind CD147-expressed EV isolated from a plasma sample. In this case, the magnetic LFIA techniques have been coupled to an inductive sensor to quantify the concentration of CD147 biomarkers in the isolated EV from human plasma.
The bio-barcode assay concept has been developed for a long time, combining barcoded DNA and nanotechnology approaches to detect biomarkers, proteins, and various antigens in clinical samples (Yu et al., 2018). The incorporation of monoclonal antibodies into the magnetic nanoparticles (MNP) followed by a mixture with the samples would enable the capture of a specific protein of interest. This MNP was combined with AuNPs that carry the target binding molecules’ DNA barcodes. This technique was proven to be more sensitive than other established methods such as enzyme-linked immunoassay (ELISA) and polymerase chain reaction (PCR). AuNP have two forms of bio-barcode that connect to the linker DNA as a reporter and the other as a signal creating a “sandwich-like” structure between MNP-target DNA barcode-AuNP. Dehybridization will occur on the oligonucleotides of the NP surface allowing the detection of the target DNA (Munir et al., 2020).
Mercadal et al. (2018) proposed the intensity depletion immunolinked assay as an alternative to ELISA. Silver NPs coated with antibodies were used to detect gliadin, a marker for celiac disease. The result itself was comparable to ELISA and provided an alternative to fill the need for an enzyme-free analytical assay with improved stability and smaller sample size.
NP PC-BASED DIAGNOSTIC CHALLENGES
Some challenges that occurred during PC analysis were mostly due to technical issues, such as lack of standardized research protocols, complexities of LC-MS analysis preparations, and variations in the biological mediums and the NP characteristics (Hajipour et al., 2015). This could cause both inconsistent experimental results and data misinterpretation leading to reproducibility problems. Different instrument types utilized among laboratories also contributed to data reproducibility issues. A previous study showed that the LC-MS instrument type employed gives a 30%–60% impact on the peptide’s repeatability in technical replicates. For example, the Orbitrap instrument had better repeatability and stability for protein analysis than Thermo LTQ (Tabb et al., 2010). Additionally, poor sample preparation may introduce impurities and contamination during PC analysis, reducing the proteomics data validity.
Before analysis, a sample authentication process is an important step. All biological samples must have proper identification including sample collection methods, patient demographics data, and medical information. This information is crucial since it could affect the PC compositions and interpretations (Mahmoudi, 2022). The PPC concept showed that NP impacts on each individual are different depending on their health status, age, gender, and genetic background creating a unique PPC characteristic. This unique PPC identity of each patient will influence its diagnostic and personalized therapeutic utilization (Corbo et al., 2017). A deeper understanding of NP-proteins binding sites and PC structural organization and formation behavior will be beneficial to creating new strategies to control NP interaction with the biological system (Mahmoudi, 2022).
PC structure consists of several layers, not only one layer, which means the PC’s outermost layer most likely has the largest influence on the NP behavior in vivo. Therefore, the study design must separate inner and outer PC layers to get a more accurate interpretation instead of combining the total PC (Bai et al., 2021). Additionally, the widely employed PC characterization method using LC-MS analysis is only effective in detecting hard PC since most of the soft PC was removed during the sample purification step. Soft PC’s unstable and dynamic nature becomes a huge challenge for its detection, making biomarker discoveries research challenging. Most disease biomarkers are low-abundance proteins that do not have a high affinity toward the NP (Blume et al., 2020). Besides, most small proteins are also more prone to enzymatic degradation after their collection, presenting more challenges for sample collection and storage (Marshall et al., 2003).
However, some studies reported successful soft PC detection by utilizing differential centrifugal sedimentation and asymmetric flow field-flow fractionation (AF4) techniques (Davidson et al., 2017; Weber et al., 2018). The utilization of advanced imaging techniques such as fluorescence microscopy or Forster resonance energy transfer spectroscopy enables PC formation observation in real time (Zhang et al., 2020). In the future, this real-time analysis will be useful for NP theranostic applications, where disease diagnostics and therapeutics can be done simultaneously.
Other challenges would be discrepancies between the static in vitro PC experimental results and the dynamic condition in vivo due to environmental factors that are difficult to replicate in vitro such as blood shear stress and other hemodynamic conditions in the vascular system (Pozzi et al., 2015). Caracciolo et al. (2014) utilized peptide mass fingerprinting to identify and classify the PC components from both human and mice plasma based on their functionality. They found significant differences in the mice and human PC identity that affect its biological behavior differently. This indicates that results from mice experiments might not be directly applicable to humans due to differences in the physiological environment between both species. Thus, developing new types of screening that better mimic human body systems will be beneficial to reducing discrepancies which hinder translational research.
In the bloodstream, uncoated NP interaction with blood proteins may form opsonin-rich PC, leading to opsonin-mediated phagocytosis and faster elimination, as shown in Figure 3. Many factors play roles in PC formation including the presence of opsonins, coagulation factors, and white blood cells. Therefore, surface precoating might be one possible strategy to reduce the unwanted protein binding to the NP surface (Hadjidemetriou, 2019). It should be noted that modifying the NP PC can affect its interaction with other NP and the cells by influencing its aggregation tendency, payload efficiency, or cellular uptakes (Qu et al., 2020). A deeper analysis of PC formation behavior and characteristics is crucial to creating an effective NP design for the theranostic application. Data accuracy, reproducibility, and scalability remain the core issues for PC-based NP technologies clinical translations.
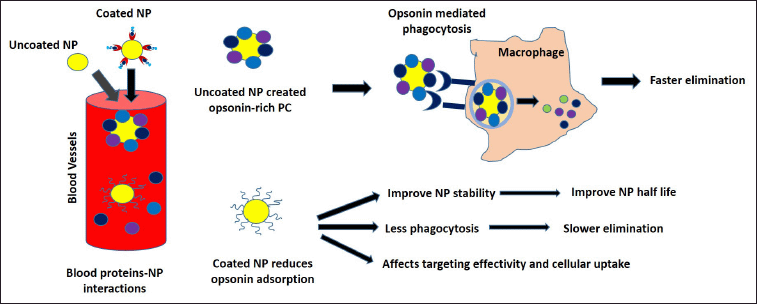 | Figure 3. NPs surface pre-coating strategy. NP surface pre-coating could affect its PC compositions. Uncoated NP will interact with opsonin (IgG, complement, and fibrinogen) in the blood, creating an opsonin-rich PC. A surface modification strategy is often employed to reduce NP–opsonin interaction to improve NP half-life, and stability while preventing opsonin-mediated phagocytosis. However, high-density surface coating might hinder cellular uptakes, affecting its targeting effectivity. [Click here to view] |
PPC STRATEGIES AND FUTURE OUTLOOK
Biomarkers development can be classified into three stages, namely, discovery, verification, and validation. Discovery is the first step, in which numerous biomarkers candidates are screened and identified through the untargeted MS approach. In the verification phase, the target peptides are identified and quantified using the targeted MS technology. For the last validation step, the sample analysis is focused on a small number of selected peptides to ensure their sensitivity, specificity, stability, and reproducibility (Nakayasu et al., 2021). Blood plasma sample analysis is challenging due to the presence of various high-abundance proteins that could mask the biomarkers proteins. Some approach that is often employed to remove the high-abundance proteins is either immunodepletion or fractionation by chromatography. Despite its effectivity, immunodepletion could codeplete the low-abundance proteins due to protein–protein interactions (Keshishian et al., 2017). Alternatively, fractionation and chromatography approaches are also successful in reducing the sample complexities, yet they need multiple runs of chromatography. To enhance the visibility of low-abundance proteins, isobaric labeling quantification techniques such as Isobaric tags for relative and absolute quantitation (iTRAQ) or tandem mass tags (TMT) can be performed prior to fractionation and MS analysis (Nakayasu et al., 2021). Thus, a more advanced proteomics platform and integration with machine learning algorithms may potentially improve biomarker development and data analysis capabilities.
Several strategies have been explored to improve PC identification and quantification efficiency and scalability, as summarized in Figure 4. Blume et al. (2020) developed a five panels of iron oxide superparamagnetic NPs automated Proteograph platform that has been commercialized, called Proteomics Product Suite. Each of these NP has a superparamagnetic core enabling it to quickly separate the PC from blood plasma in less than 30 seconds, replacing the need for SDS PAGE and in-gel digestion processes prior to LC-MS. PC protein separation was conducted in the plate by the magnetic properties; hence, unbound protein can be washed, followed by automated in-plate digestion and protein purification. This also demonstrated that magnetic NP functionalization could be tailored for proteomics analysis purposes. The Proteograph workflow has a wide proteome coverage, as shown by its capability to detect both high- and low-abundance proteins. It is also integrated with the cloud-based Proteograph software, called Proteograph Analytics Suite (PAS), which could analyze both the Proteograph and NGS data from the proteogenomics platform. Various visualizations were incorporated into the software such as proteomics heatmaps, protein interaction maps, protein intensity comparison and volcano mapping, hierarchical clustering, and peptide relation to gene structure analysis for large-scale proteogenomics study. All of these data analytics tools will help to create a comprehensive database of peptide data mapping for PC profiling and quantification (Gajadhar et al., 2022). The compatibility of the Proteograph approach with large-scale genomics opens up multiomic study opportunities for plasma PC profiling.
Furthermore, Hornburg et al. (2022) calculated the protein-to-NP-surface ratio (P/NP ratio) attributable to the competitive binding nature among proteins that affect PC formation. Thus, optimization of the P/NP ratio can improve the Proteograph system workflow performance. For instance, by increasing the competitive binding and reducing the NP binding surface, there will be 1.2–1.7 times more proteins in the PC than in the references, indicating the presence of a more diverse set of proteins. It was also proven to be three times more accurate than the conventional neat proteomics workflow.
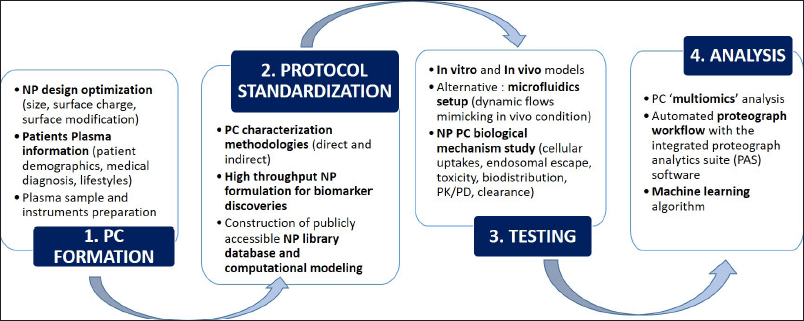 | Figure 4. PPC analysis strategies for translational research. The first step is PC formation which depends on the NP physicochemical characteristics and patients’ plasma sample preparations. Then, protocol standardization for large-scale PPC analysis will be employed, including the PC characterization methodologies, high throughput NP formulations, and construction of NP library database and computational modeling. Automated high throughput screening will be ideal for biomarker discoveries since it has better efficiency, speed, and reproducibility (Ferdosi et al., 2022; Yan et al., 2020). The third step would be preclinical testing to examine the PC biological mechanisms using in vitro and in vivo modeling. Several aspects such as cellular uptakes, endosomal escape, toxicity, and clearance will be examined. To tackle the discrepancies between in vitro and in vivo data, a microfluidics setup was developed as an alternative model that heavily mimics the real in vivo conditions (Digiacomo et al., 2019). The last step is PC analysis, consisting of multi-omics analysis, automated Proteograph workflow, and machine learning algorithm. The integrated PAS software can create heatmaps, protein interaction maps, protein intensity comparison, and integrated analysis of genomics and proteomics data (Blume et al., 2020). These strategies’ overall aim is to improve PPC analysis performance, scalability, accuracy, and reproducibility. [Click here to view] |
Thus, the Proteograph approach simplifies LC-MS sample preparation and improves PC detection scalability by employing an automated high-throughput proteomics system. This method also has been employed to diagnose the early non-small-cell lung cancer (NSCLC) subjects from age- and gender-matched healthy controls, resulting in the detection of 53 protein biomarkers for NSCLC in the patients’ plasma, indicating its huge potential to be used as a biomarker discovery tool (Blume et al., 2020; Donovan et al., 2022). Simulations of PC-biological components molecular dynamics allow a deeper understanding of atomic-level phenomena that are currently difficult to observe.
However, there is still a lack of NP PC database libraries, making the creation of standardized protocols and large-scale analysis difficult. Kamaly et al. (2022) employed a machine learning algorithm to improve PC identification from Alzheimer’s patients’ blood samples. To train the algorithm, they employed six NP for Alzheimer’s patient PC profiling, which consisted of two different NP types (silica and polystyrene) and three different functions (none, carboxy-, and ammino-), resulting in a unique PC fingerprint for Alzheimer’s disease. The algorithm can accurately predict Alzheimer’s onset even before official clinical diagnosis with 100% sensitivity and >93% specificity, proving its significant role in PC profiling for disease diagnosis and PC fingerprinting.
The integration of machine learning modeling, such as random forest (RF), raises the need for further improvement in the data extraction and data mining processes to better examine the relationship between NP and PC and its biological responses based on published pieces of evidence. Strict criteria must be applied during literature extraction and data mining to reduce biases. Even though machine learning models can explain observation using available training data, new data on factor-response dependence models is required to achieve higher accuracy on PC formation and behavior assessment. Ban et al. (2020) compared 40 types of unmodified NP with 50 types of surface-modified NP to evaluate the accuracy of this model prediction by tenfold cross-validation. The RF statistical model was able to classify the PC compositions according to the pI, mass, and Grand Average of Hydropathicity Index score, making it suitable to assess large-scale heterogeneous NP PC data. Thus, machine learning exhibited tremendous capability to predict NP–cell interactions based on the PC compositions, which is very useful for NP design strategy.
The construction of a publicly accessible online NP database and computational modeling would be useful to aid NP design optimization and biological response prediction based on individual NP nanostructures. It has the potential to significantly accelerate PPC translational research globally (Yan et al., 2020). Some other modeling approaches, namely, nano-quantitative structure-activity relationship, have been utilized by Buglak et al. (2019) to analyze the correlation between PC structure and activity using machine learning methods, resulting in a more accurate prediction of the PC effects on cells including its toxicity.
Another major challenge that needs to be addressed in the PPC field is the highly dynamic nature of in vivo modeling. Besides, the PC component in different species samples may vary, for instance, between murine and human. This leads to discrepancies between in vivo and clinical data, hindering its translational research progress. The current advances in the in vitro microfluidics setup have the potential to bridge the gap since it closely mimics the realistic biological in vivo condition by precise control of the physiological factors such as shear stress, hydrostatic pressure, and nutrients flow. The concept itself has been utilized for high-throughput drug screening and NP-targeted drug delivery (Ozcelikkale et al., 2017). Digiacomo et al. (2019) injected AuNP and human plasma into each islet of a remote-controlled microfluidic device and compared it with the static system as a reference. It was found that the PC components in the microfluidic environment were dominated by immunoglobulins, whereas tissue leakage proteins were the PC’s majority in the static systems raising the need for further investigation. Additionally, a high-throughput real-time endosomal escape imaging assay developed by Munson et al. (2021) enables researchers to observe real-time nano-bio-interactions starting from cellular uptakes and endosomal escape until protein translation and clearance.
Overall, the recent growth of the multi-NP Proteograph assay combined with an advanced proteomics data processing workflow strategy exhibited better performance than a conventional data processing workflow (such as a depleted plasma or neat plasma workflow). It gives superior PC profiling scalability, reproducibility, and protein group and dynamic range coverage. This field is still continually evolving, with multidisciplinary “omics” and machine learning algorithms playing increasingly important roles in the PC-based biomarker discoveries analysis (Liu et al., 2022).
CONCLUSION
The interaction between NPs and biological components form PC that might change the NP identity in biological settings. NP–protein interaction is dynamic and multifactorial, depending on the NP physicochemical characteristics and the surrounding environment. However, the complex and dynamic nature of PC makes characterization quite challenging. High-affinity proteins on the NP surface will form hard corona that can be characterized easily, whereas low-affinity proteins form soft corona which is more difficult to analyze. The lack of standardized PC research protocols and the discrepancy between the static in vitro and in vivo experimental conditions hinder its translational research. Therefore, more advanced methodologies were developed, including the automated Proteograph workflow, machine learning algorithms, microfluidics models, and multiomics analysis, to improve PC analysis performance, accuracy, reproducibility, and scalability. NP PC characterization patterns vary in different disease plasma samples indicating its potential use for disease screening and detection. The application of PPC is very promising since it offers a less invasive, simpler, and more reliable diagnostic tool than conventional methods, for instance, by using -NEB test and NP immunoassay strategies. The theranostic approach would be the next step, where NP-based treatment and diagnostics can be done simultaneously in real time. Additionally, a more comprehensive pathway analysis using a multiomics approach can be explored.
ACKNOWLEDGEMENTS
Declared none.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
FUNDING
There is no funding to report.
ETHICAL APPROVAL
Not applicable.
AUTHORS’ CONTRIBUTIONS
FSM designed, wrote, and edited the manuscript.
REFERENCES
Ahsan SM, Rao CM, Ahmad MF. Nanoparticle-protein interaction: the significance and role of protein corona. Adv Exp Med Biol, 2018; 1048:175–98; doi:10.1007/978-3-319-72041-8_11. CrossRef
Amin F, Yushchenko DA, Montenegro JM, Parak WJ. Integration of organic fluorophores in the surface of polymer-coated colloidal nanoparticles for sensing the local polarity of the environment. Chemphyschem, 2012; 13(4):1030–5; doi:10.1002/cphc.201100901. CrossRef
Bai X, Wang J, Mu Q, Su G. In vivo protein corona formation: characterizations, effects on engineered nanoparticles’ Biobehaviors, and applications. Front Bioeng Biotechnol, 2021; 9:646708; doi:10.3389/fbioe.2021.646708. CrossRef
Ban Z, Yuan P, Yu F, Peng T, Zhou Q, Hu X. Machine learning predicts the functional composition of the protein corona and the cellular recognition of nanoparticles. Proc Natl Acad Sci USA, 2020; 117(19):10492–9; doi:10.1073/pnas.1919755117. CrossRef
Beal SG, Ciurca J, Smith G, John J, Lee F, Doern CD, Gander RM. Evaluation of the nanosphere verigene Gram-positive blood culture assay with the VersaTREK blood culture system and assessment of possible impact on selected patients. J Clin Microbiol, 2013; 51(12):3988–92; doi:10.1128/JCM.01889-13. CrossRef
Blume JE, Manning WC, Troiano G, Hornburg D, Figa M, Hesterberg L, Platt TL, Zhao X, Cuaresma RA, Everley PA, Ko M, Liou H, Mahoney M, Ferdosi S, Elgierari EM, Stolarczyk C, Tangeysh B, Xia H, Benz R, Siddiqui A, Carr SA, Ma P, Langer R, Farias V, Farokhzad OC. Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona. Nat Commun, 2020; 11(1):3662; doi:10.1038/s41467-020-17033-7. CrossRef
Boulos SP, Davis TA, Yang JA, Lohse SE, Alkilany AM, Holland LA, Murphy CJ. Nanoparticle-protein interactions: a thermodynamic and kinetic study of the adsorption of bovine serum albumin to gold nanoparticle surfaces. Langmuir, 2013; 29(48):14984–96; doi:10.1021/la402920f. CrossRef
Buglak AA, Zherdev AV, Dzantiev BB. Nano-(Q)SAR for cytotoxicity prediction of engineered nanomaterials. Molecules, 2019; 24(24):4537; doi:10.3390/molecules24244537. CrossRef
Caputo D, Papi M, Coppola R, Palchetti S, Digiacomo L, Caracciolo G, Pozzi D. A protein corona-enabled blood test for early cancer detection. Nanoscale, 2017; 9(1):349–54; doi:10.1039/c6nr05609a. CrossRef
Caracciolo G, Pozzi D, Capriotti AL, Cavaliere C, Piovesana S, La Barbera G, Amici A, Laganà A. The liposome-protein corona in mice and humans and its implications for in vivo delivery. J Mater Chem B, 2014; 2(42):7419–28; doi:10.1039/c4tb01316f. CrossRef
Cedervall T, Lynch I, Lindman S, Berggård T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci USA, 2007; 104(7):2050–5; doi:10.1073/pnas.0608582104. CrossRef
Chakraborty D, Chauhan P, Alex SA, Chaudhary S, Ethiraj KR, Chandrasekaran N, Mukherjee A. Comprehensive study on biocorona formation on functionalized selenium nanoparticle and its biological implications. J Mol Liq, 2018; 268:335–42; doi:10.1016/j.molliq.2018.07.070 CrossRef
Chetwynd AJ, Lynch I. The rise of the nanomaterial metabolite corona, and emergence of the complete corona. Environ Sci Nano, 2020; 7:1041–60; doi:10.1039/C9EN00938H CrossRef
Chellat F, Merhi Y, Moreau A, Yahia L. Therapeutic potential of nanoparticulate systems for macrophage targeting. Biomaterials, 2005; 26(35):7260–75; doi:10.1016/j.biomaterials.2005.05.044. CrossRef
Cheng X, Tian X, Wu A, Li J, Tian J, Chong Y, Chai Z, Zhao Y, Chen C, Ge C. Protein corona influences cellular uptake of gold nanoparticles by phagocytic and nonphagocytic cells in a size-dependent manner. ACS Appl Mater Interfaces, 2015; 7(37):20568–75; doi:10.1021/acsami.5b04290. CrossRef
Colapicchioni V, Tilio M, Digiacomo L, Gambini V, Palchetti S, Marchini C, Pozzi D, Occhipinti S, Amici A, Caracciolo G. Personalized liposome-protein corona in the blood of breast, gastric and pancreatic cancer patients. Int J Biochem Cell Biol, 2016; 75:180–7; doi:10.1016/j.biocel.2015.09.002. CrossRef
Corbo C, Molinaro R, Tabatabaei M, Farokhzad OC, Mahmoudi M. Personalized protein corona on nanoparticles and its clinical implications. Biomater Sci, 2017; 5(3):378–87; doi:10.1039/c6bm00921b. CrossRef
Dai Q, Yan Y, Ang CS, Kempe K, Kamphuis MM, Dodds SJ, Caruso F. Monoclonal antibody-functionalized multilayered particles: targeting cancer cells in the presence of protein coronas. ACS Nano, 2015; 9(3):2876–85; doi:10.1021/nn506929e. CrossRef
Davidson AM, Brust M, Cooper DL, Volk M. Sensitive analysis of protein adsorption to colloidal gold by differential centrifugal sedimentation. Anal Chem, 2017; 89(12):6807–14; doi:10.1021/acs.analchem.7b01229. CrossRef
Debayle M, Balloul E, Dembele F, Xu X, Hanafi M, Ribot F, Monzel C, Coppey M, Fragola A, Dahan M, Pons T, Lequeux N. Zwitterionic polymer ligands: an ideal surface coating to totally suppress protein-nanoparticle corona formation? Biomaterials, 2019; 219:119357; doi:10.1016/j.biomaterials.2019.119357. CrossRef
Digiacomo L, Palchetti S, Giulimondi F, Pozzi D, Zenezini Chiozzi R, Capriotti AL, Laganà A, Caracciolo G. The biomolecular corona of gold nanoparticles in a controlled microfluidic environment. Lab Chip, 2019; 19(15):2557–67; doi:10.1039/c9lc00341j. CrossRef
Digiacomo L, Caputo D, Coppola R, Cascone C, Giulimondi F, Palchetti S, Pozzi D, Caracciolo G. Efficient pancreatic cancer detection through personalized protein corona of gold nanoparticles. Biointerphases, 2021a; 16(1):011010; doi:10.1116/6.0000540. CrossRef
Digiacomo L, Giulimondi, F, Pozzi D, Coppola A, La Vaccara V, Caputo D, Caracciolo G. A proteomic study on the personalized protein corona of liposomes. relevance for early diagnosis of pancreatic ductal adenocarcinoma and biomarker detection. J Nanotheranostics, 2021b; 2(2):82–93; doi:10.3390/jnt2020006 CrossRef
Docter D, Westmeier D, Markiewicz M, Stolte S, Knauer SK, Stauber RH. The nanoparticle biomolecule corona: lessons learned—challenge accepted? Chem Soc Rev, 2015; 44(17):6094–121; doi:10.1039/c5cs00217f. CrossRef
Donovan MKR, Huang Y, Blume JE, Wang J, Hornburg D, Mohtashemi I, Kim S, Ko M, Benzi RW, Platt TL, Batzoglou S, Farokhzad OC, Siddiqui OC. Peptide-centric analyses of human plasma enable increased resolution of biological insights into non-small cell lung cancer relative to protein-centric analysis. bioRxiv, 2022; doi:10.1101/2022.01.07.475393. CrossRef
Durán N, Silveira CP, Durán M, Martinez DS. Silver nanoparticle protein corona and toxicity: a mini-review. J Nanobiotechnol, 2015; 13:55; doi:10.1186/s12951-015-0114-4. CrossRef
European Patent Office. Espacenet Patent Database. 2022. Available via https://worldwide.espacenet.com/ (Accessed 5 July 2022).
Farmer A, Lanteri CA, Steele E, Mende K, Pamplin J, Akers KS. Utility of the nanosphere Verigene in the identification of bacteremia isolates from the burn intensive care unit. Mil Med, 2017; 182(9):e1779–84; doi:10.7205/MILMED-D-17-00018. CrossRef
Ferdosi S, Tangeysh B, Brown TR, Everley PA, Figa M, McLean M, Elgierari EM, Zhao X, Garcia VJ, Wang T, Chang MEK, Riedesel K, Chu J, Mahoney M, Xia H, O’Brien ES, Stolarczyk C, Harris D, Platt TL, Ma P, Goldberg M, Langer R, Flory MR, Benz R, Tao W, Cuevas JC, Batzoglou S, Blume JE, Siddiqui A, Hornburg D, Farokhzad OC. Engineered nanoparticles enable deep proteomics studies at scale by leveraging tunable nano-bio interactions. Proc Natl Acad Sci USA, 2022; 119(11):e2106053119; doi:10.1073/pnas.2106053119. CrossRef
Gagner JE, Lopez MD, Dordick JS, Siegel RW. Effect of gold nanoparticle morphology on adsorbed protein structure and function. Biomaterials, 2011; 32(29):7241–52; doi:10.1016/j.biomaterials.2011.05.091. CrossRef
Gajadhar AS, Donovan MK, Auluck H, Berk Y, Lou Y, Platt T, Batzoglou S. A cloud-scalable software suite for large-cohort proteogenomics data analysis and visualization [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2022, 2022 Apr 8–13, AACR, Philadelphia, PA; Cancer Res, 2022; 82(12_Suppl):Abstract nr 6348. CrossRef
Giulimondi F, Digiacomo L, Pozzi D, Palchetti S, Vulpis E, Capriotti AL, Chiozzi RZ, Laganà A, Amenitsch H, Masuelli L, Peruzzi G, Mahmoudi M, Screpanti I, Zingoni A, Caracciolo G. Interplay of protein corona and immune cells controls blood residency of liposomes. Nat Commun, 2019; 10(1):3686; doi:10.1038/s41467-019-11642-7 CrossRef
Goossens N, Nakagawa S, Sun X, Hoshida Y. Cancer biomarker discovery and validation. Transl Cancer Res, 2015; 4(3):256–69; doi:10.3978/j.issn.2218-676X.2015.06.04.
Gupta MN, Roy I. How corona formation impacts nanomaterials as drug carriers. Mol Pharm, 2020; 17(3):725–37; doi:10.1021/acs.molpharmaceut.9b01111. CrossRef
Hadjidemetriou M, Al-Ahmady Z, Kostarelos K. Time-evolution of in vivo protein corona onto blood-circulating PEGylated liposomal doxorubicin (DOXIL) nanoparticles. Nanoscale, 2016; 8(13):6948–57; doi:10.1039/c5nr09158f. CrossRef
Hadjidemetriou M, Al-Ahmady Z, Buggio M, Swift J, Kostarelos K. A novel scavenging tool for cancer biomarker discovery based on the blood-circulating nanoparticle protein corona. Biomaterials, 2019; 188:118–29; doi:10.1016/j.biomaterials.2018.10.011. CrossRef
Hajipour MJ, Raheb J, Akhavan O, Arjmand S, Mashinchian O, Rahman M, Abdolahad M, Serpooshan V, Laurent S, Mahmoudi M. Personalized disease-specific protein corona influences the therapeutic impact of graphene oxide. Nanoscale, 2015; 7(19):8978–94; doi:10.1039/c5nr00520e. CrossRef
Hornburg D, Ferdosi S, Hasan M, Tangeysh B, Brown TR, Wang T, Elgierari EM, Zhao X, Alavi A, Chu J, Figa M, Tao W, Wang J, Goldberg M, Xia H, Stolarczyk C, Batzoglou S, Siddiqui A, Farokhzad OC. Enhanced competitive protein exchange at the nano-bio interface enables ultra-deep coverage of the human plasma proteome. bioRxiv, 2022; doi:10.1101/2022.01.08.475439. CrossRef
Hühn D, Kantner K, Geidel C, Brandholt S, De Cock I, Soenen SJ, Rivera Gil P, Montenegro JM, Braeckmans K, Müllen K, Nienhaus GU, Klapper M, Parak WJ. Polymer-coated nanoparticles interacting with proteins and cells: focusing on the sign of the net charge. ACS Nano, 2013; 7(4):3253–63; doi:10.1021/nn3059295. CrossRef
Huo Q, Colon J, Cordero A, Bogdanovic J, Baker CH, Goodison S, Pensky MY. A facile nanoparticle immunoassay for cancer biomarker discovery. J Nanobiotechnol, 2011; 9:20; doi:10.1186/1477-3155-9-20. CrossRef
Kamaly N, Farokhzad OC, Corbo C. Nanoparticle protein corona evolution: from biological impact to biomarker discovery. Nanoscale, 2022; 14(5):1606–20; doi:10.1039/d1nr06580g. CrossRef
Ke PC, Lin S, Parak WJ, Davis TP, Caruso F. A decade of the protein corona. ACS Nano, 2017; 11(12):11773–6; doi:10.1021/acsnano.7b08008. CrossRef
Keshishian H, Burgess MW, Specht H, Wallace L, Clauser KR, Gillette MA, Carr SA. Quantitative, multiplexed workflow for deep analysis of human blood plasma and biomarker discovery by mass spectrometry. Nat Protoc, 2017; 12(8):1683–701; doi:10.1038/nprot.2017.054. CrossRef
Kuppusamy P, Mashitah MY, Maniam GP, Govindan N. Biosynthesized gold nanoparticle developed as a tool for detection of HCG hormone in pregnant women urine sample. Asian Pac J Trop Dis, 2014; 4(3):237; doi:10.1016/S2222-1808(14)60538-7. CrossRef
Lindman S, Lynch I, Thulin E, Nilsson H, Dawson KA, Linse S. Systematic investigation of the thermodynamics of HSA adsorption to N-iso-propylacrylamide/N-tert-butylacrylamide copolymer nanoparticles. Effects of particle size and hydrophobicity. Nano Lett, 2007; 7(4):914–20; doi:10.1021/nl062743. CrossRef
Li Y, Lee JS. Insights into characterization methods and biomedical applications of nanoparticle-protein corona. Materials (Basel), 2020; 13(14):3093; doi:10.3390/ma13143093. CrossRef
Liu K, Salvati A, Sabirsh A. Physiology, pathology and the biomolecular corona: the confounding factors in nanomedicine design. Nanoscale, 2022; 14(6):2136–54; doi:10.1039/d1nr08101b. CrossRef
Lu X, Xu P, Ding HM, Yu YS, Huo D, Ma YQ. Tailoring the component of protein corona via simple chemistry. Nat Commun, 2019; 10(1):4520; doi:10.1038/s41467-019-12470-5. CrossRef
Lu Z, Wu J, Li Y, Chen W, Yu Q, Zhou, J. FCN3 is a prognostic biomarker correlated with immune response in liver cancer. Res Sq, 2020 PREPRINT (Version 1); doi:10.21203/rs.3.rs-90864/v1 CrossRef
Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci USA, 2008; 105(38):14265–70; doi:10.1073/pnas.0805135105. CrossRef
Mahmoudi M. The need for improved methodology in protein corona analysis. Nat Commun, 2022; 13(1):49; doi:10.1038/s41467-021-27643-4. CrossRef
Marshall J, Kupchak P, Zhu W, Yantha J, Vrees T, Furesz S, Jacks K, Smith C, Kireeva I, Zhang R, Takahashi M, Stanton E, Jackowski G. Processing of serum proteins underlies the mass spectral fingerprinting of myocardial infarction. J Proteome Res, 2003; 2(4):361–72; doi:10.1021/pr030003l. CrossRef
Mercadal PA, Fraire JC, Motrich RD, Coronado EA. Enzyme-free immunoassay using silver nanoparticles for detection of gliadin at ultralow concentrations. ACS Omega, 2018; 3(2):2340–50; doi:10.1021/acsomega.7b01840. CrossRef
Moghimi SM, Hunter AC. Recognition by macrophages and liver cells of opsonized phospholipid vesicles and phospholipid headgroups. Pharm Res, 2001; 18(1):1–8; doi:10.1023/a:1011054123304. CrossRef
Moyano A, Serrano-Pertierra E, Duque JM, Ramos V, Teruel-Barandiarán E, Fernández-Sánchez MT, Salvador M, Martínez-García JC, Sánchez L, García-Flórez L, Rivas M, Blanco-López MDC. Magnetic lateral flow immunoassay for small extracellular vesicles quantification: application to colorectal cancer biomarker detection. Sensors (Basel), 2021; 21(11):3756; doi:10.3390/s21113756. CrossRef
Moyer VA, US Preventive Services Task Force. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med, 2012; 157(2):120–34; doi:10.7326/0003-4819-157-2-201207170-00459. CrossRef
Mozar FS, Chowdhury EH. Surface-modification of carbonate apatite nanoparticles enhances delivery and cytotoxicity of gemcitabine and anastrozole in breast cancer cells. Pharmaceutics, 2017; 9(2):21; doi:10.3390/pharmaceutics9020021. CrossRef
Mozar FS, Chowdhury EH. Impact of PEGylated nanoparticles on tumor targeted drug delivery. Curr Pharm Des, 2018; 24(28):3283–96; doi:10.2174/1381612824666180730161721. CrossRef
Munir S, Ahmed S, Ibrahim M, Khalid M, Ojha SC. A spellbinding interplay between biological barcoding and nanotechnology. Front Bioeng Biotechnol, 2020; 8:883; doi:10.3389/fbioe.2020.00883. CrossRef
Munson MJ, O’Driscoll G, Silva AM, Lázaro-Ibáñez E, Gallud A, Wilson JT, Collén A, Esbjörner EK, Sabirsh A. A high-throughput Galectin-9 imaging assay for quantifying nanoparticle uptake, endosomal escape and functional RNA delivery. Commun Biol, 2021; 4(1):211; doi:10.1038/s42003-021-01728-8 CrossRef
Nakayasu ES, Gritsenko M, Piehowski PD, Gao Y, Orton DJ, Schepmoes AA, Fillmore TL, Frohnert BI, Rewers M, Krischer JP, Ansong C, Suchy-Dicey AM, Evans-Molina C, Qian WJ, Webb-Robertson BM, Metz TO. Tutorial: best practices and considerations for mass-spectrometry-based protein biomarker discovery and validation. Nat Protoc, 2021; 16(8):3737–60; doi:10.1038/s41596-021-00566-6. CrossRef
Nguyen VH, Lee BJ. Protein corona: a new approach for nanomedicine design. Int J Nanomed, 2017; 12:3137–51; doi:10.2147/IJN.S129300. CrossRef
Ozcelikkale A, Moon HR, Linnes M, Han B. In vitro microfluidic models of tumor microenvironment to screen transport of drugs and nanoparticles. Wiley Interdiscip Rev Nanomed Nanobiotechnol, 2017; 9(5):10.1002/wnan.1460; doi:10.1002/wnan.1460. CrossRef
Papini E, Tavano R, Mancin F. Opsonins and dysopsonins of nanoparticles: facts, concepts, and methodological guidelines. Front Immunol, 2020; 11:567365; doi:10.3389/fimmu.2020.567365. CrossRef
Pfeiffer C, Rehbock C, Hühn D, Carrillo-Carrion C, de Aberasturi DJ, Merk V, Barcikowski S, Parak WJ. Interaction of colloidal nanoparticles with their local environment: the (ionic) nanoenvironment around nanoparticles is different from bulk and determines the physico-chemical properties of the nanoparticles. J R Soc Interface, 2014; 11(96):20130931; doi:10.1098/rsif.2013.0931. CrossRef
Piella J, Bastús NG, Puntes V. Size-dependent protein-nanoparticle interactions in citrate-stabilized gold nanoparticles: the emergence of the protein corona. Bioconjug Chem, 2017; 28(1):88–97; doi:10.1021/acs.bioconjchem.6b00575. CrossRef
Pozzi D, Colapicchioni V, Caracciolo G, Piovesana S, Capriotti AL, Palchetti S, De Grossi S, Riccioli A, Amenitsch H, Laganà A. Effect of polyethyleneglycol (PEG) chain length on the bio-nano-interactions between PEGylated lipid nanoparticles and biological fluids: from nanostructure to uptake in cancer cells. Nanoscale, 2014; 6(5):2782–92; doi:10.1039/c3nr05559k. CrossRef
Pozzi D, Caracciolo G, Digiacomo L, Colapicchioni V, Palchetti S, Capriotti AL, Cavaliere C, Zenezini Chiozzi R, Puglisi A, Laganà A. The biomolecular corona of nanoparticles in circulating biological media. Nanoscale, 2015; 7(33):13958–66; doi:10.1039/c5nr03701h CrossRef
Qu S, Sun F, Qiao Z, Li J, Shang L. In situ investigation on the protein corona formation of quantum dots by using fluorescence resonance energy transfer. Small, 2020; 16(21):e1907633; doi:10.1002/smll.201907633. CrossRef
Radauer-Preimi I, Andosch A, Hawranek T, Luetz-Meindl U, Wiederstein M, Horejs-Hoeck J, Himly M, Boyles M, Duschl A. Nanoparticle-allergen interactions mediate human allergic responses: protein corona characterization and cellular responses. Part Fibre Toxicol, 2016; 13:3; doi:10.1186/s12989-016-0113-0. CrossRef
Rojanathanes R, Sereemaspun A, Pimpha N, Buasorn V, Ekawong P, Wiwanitkit V. Gold nanoparticle as an alternative tool for a urine pregnancy test. Taiwan J Obstet Gynecol, 2008; 47(3):296–9; doi:10.1016/S1028-4559(08)60127-8. CrossRef
Selleck MJ, Senthil M, Wall NR. Making meaningful clinical use of biomarkers. Biomark Insights, 2017; 12:1177271917715236; doi:10.1177/1177271917715236. CrossRef
Shannahan JH, Podila R, Aldossari AA, Emerson H, Powell BA, Ke PC, Rao AM, Brown JM. Formation of a protein corona on silver nanoparticles mediates cellular toxicity via scavenger receptors. Toxicol Sci, 2015; 143(1):136–46; doi:10.1093/toxsci/kfu217. CrossRef
Sund J, Alenius H, Vippola M, Savolainen K, Puustinen A. Proteomic characterization of engineered nanomaterial-protein interactions in relation to surface reactivity. ACS Nano, 2011; 5(6):4300–9; doi:10.1021/nn101492k. CrossRef
?wierzko AS, Michalski M, Soko?owska A, Nowicki M, Szala-Po?dziej A, Eppa ?, Mitrus I, Szmigielska-Kap?on A, Sobczyk-Kruszelnicka M, Michalak K, Go?os A, Wierzbowska A, Giebel S, Jamroziak K, Kowalski ML, Brzezi?ska O, Thiel S, Matsushita M, Jensenius JC, Gajek G, Cedzy?ski M. Associations of ficolins with hematological malignancies in patients receiving high-dose chemotherapy and autologous hematopoietic stem cell transplantations. Front Immunol, 2020; 10:3097; doi:10.3389/fimmu.2019.03097. CrossRef
Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, Kilpatrick LE, Billheimer DD, Blackman RK, Cardasis HL, Carr SA, Clauser KR, Jaffe JD, Kowalski KA, Neubert TA, Regnier FE, Schilling B, Tegeler TJ, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Fisher SJ, Gibson BW, Kinsinger CR, Mesri M, Rodriguez H, Stein SE, Tempst P, Paulovich AG, Liebler DC, Spiegelman C. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res, 2010; 9(2):761–76; doi:10.1021/pr9006365. CrossRef
Tang D, Cui Y, Chen G. Nanoparticle-based immunoassays in the biomedical field. Analyst, 2013; 138(4):981–90; doi:10.1039/c2an36500f. CrossRef
Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, Landfester K, Schild H, Maskos M, Knauer SK, Stauber RH. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat Nanotechnol, 2013; 8(10):772–81; doi:10.1038/nnano.2013.181. CrossRef
Vidaurre-Agut C, Rivero-Buceta E, Romaní-Cubells E, Clemments AM, Vera-Donoso CD, Landry CC, Botella P. Protein corona over mesoporous silica nanoparticles: influence of the pore diameter on competitive adsorption and application to prostate cancer diagnostics. ACS Omega, 2019; 4(5):8852–61; doi:10.1021/acsomega.9b00460. CrossRef
Wang A, Perera YR, Davidson MB, Fitzkee NC. Electrostatic interactions and protein competition reveal a dynamic surface in gold nanoparticle-protein adsorption. J Phys Chem C Nanomater Interfaces, 2016; 120(42):24231–9; doi:10.1021/acs.jpcc.6b08469. CrossRef
Weber C, Simon J, Mailänder V, Morsbach S, Landfester K. Preservation of the soft protein corona in distinct flow allows identification of weakly bound proteins. Acta Biomater, 2018; 76:217–24; doi:10.1016/j.actbio.2018.05.057. CrossRef
Winzen S, Schoettler S, Baier G, Rosenauer C, Mailaender V, Landfester K, Mohr K. Complementary analysis of the hard and soft protein corona: sample preparation critically effects corona composition. Nanoscale, 2015; 7(7):2992–3001; doi:10.1039/c4nr05982d. CrossRef
Wiwanitkit V, Sereemaspun A, Rojanathanes R. Gold nanoparticle as an alternative tool for urine microalbumin test: the first world report. Ren Fail, 2007; 29(8):1047–8; doi:10.1080/08860220701643575. CrossRef
Yan X, Sedykh A, Wang W, Yan B, Zhu H. Construction of a web-based nanomaterial database by big data curation and modeling friendly nanostructure annotations. Nat Commun, 2020; 11(1):2519; doi:10.1038/s41467-020-16413-3. CrossRef
Yin H, Chen R, Casey PS, Ke PC, Davis TP, Chen C. Reducing the cytotoxicity of ZnO nanoparticles by a pre-formed protein corona in a supplemented cell culture medium. RSC Adv, 2015; 5(90):73963–73; doi:10.1039/C5RA14870G CrossRef
Yu YY, Chen YY, Gao X, Liu YY, Zhang HY, Wang TY. Nanoparticle based bio-bar code technology for trace analysis of aflatoxin B1 in Chinese herbs. J Food Drug Anal, 2018; 26(2):815–22; doi:10.1016/j.jfda.2017.11.003. CrossRef
Zhang D, Yang J, Guan J, Yang B, Zhang S, Sun M, Yang R, Zhang T, Zhang R, Kan Q, Zhang H, He Z, Shang L, Sun J. In vivo tailor-made protein corona of a prodrug-based nanoassembly fabricated by redox dual-sensitive paclitaxel prodrug for the superselective treatment of breast cancer. Biomater Sci, 2018; 6(9):2360–74; doi:10.1039/c8bm00548f. CrossRef
Zhang Y, Wu JLY, Lazarovits J, Chan WCW. An analysis of the binding function and structural organization of the protein corona. J Am Chem Soc, 2020; 142(19):8827–36; doi:10.1021/jacs.0c01853. CrossRef
Zheng T, Pierre-Pierre N, Yan X, Huo Q, Almodovar AJ, Valerio F, Rivera-Ramirez I, Griffith E, Decker DD, Chen S, Zhu N. Gold nanoparticle-enabled blood test for early stage cancer detection and risk assessment. ACS Appl Mater Interfaces, 2015; 7(12):6819–27; doi:10.1021/acsami.5b00371. CrossRef