INTRODUCTION
Lipases are one of biocatalysts that act in various ranges of bioconversion, including hydrolysis, esterification, interesterification, alcoholysis, acidolysis, and aminolysis (Soliman et al., 2007). It works at the hydrophilic-lipophilic interfaces and is tolerant to the presence of organic solvents in a reaction mixture (Hasan et al., 2006). Furthermore, lipases show potential applications in industries. Thermostable lipase from thermophilic microbes was used commercially such as on modification of fats, flavor enrichment in food processing, organic synthesis, hydrolysis of fats and oils, resolution of racemic mixtures, and other chemical analysis industries (Contesini et al., 2020; Li and Zhang, 2005).
Thermophilic microorganisms are able to produce thermozymes or also known as thermostable enzymes such as thermostable lipases exhibiting extreme stability at elevated temperatures and in organic solvents (Bornscheuer et al., 1994). Thermal denaturation in most enzymes will negatively affect the activity (Maciver et al., 1994). The properties of thermostable lipases are essential in industrial applications and biotechnology, especially in converting mixtures at a higher temperature.
Most thermostable lipases are generally expressed at low levels by wild-type hosts. In recent years, they have been produced using bacteria and yeast expression systems. Studies on the interesting enzyme have been extensively explored to express in Escherichia coli; however, some of the enzymes show low levels of lipolytic activity. A local thermophilic microorganism from Manuk hot spring, Kamojang, West Java, has been isolated and cultivated and the gene encoding thermostable lipase Itb1.1 was cloned and expressed into E. coli expression vector [pET-30a(+)] with 1,248 base pairs (bp) coding for 416 amino acids (Widhiastuty et al., 2011). The enzyme showed low specific activity under the condition at pH 8.0 and 70°C using p-nitrophenyl palmitate (pNPP) as substrate (Brilliantoro et al., 2015). Another thermostable lipase Lk3 was obtained from compost based on metagenome exploration. The highest homology of thermostable lipase Lk3 was closest to Pseudomonas stutzeri. Gene encoding thermostable lipase Lk3 was cloned and expressed into E. coli with 843 bp coding for 280 amino acids (Nurhasanah et al., 2015). The enzyme showed a low specific activity under the condition at pH 8.0 and 50°C using p-nitrophenyl laurate (pNPL) as substrate (Nurhasanah et al., 2017).
Nowadays, other expression systems such as yeast expression systems have been widely used to establish more efficient production of thermostable recombinant lipases. Pichia pastoris is one of the yeast expression systems that have the potential as a host to express typical and thermostable lipases (Furqan and Akhmaloka, 2020; Lan et al., 2016; Sabri et al., 2009). The methylotrophic yeast, P. pastoris, was an attractive host for producing heterologous protein growing on methanol as the sole carbon and energy source, and the proteins involved in methanol metabolism are strongly induced (Gasser et al., 2013; Yurimoto et al., 2011).
Pichia pastoris was introduced 40 years ago by Philips Petroleum to produce single-cell protein commercially as animal food additive ingredients because of the fermentation process in high-density cells (Ahmad et al., 2014). Pichia pastoris genome contains four chromosomes with a total size of about 9.43 Mbp. Genome sequences of yeast strain GS115 have been successfully determined in recent years (Fickers, 2014). The strain was the most valuable for industrial and research development used to study the gene expression and regulation through methanol. The alcohol oxidase (AO) promotor was the first methanol metabolism pathway regulated tightly. When the yeast was grown in medium containing glucose or ethanol, AO was not detected in cells, but when the cell was grown in medium-containing methanol, AO highly detected up to 35% of cellular protein. In addition, P. pastoris has many advantages such as being able to grow to very high cell densities and produce large amounts of protein, posttranslational modification, strong methanol-inducible AOX1 promoter, extracellular secretion for novel protein, and stable integration events in host chromosomal DNA (Cereghino and Cregg, 2000; Korona et al., 2006; Ol?dzka et al., 2003; Yamada et al., 2016).
Some studies have reported using P. pastoris expression systems such as thermostable lipase from Bacillus thermocatenulatus (Quyen et al., 2003), recombinant thermostable lipase B from Candida antarctica (Robert et al., 2017), highly Ca2+-activated thermostable L2 lipase (Sabri et al., 2009), thermostable lipase from Pseudomonas fluorescens 26-2 (Yang et al., 2009), thermostable T1 lipase from Geobacillus zalihae (Oslan et al., 2013), and thermostable lipase Lk1 from bacterial compost using metagenome technique (Furqan and Akhmaloka, 2020).
This paper reports the heterologous expression of thermostable recombinant lipases Itb1.1 and Lk3 in E. coli BL21 (DE3) under the control of T7 promoter and in P. pastoris GS115 under the control of AOX1 promoter. Following purification of the enzyme using Immobilized Affinity Chromatography (IMAC) nickel-nitrilotriacetic acid (Ni-NTA) the activity of both proteins expressed in E. coli and in P. pastoris was compared.
MATERIALS AND METHODS
Host strain and plasmids
The host yeast strain used was P. pastoris GS115; cloning plasmid pJET1.2/blunt and expression plasmid pPICZαA were purchased from Invitrogen, USA. E. coli TOP10F’ was used for subcloning and plasmid propagation. The gene encoding thermostable lipase Itb1.1 was amplified from the recombinant plasmids pET30a(+)-Itb1.1 (pITBLip1.2) and Lk3 from the recombinant plasmids pET30a(+)-lk3 (pITBLip2.1) in E. coli BL21 (DE3) cells.
Recombinant plasmid construction in pPICZαA
Recombinant plasmids pITBlip1.2 and pITBLip2.1 in E. coli BL21 (DE3) cells first analyzed the restriction sites using EcoRI and HindIII to confirm the existence of gene encoding thermostable lipases Itb1.1 and Lk3. Then mature Itb1.1 and Lk3 lipases were subcloned into pJET1.2/blunt (pJET1.2-itb1.1 or pITBLip1.3 and pJET1.2-lk3 or pITBLip2.2), and increasing of plasmid was conducted in E. coli TOP10F’. Furthermore, mature Itb1.1 lipase was amplified by using a forward primer incorporated with restriction sites. EcoRI was for ITB1.1 (5′-CTTAGAATTCGCATCCCCACG-3′), and reverse primer incorporated with XbaI was revITB1.1 (5′-GAAGTCTAGATAAGGCCGCAAAC-3′).
Moreover, mature Lk3 lipase was amplified using forward primer forLK3 (5′-CGTCGAATTCATGAACAAGAACAAAACCTTGC-3′) and reverse primer revLK3 (5′-GAAGTCTAGATAGAGCCCCGCGTTCTTC-3′) with restriction endonucleases EcoRI and XbaI being underlined.
The purified amplicon and pPICZαA plasmids were digested with EcoRI and XbaI. The insert and plasmid were ligated and transformed in E. coli TOP10F’. The recombinant plasmid construction for Itb1.1 gene was designed as pITBLip1.4 (pPICZαA-itb1.1) and for lk3 gene was designed as pITBLip2.3 (pPICZαA-lk3). The amplified polymerase chain reaction (PCR) product encoded 404 amino acids for mature Itb1.1 lipase, and 296 amino acids for Lk3 lipase have been fused with poly histidine-tag at C-terminal for further purification steps and fused with α-factor secretion signal at N-terminal. Recombinant plasmids pITBLip1.2 and pITBLip1.4 were then confirmed by sequencing through First Base Inc., Singapore.
Yeast transformation
Transformation of P. pastoris GS115 was performed by electroporation method prepared according to EasySelectTM Pichia Expression Kit User Manual, Cat. No. K1740-01 (Invitrogen, USA) (Invitrogen Life Technologies, 2010). SacI linearized the recombinant plasmid pITBLip1.4 and pITBLip2.3 before gene integration into the yeast genome. To confirm positive colonies, colony PCR was conducted using AOX1 forward-reverse primers and size screening. All positive transformants were selected in yeast peptone dextrose sorbitol agar medium (1% yeast extract, 2% peptone, 2% dextrose, 1 M sorbitol, and 2% agar) at 30°C with the addition of 100 μg/ml zeocin and further selection on higher zeocin concentration 1,000–2,000 μg/ml for analysis multicopy of cassette expression (Fig. 1).
Heterologous expression in E. coli and P. pastoris
For the production of thermostable lipase Itb1.1 and Lk3 through E. coli, an overnight culture of E. coli BL21 (DE3) harboring pITBlip1.2 and pITBLip2.1 was inoculated in Luria Bertani liquid medium, which contains antibiotic of 0.1% kanamycin until it reached the optical density of 0.6; then, it was induced by 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) within 4 hours and shaken at 180 rpm with temperature 37°C. After 4 hours, the culture was cultivated by centrifugation at 6,000 g for 20–30 minutes. All supernatants were removed, and all pellets were collected and then mixed with lysis buffer [50 mM phosphate buffer saline (PBS) buffer at pH 8 with 0.1% sodium dodecyl sulfate (SDS)]. The cell pellets containing Itb1.1 lipase were incubated for 30 minutes at 70°C and lipase Lk3 at 50°C. Furthermore, all supernatants as crude extract of Itb1.1 and Lk3 thermostable lipases were collected by centrifugation at 8,000 g for 30 minutes and then concentrated through Amicon diafiltration with 30 and 10 kDa cutoff (Permana et al., 2017).
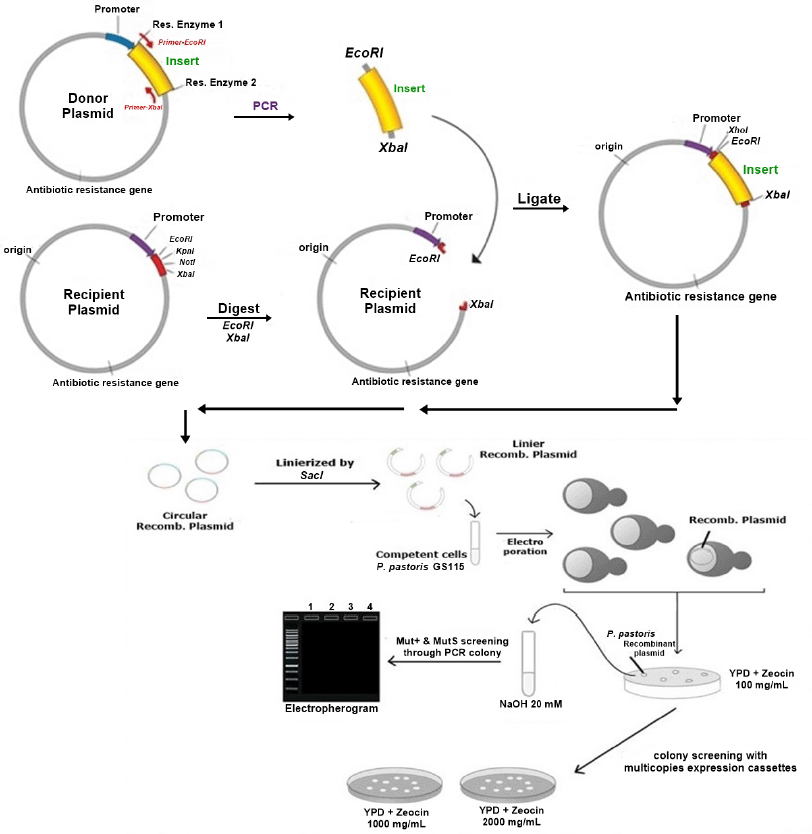 | Figure 1. Illustration for construction of recombinant vector and yeast transformation. [Click here to view] |
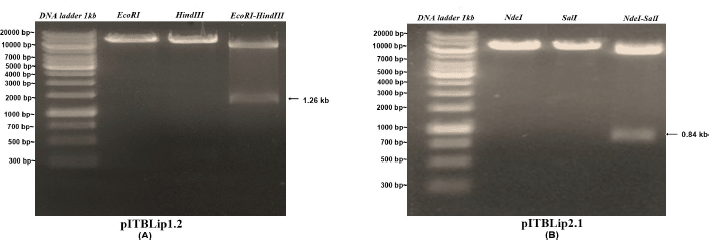 | Figure 2. Electropherogram of single and double digest from recombinant E. coli expression vectors. [Click here to view] |
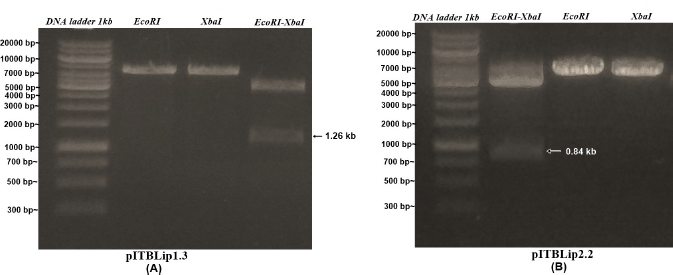 | Figure 3. Electropherogram of single and double digest from recombinant E. coli cloning vectors. [Click here to view] |
To produce Itb1.1 and Lk3 thermostable lipases through P. pastoris, one positive transformant from each isolate was used for recombinant thermostable lipase expression. A single colony from YPDz (1% yeast extract, 2% peptone, and 2% dextrose) agar supplemented with 100 μg/ml zeocin was inoculated into 10 ml YPDz broth for 24 hours at 30°C and agitation rate 250 rpm. Then, 500 μl of culture was transferred into 50 ml buffered glycerol-complex medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate buffer pH 6.0, 1.34% yeast nitrogen base without amino acid, and 1% glycerol) in 500 ml conical flask and agitation with 250 rpm at 30°C until the culture reached an OD600 = 4–6 (approximately 16–20 hours). The cells were harvested by centrifuging 3,000 g for 10–15 minutes at 4°C and then resuspended the cells to an OD600 of 1.0 in 50 ml buffered methanol-complex medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate buffer with pH 6.0, 1.34% yeast nitrogen base without amino acid, and 0.5% methanol) to induce expression. The cultures were induced with 0.5% methanol at 24 hours for 6 days. Afterward, the cultures were cultivated by centrifugation at 8,000 g for 30 minutes and then concentrated through Amicon diafiltration with 30 and 10 kDa cutoff (Invitrogen Life Technologies, 2010). To get the optimum methanol concentration for the inducer, the same procedure was carried out as before but using various methanol concentrations from 1% to 5% (v/v).
The lipolytic test was performed three times according to the colorimetric method (Govindappa et al., 2014). One unit (U) of thermostable lipase activity was defined as the rate of p-nitrophenol from p-NPP for Itb1.1 lipase and p-NPL for Lk3 lipase as substrate in μmole per minutes.
Protein purification
Lipase purification was carried out by using Ni-NTA affinity chromatography in cold conditions (4°C). As much as 2 ml of Ni-NTA agarose resin was put into a 10 ml chromatography column, and then we waited until all resin had been settled and separated from the solvent. The resin was washed with 3 × 6 ml sterile mili-Q aqua. Next, the resin was irrigated with 4 × 6 ml binding buffer solution (50 mM PBS buffer pH 8, 100 M NaCl, and 0.1% (v/v) Triton-X 100) frequently. Furthermore, a 5 ml protein solution (concentrated crude extract of ITB1.1 lipase) was put into the column and let pass through the resin continuously. Then, the resin was washed with 2 × 10 ml buffer solution (50 mM PBS buffer pH 8, 0.1 M NaCl, and 10 mM Imidazole). The protein already bound with resin was eluted with 5 ml elution buffer (50 mM PBS buffer pH 8, 300 mM NaCl, and 100 mM Imidazole). After that, the resin was rinsed with the final buffer (50 mM PBS buffer pH 8, 300 mM NaCl, and 250 mM Imidazole). Dialysis filtration using Amicon with 30 and 10 kDa cutoff was performed by centrifugation 4,000 g for 30 minutes to remove impurities solutions, and pure protein was analyzed using SDS-poly-acrylamide gel electrophoresis (PAGE) electrophoresis (Furqan and Akhmaloka, 2020).
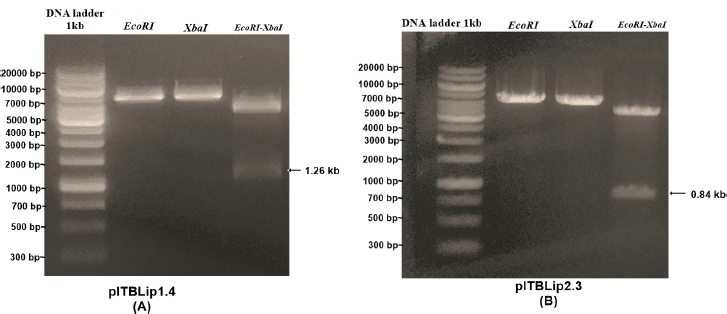 | Figure 4. Electropherogram of single and double digest from recombinant P. pastoris expression vectors. [Click here to view] |
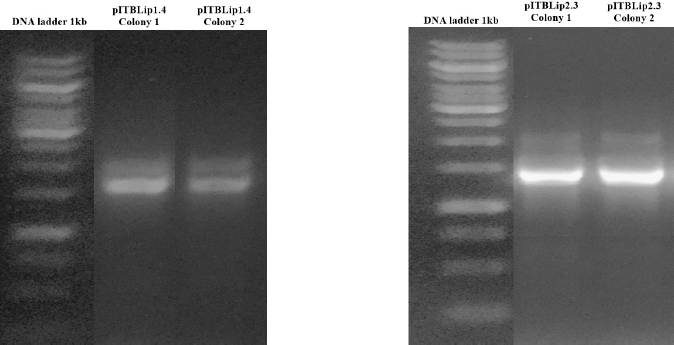 | Figure 5. Electropherogram of PCR colony P. pastoris GS115. [Click here to view] |
RESULTS AND DISCUSSION
Construction of recombinant vector and transformation
Recombinant yeast expression vectors were constructed using the pPICZαA vector through EcoRI and XbaI restriction sites. The vectors were linearized, followed by yeast transformation (Fig. 1). For Itb1.1 gene, pITBlip1.2 plasmid was digested with EcoRI and HindIII resulting in fragment gene at around 1.26 kb (Fig. 2A); meanwhile, for lk3 gene, pITBlip2.1 plasmid was digested with NdeI and SalI resulting in lk3 fragment at around 0.84 kb (Fig. 2B). The fragments were then ligated at the pJET1.2/blunt cloning vector resulting in pITBlip1.3 and pITBlip2.2 plasmids. The recombinant plasmids, pITBlip1.3 and pITBlip2.2, were then digested by EcoRI and XbaI resulting in 1.26 and 0.84 kb fragments for pITBlip1.3 pITBlip2.2, respectively (Fig. 3). The fragments were then inserted into the yeast expression vector (pPICZαA) through EcoRI and XbaI restriction sited resulting in pITBlip1.4 and pITBlip2.3 expression vectors, respectively. To confirm that the lipase gene was inserted on pPICZαA, the vectors were digested and sequenced. Following digestion by EcoRI and XbaI, agarose gel electrophoresis showed that the genes were correctly inserted (Fig. 4). Furthermore, the sequencing results showed that the genes were correctly fused with an α-factor signal sequence and 6x His-tag. The α-factor has a role in the secretion of the protein in the media (Furqan and Akhmaloka, 2020)
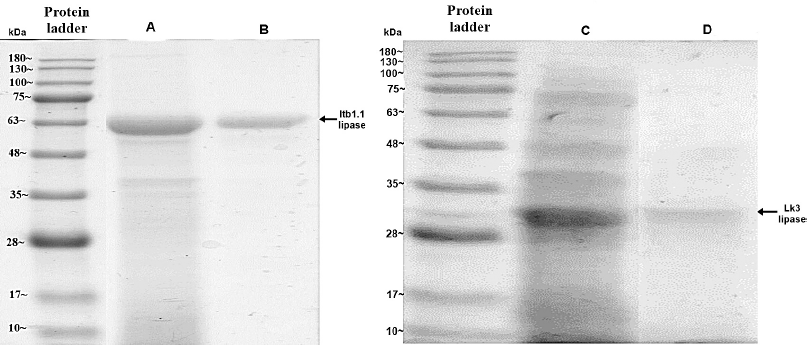 | Figure 6. Electropherogram of IMAC Ni-NTA chromatography for Itb1.1 and Lk3 expressed in E. coli (line A for Itb1.1 crude extract, line B for purified Itb1.1, line C for Lk3 crude extract, and line D for purified Lk3) [Click here to view] |
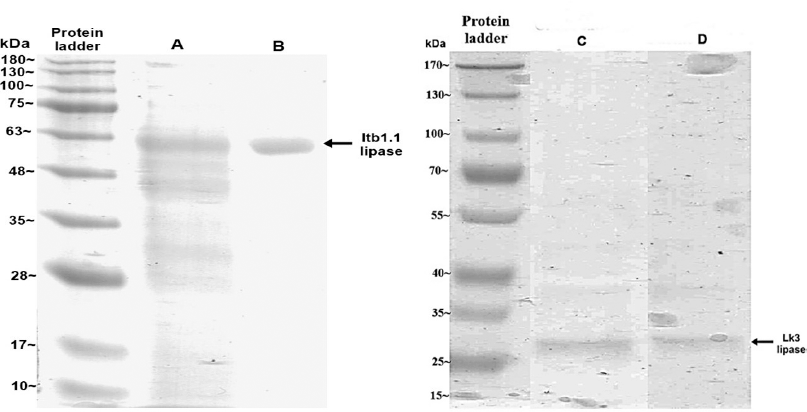 | Figure 7. Electropherogram of IMAC Ni-NTA chromatography for Itb1.1 and Lk3 expressed in P. pastoris (line A for Itb1.1 crude extract, line B for purified Itb1.1, line C for Lk3 crude extract, and line D for purified Lk3). [Click here to view] |
 | Table 1. Lipolytic test of thermostable lipase crude extract. [Click here to view] |
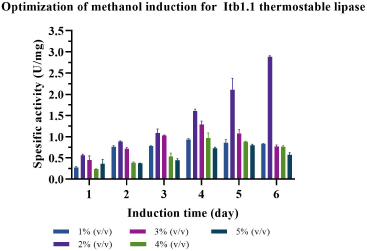 | Figure 8. Optimization of methanol concentration to express Itb1.1 thermostable lipase. [Click here to view] |
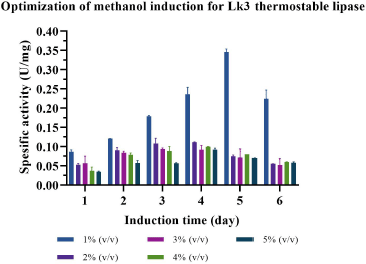 | Figure 9. Optimization of methanol concentration to express Lk3 thermostable lipase. [Click here to view] |
Linear plasmids of pITBLip1.4 and pITBlip2.3 were used to transform P. pastoris GS115. Positive colonies were confirmed through PCR colony. Two bands appeared on agarose electropherogram (Fig. 5), indicating that Mut+ phenotypic cells or methanol utilization plus means that the AOX1 gene was not deleted from the genome. Positive colony transformants were chosen for further analysis.
Expression of thermostable lipase in E. coli and P. pastoris
Thermostable lipases of Itb1.1 and Lk3 were successfully expressed in E. coli and P. pastoris. Expression of Itb1.1 and Lk3 lipases in E. coli was induced by 1 mM IPTG. The enzymes were expressed as intracellular proteins. The crude extracts of the enzymes were electrophorized on SDS-PAGE following lysis cells. Dominant bands on SDS-PAGE with the size at around 50 kDa for Itb1.1 lipase and 32 kDa for Lk3 lipase, respectively, appeared, showing that the proteins were overexpressed (Fig. 6). Meanwhile, the expression of the proteins on P. pastoris was induced by 0.5% (v/v) methanol and expressed as extracellular proteins. The cell-free supernatants were electrophorized on SDS-PAGE. The protein size was around 50 kDa for Itb1.1 lipase and 32 kDa for Lk3 lipase, respectively (Fig. 7). All the enzymes expressed in E. coli and P. pastoris showed lipolytic activity (Table 1).
Methanol concentration and incubation time varied to probe the best expression of Itb1.1 and Lk3 lipases in P. pastoris. The result showed that 2% (v/v) methanol concentration and 6-day incubation were the best conditions to express Itb1.1 lipase (Fig. 8); meanwhile, Lk3 lipase appeared at 1% (v/v) methanol concentration for 5-day incubation (Fig. 9).
Purification and lipolytic activity of Itb1.1 and Lk3 lipases
Itb1.1 and Lk3 thermostable lipases were produced at their best condition using E. coli and P. pastoris host cells. pastoris. The crude extract (E. coli) and the cell-free supernatant (P. pastoris) were purified using IMAC Ni-NTA chromatography. For crude extract enzymes from E. coli, the samples were directly purified by IMAC Ni-NTA. Meanwhile, cell-free supernatant from P. pastoris was needed to pretreat the samples by washing with 50 mM PBS pH 8 using Amicon diafiltration with 30 kDa cutoff. The samples were then centrifuged until yellow color brownish became yellow.
The purified proteins were electrophorized by SDS-PAGE to determine the molecular weights. The result showed that the molecular weight of Itb1.1 and Lk3 lipases is 50 and 32 kDa, respectively (Figs. 6 and 7). A sharp single band on the SDS-PAGE electropherogram showed that each protein was successfully purified.
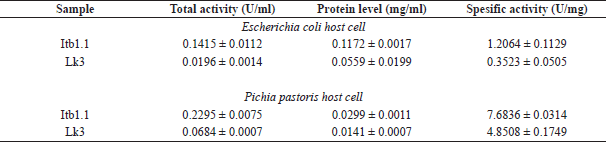 | Table 2. Lipolytic test of purified thermostable lipase. [Click here to view] |
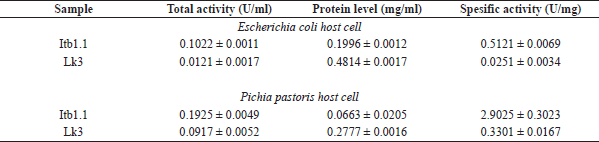
The lipolytic activity of the purified protein was carried out using the colorimetric method (Govindappa et al., 2014). The specific activity of Itb1.1 lipase expressed in E. coli was two times higher than that in the crude extract. Meanwhile, the specific activity of the enzyme expressed in P. pastoris was three times higher than that in the cell-free supernatant (Table 2). The specific activity of purified Itb1.1 lipase expressed in E. coli was six times higher than that expressed in P. pastoris. Furthermore, the specific activity of purified Lk3 lipase expressed in E. coli was 14 times higher than that in the crude extract. Meanwhile, the specific activity of the purified enzyme expressed in P. pastoris was 15 times higher than that in the cell-free supernatant (Table 2). The specific activity of purified Lk3 lipase expressed in E. coli was 14 times higher than that expressed in P. pastoris. Therefore, the specific activity of Itb1.1 and Lk3 thermostable lipases expressed in P. pastoris host was higher than expressed in E. coli. Thus, it suggests that P. pastoris was a better host to express Itb1.1 and Lk3 thermostable lipase.
CONCLUSION
In conclusion, the specific activity of purified Itb1.1 lipase expressed in P. pastoris host cell was six times higher than expressed in E. coli. Additionally, the specific activity of purified Lk3 lipase expressed in P. pastoris host cell was 14 times higher than that expressed in E. coli. Finally, it could be concluded that the secretory expression of Itb1.1 and Lk3 thermostable lipases expressed in P. pastoris system was significantly better than that expressed in E. coli.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
CONFLICT OF INTERESTS
The authors report no financial or any other conflicts of interest in this work.
FUNDING
The authors would like to acknowledge the PDUPT research grant for ITB from the Ministry of Research and Technology/BRIN Indonesia with a contract number of 2/E1/KP.PTNBH/2021 and LPDP scholarships for DFS to make this research possible to be carried out.
ETHICAL APPROVAL
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Ahmad M, Hirz M, Pichler H, Schwab H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol, 2014; 98(12):5301–17. CrossRef
Bornscheuer U, Reif OW, Lausch R, Freitag R, Scheper T, Kolisis FN, Menge U. Lipase of Pseudomonas cepacia for biotechnological purposes: purification, crystallization and characterization. Biochim Biophys Acta, 1994; 1201(1):55–60. CrossRef
Brilliantoro R, Zidny R, Widhiastuty MP, Akhmaloka A. Hydrolytic and transesterification activities of thermostable lipase ITB1.1. Biosci Biotechnol Res Asia, 2015; 12(1):1–6. CrossRef
Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev, 2000; 24(1):45–66. CrossRef
Contesini FJ, Davanço MG, Borin GP, Vanegas KG, Cirino JPG, de Melo RR, Mortensen UH, Hildén K, Campos DR, Carvalho PDO. Advances in recombinant lipases: application in the pharmaceutical industry. Catalysts, 2020; 10:1–33. CrossRef
Fickers P. Pichia pastoris: a workhorse for recombinant protein production. Curr Res Microbiol Biotechnol, 2014; 2(3):354–63.
Furqan BRN, Akhmaloka A. Heterologous expression and characterization of thermostable lipase (Lk1) in Pichia pastoris GS115. Biocatal Agric Biotechnol, 2020; 23:101448. CrossRef
Gasser B, Prielhofer R, Marx H, Maurer M, Nocon J, Steiger M, Puxbaum V, Sauer M, Mattanovich D. Pichia pastoris: protein production host and model organism for biomedical research. Future Microbiol, 2013; 8(2):191–208. CrossRef
Govindappa N, Hanumanthappa M, Venkatarangaiah K, Periyasamy S, Sreenivas S, Soni R, Sastry K. A new signal sequence for recombinant protein secretion in Pichia pastoris. J Microbiol Biotechnol, 2014; 24(3):337–45. CrossRef
Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzyme Microb Technol, 2006; 39(2):235–51. CrossRef
Invitrogen Life Technologies. User manual - EasySelectTM Pichia Expression Kit. Invitrogen Life Technologies, Carlsbad, CA, 2010.
Korona B, Korona D, Bielecki S. Efficient expression and secretion of two co-produced xylanases from Aspergillus niger in Pichia pastoris directed by their native signal peptides and the Saccharomyces cerevisiae α-mating factor. Enzyme Microb Technol, 2006; 39(4):683–9. CrossRef
Lan D, Qu M, Yang B, Wang Y. Enhancing production of lipase MAS1 frommarine Streptomyces sp. strain in Pichia pastoris by chaperones co-expression. Electron J Biotechnol, 2016; 22:62–7. CrossRef
Li H, Zhang X. Characterization of thermostable lipase from thermophilic Geobacillus sp. TW1. Protein Expr Purif, 2005; 42(1):153–9. CrossRef
Maciver B, McHale RH, Saul DJ, Bergquist PL. Cloning and sequencing of a serine proteinase gene from a thermophilic Bacillus species and its expression in Escherichia coli. Appl Environ Microbiol, 1994; 60(11):3981–8. CrossRef
Nurhasanah, Nurbaiti S, Madayanti F, Akhmaloka A. Heterologous expression of gene encoded thermostable lipase and lipolytic activity. J Pure Appl Microbiol, 2017; 11(1):135–9. CrossRef
Nurhasanah, Nurbaiti S, Warganegara FM, Akhmaloka A. Diversity of gene encoding thermostable lipase from compost based on metagenome analysis. Int J Integr Biol, 2015; 16(1):7–12.
Ol?dzka G, Da?browski S, Kur J. High-level expression, secretion, and purification of the thermostable aqualysin I from Thermus aquaticus YT-1 in Pichia pastoris. Protein Expr Purif, 2003; 29(2):223–9. CrossRef
Oslan SN, Salleh AB, Rahman RNZRAR, Leow TC, Basri M. Pichia pastoris as a host to overexpress the thermostable T1 lipase from Geobacillus zalihae. GSTF Int J Biosci, 2013; 3(1):7–13. CrossRef
Permana AH, Warganegara FM, Wahyuningrum D, Widhiastuty MP, Akhmaloka A. Heterologous expression and characterization of thermostable lipases from Geobacillus thermoleovorans PPD2 through Escherichia coli. Biosci Biotechnol Res Asia, 2017; 14(3):1081–8. CrossRef
Quyen DT, Schmidt-Dannert C, Schmid RD. High-level expression of a lipase from Bacillus thermocatenulatus BTL2 in Pichia pastoris and some properties of the recombinant lipase. Protein Expr Purif, 2003; 28(1):102–10. CrossRef
Robert JM, Lattari FS, Machado AC, de Castro AM, Almeida RV, Torres FAG, Valero F, Freire DM. Production of recombinant lipase B from Candida antarctica in Pichia pastoris under control of the promoter PGK using crude glycerol from biodiesel production as carbon source. Biochem Eng J, 2017; 118:123–31. CrossRef
Sabri S, Rahman RNZRA, Leow TC, Basri M, Salleh AB. Secretory expression and characterization of a highly Ca2+-activated thermostable L2 lipase. Protein Expr Purif, 2009; 68(2):161–6. CrossRef
Soliman NA, Knoll M, Abdel-Fattah YR, Schmid RD, Lange S. Molecular cloning and characterization of thermostable esterase and lipase from Geobacillus thermoleovorans YN isolated from desert soil in Egypt. Process Biochem, 2007; 42(7):1090–100. CrossRef
Widhiastuty MP, Febriani, Moeis MR, Akhmaloka, Madayanti F. Cloning, homological analysis and expression of lipase gene from hot spring isolate. Int J Integr Biol, 2011; 11(1):8–13.
Yamada R, Kimoto Y, Ogino H. Combinatorial library strategy for strong overexpression of the lipase from Geobacillus thermocatenulatus on the cell surface of yeast Pichia pastoris. Biochem Eng J, 2016; 113:7–11. CrossRef
Yang J, Zhang B, Yan Y. Cloning and expression of pseudomonas fluorescens 26-2 lipase gene in Pichia pastoris and characterizing for transesterification. Appl Biochem Biotechnol, 2009; 159(2):355–65. CrossRef
Yurimoto H, Oku M, Sakai Y. Yeast methylotrophy: metabolism, gene regulation and peroxisome homeostasis. Int J Microbiol, 2011; 2011:1–8. CrossRef