INTRODUCTION TO THE NEURAL NETWORK
The development of various machine learning algorithms, i.e., naive Bayes classifiers, logistic regression, vector machines, and artificial intelligence systems, have contributed in the revolution of a new drug design which is also supported via in silico molecular docking and simulation, protein structure validation, molecular energy prediction, and virtual screening (Yang et al., 2019a, 2019b). In addition, the rapid development of neural network models due to huge data has improved multiple calibration techniques (Lopes et al., 2019; Schmidt et al., 2019). Furthermore, utilizing a large set of data series, the artificial neural network (ANN) models are adopted to correlate dependent and independent variables (Pasini, 2015).
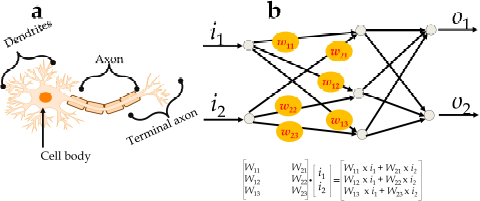
A basic unit of the neural network comprises an input, hidden, and output layer and is similar to a human neuronal cell (Jothilakshmi and Gudivada, 2016). The input layer connects the hidden layer and transmits the signal, termed as X1, X2, X3, X4..... Xn, with their respective weights W1, W2, W3, W4.....Wn. The weight of each signal represents an input signal as they depend on the function and magnitude of signals. The summation of input signals with their respective weights, i.e., X1 W1 + X2 W2 + X3 W3 + X4 W4 +..... Xn Wn, is processed within the hidden layer and the output is obtained. The concept of the basic node of the neural network can be represented mathematically as y = f (I) = f (b + ∑ni xi. wi), where b = bias, x = input to the neuron, w = weights, n = the number of inputs from the incoming layer, and i = counter from 0 to n and is similar to the human neural system (Figure 1).
Weights
A weight represents a connecting link between neurons with a numerical value (Dudek, 2017; Georgevici and Terblanche, 2019) in which the value is directly proportional to the weight. If the value of weight is higher, it may possess an important input signal that can be visualized in the matrix format, as shown in Figure 1.
Bias
A bias is a number that helps to understand the situation. This can be explained as if one tries to make a decision, he/she needs to focus on all the possible/observable factors (Dudek, 2017). However, there could be multiple parameters or variables that may not be noticed. These unnoticed parameters or variables are tried to be incorporated in the neural network in terms of bias.
Activation
A neuron decides on its output and also makes a tiny decision for its output which is called activation. It can be represented as f (z), where z is the cluster of all inputs and can be categorized as binary, linear, and nonlinear (Dudek, 2017; Georgevici and Terblanche, 2019). Suppose, if an input value is above or below a basal threshold (also considered as a resting state of the neuron), the neuron gets activated and sends a signal to the next layer which is termed as binary activation. This function is limited with the multi-value output; categorization of multiple inputs is not possible with this activation. Therefore, it can be explained as a qualitative analysis as it provides the results as “yes” or “no” (Feng and Lu, 2019).
In linear activation function, inputs are multiplied with respective weights to produce an output signal and are proportional to the input. Unlike binary activation, it provides multiple outputs. Mathematically, it can be explained as if f (z) = z, then f (z) is called linear activation, meaning nothing happens. However, it is limited with two major problems, i.e., it cannot be used in backpropagation to train the model and all the layers are collapsed into one (Feng and Lu, 2019).
Similarly, nonlinear neural activation produces the complex plot between the inputs and outputs to model the complex data. Nonlinear activation addresses the problems associated with linear activation via the backpropagation and creates the deep neural network. Some of the activation functions widely used to produce the complex plot in the ANN include sigmoid, TanH, ReLU, Leaky ReLU, parametric ReLU, softmax, and swish (Feng and Lu, 2019).
APPLICATION IN MODERN DRUG DISCOVERY
ANNs are defined as “digitalized models of the brain” as they are complex; utilize the nonlinear relationship; and the basic anatomy is similar to the human neurons (Zador, 2019). Thus, they have their importance in drug discovery and development with the proper utilization of virtual screening, quantitative structure-activity relationship (QSAR) study, mathematical modeling, pharmacophore identification, in silico molecular docking, and ADMET prediction. Virtual screening may help to predict the biological spectrum of lead molecules (Ekins et al., 2007; Tang and Marshall, 2011). Furthermore, machine learning is also successfully implemented in the discovery of modern medicine based on target identification (gene–disease association, identification of splice variants, and target druggability prediction), compound design (reaction plan, ligand-based drug design), prediction of biomarkers (tissue-specific biomarkers, drug–response signature), and determination of drug response (cellular phenotyping and microenvironment measurement) (Vamathevan et al., 2019). In this regard, Bayesian neural networks can be used to identify the biomolecules that act on the brain and cardiovascular-related pathogenesis. Binding site identification of the receptor can be predicted via the pharmacophore modeling (Huang et al., 2018) in which the active site and its geometry play an important role and can be incorporated to predict molecular surface and create a 2D feature map.
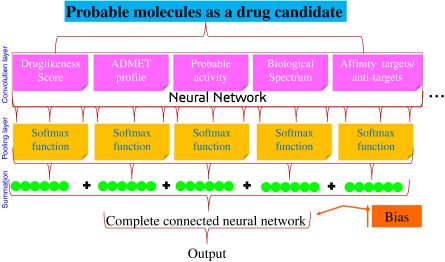
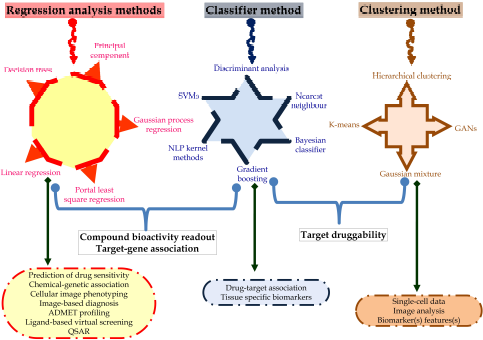
In silico molecular docking also helps to identify the suitable pose of the ligand with its target (Meng et al., 2011). The ligand binds with the given target with some energy or affinity; explained in terms of the kcal/mol for AutoDock tools and also interacts with amino acid residues, i.e., hydrogen bond interactions or pi bond interactions. After docking, different poses of the ligand molecules are obtained from which the pose scoring the lowest binding energy is chosen to identify the ligand–protein interaction. Hence, this confirmation and the binding affinity can be trained in the neural network to identify the regulators of the protein. So, based on the binding energy, the pose with minimum binding energy can be considered as the input signals to the ANN. Furthermore, the ADMET profile also plays an important role in the drug development steps to evaluate the probable pharmacokinetic profile of lead candidates (Morgan, 2011). Multiple in silico tools can be utilized to identify the probable toxicity to assess the probable activity of the biomolecules for ADMET which may be used in predicting the drug sensitivity, chemical-genetic association, assessing the structure–activity relationship via the multiple regression analysis methods using decision trees, principal components, and linear, portal least square, and Gaussian process regression. Likewise, drug–target association and tissue-specific biomarkers can be traced via the classifier methods via natural language processing kernel methods, gradient boosting, Bayesian classifier, nearest neighbor, and discriminant analysis. Additionally, single-cell information, image analysis, and biomarker assessment can help in target druggability via the clustering method through a generative adversarial network, Gaussian mixture, k-means, and hierarchical clustering (Fig. 3)
Due to the ANN efficacy to task concerning the trained dataset, it can self-correct the errors, organize and store the learned information, and compute faster data integration and retrieval (Mandlik et al., 2016). Additionally, ANN can investigate the complex and nonlinear relationship and find the application in various fields including modern drug discovery. Also, they are used in the discriminant and regression data analysis which benefits in screening the huge inhibitor libraries and ligand properties based on pharmacophore features, QSAR, docking outputs, and ADMET profile (Mandlik et al., 2016). However, machine learning needs large data with specific characters like the requirement of the standardized high-dimensional drug–target–disease dataset, comprehensive omics data, successful and unsuccessful metadata from clinical trials, training dataset, compound reaction models, gold standard ADME data, and various protein structures (Vamathevan et al., 2019).
REFERENCES
Agatonovic-Kustrin S, Beresford R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J Pharm Biomed Anal, 2000; 22(5):717–27. CrossRef
Ahmadi MA, Ebadi M, Shokrollahi A, Majidi SM. Evolving artificial neural network and imperialist competitive algorithm for prediction oil flow rate of the reservoir. Appl Soft Comput, 2013; 13(2):1085–98. CrossRef
Ajam N. Heart diseases diagnoses using artificial neural network. IISTE Netw Complex Syst, 2015; 5(4):7–10.
Azati Team. Disease prediction and classification with artificial neural networks. 2021. [Internet]. Available via https://azati.ai/disease-prediction-and-classification-with-neural-networks/ (Accessed 1 October 2021).
Barr WH, Zola EM, Candler EL, Hwang SM, Tendolkar AV, Shamburek R, Parker B, Hilty MD. Differential absorption of amoxicillin from the human small and large intestine. Clin Pharmacol Ther, 1994; 56(3):279–85. CrossRef
Caramella C, Ferrari F, Bonferoni MC, Sangalli ME, De Bernardi di Valserra M, Feletti F, Galmozzi MR. In vitro/in vivo correlation of prolonged release dosage forms containing diltiazem HCI. Biopharm Drug Dispos, 1993; 14(2):143–60. CrossRef
Chen Y, Zhu QJ, Pan J, Yang Y, Wu XP. A prediction model for blood–brain barrier permeation and analysis on its parameter biologically. Comput Methods Programs Biomed, 2009; 95(3):280–7. CrossRef
de Matas M, Shao Q, Richardson CH, Chrystyn H. Evaluation of in vitro in vivo correlations for dry powder inhaler delivery using artificial neural networks. Eur J Pharm Sci, 2008; 33(1):80–90. CrossRef
de Matas M, Shao Q, Silkstone VL, Chrystyn H. Evaluation of an in vitro in vivo correlation for nebulizer delivery using artificial neural networks. J Pharm Sci, 2007; 96(12):3293–303. CrossRef
Dowell JA, Hussain A, Devane J, Young D. Artificial neural networks applied to the in vitro—in vivo correlation of an extended-release formulation: initial trials and experience. J Pharm Sci, 1999; 88(1):154–60. CrossRef
Dudek G. A method of generating random weights and biases in feedforward neural networks with random hidden nodes. arXiv preprint, 2017; arXiv:1710.04874.
Ekins S, Mestres J, Testa B. In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling. Br J Pharmacol, 2007; 152(1):9–20. CrossRef
Elçiçek H, Akdo?an E, Karagöz S. The use of the artificial neural network for prediction of dissolution kinetics. Sci World J, 2014; 2014:194874. CrossRef
Feng J, Lu S. Performance analysis of various activation functions in artificial neural networks. J Phys Conf Ser, 2019; 1237(2):022030. CrossRef
Garg P, Verma J. In silico prediction of blood brain barrier permeability: an arti?cial neural network model. J Chem Inf Model, 2006; 46(1):289–97. CrossRef
Georgevici AI, Terblanche M. Neural networks and deep learning: a brief introduction. Intensive Care Med, 2019; 45:712–4. CrossRef
Graffner C, Nicklasson M, Lindgren JE. Correlations between in vitro dissolution rate and bioavailability of alaproclate tablets. J Pharmacokinet Biopharm, 1984; 12(4):367–80. CrossRef
Grossi E. How artificial intelligence tools can be used to assess individual patient risk in cardiovascular disease: problems with the current methods. BMC Cardiovasc Disord, 2006; 6:20. CrossRef
Huang H, Zhang G, Zhou Y, Lin C, Chen S, Lin Y, Mai S, Huang Z. Reverse screening methods to search for the protein targets of chemopreventive compounds. Front Chem, 2018; 6:138. CrossRef
Hussain AS. Artificial neural network based in vitro-in vivo correlations. In: Young D, Devane JG, Butler J (eds.). In vitro-in vivo correlations. Advances in experimental medicine and biology, Springer, Boston, MA, vol. 423, 1997. CrossRef
Jothilakshmi S, Gudivada VN. Large scale data-enabled evolution of spoken language research and applications. Handbook of Statistics, Elsevier, India, vol. 35, pp 301–40, 2016. CrossRef
Kim T. Pattern recognition using artificial neural network: a review. In: Bandyopadhyay SK, Adi W, Kim T, Xiao Y, Information Security, and Assurance. ISA 2010 (eds.). Communications in computer and information science, Springer, Berlin, Germany, vol. 76, 2010. CrossRef
Lagunin A, Stepanchikova A, Filimonov D, Poroikov V. PASS: prediction of activity spectra for biologically active substances. Bioinformatics, 2000; 16(8):747–8. CrossRef
Lavé T, Coassolo P, Reigner B. Prediction of hepatic metabolic clearance based on interspecies allometric scaling techniques and in vitro-in vivo correlations. Clin Pharmacokinet, 1999; 36(3):211–31. CrossRef
Levy G, Hollister LE. Inter and intra subject variations in drug absorption kinetics. J Pharm Sci, 1964; 53:1446–52. CrossRef
Lisboa PJG. A review of evidence of health benefit from artificial neural networks in medical intervention. Neural Netw, 2002; 15:11–39. CrossRef
Lopes BT, Eliasy A, Ambrosio R. Artificial intelligence in corneal diagnosis: where are we? Curr Ophthalmol Rep, 2019; 7:204–11. CrossRef
Mendyk A, Tuszy?ski PK, Polak S, Jachowicz R. Generalized in vitro-in vivo relationship (IVIVR) model based on artificial neural networks. Drug Des Dev Ther, 2013; 7:223–32. CrossRef
Mandlik V, Bejugam PR, Singh S. Application of artificial neural networks in modern drug discovery, Chapter 6. In: Puri M, Pathak Y, Sutariya VK, Tipparaju S, Moreno W (eds.). Artificial neural network for drug design, delivery and disposition, Academic Press, USA, pp 123–39, 2016. CrossRef
Meng XY, Zhang HX, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des, 2011; 7(2):146–57. CrossRef
Michael V, George M, Konstantinos P Plagianakos, V. Introduction to artificial neural network training and applications. In: 15th Annual Conference of Hellenic Society for Neuroscience. Patras, Greece, pp 27–9, 2000; doi:10.13140/2.1.1755.2322.
Morgan D. The importance of HT-ADME in drug discovery. Bioanalysis, 2011; 3(21):2385–7. CrossRef
Paroj?i? J, Ibric S, Djuri? Z, Jovanovi? M, Corrigan OI. An investigation into the usefulness of generalized regression neural network analysis in the development of level A in vitro–in vivo correlation. Eur J Pharm Sci, 2007; 30:264–72. CrossRef
Pasini A. Artificial neural networks for small dataset analysis. J Thorac Dis, 2015; 7(5):953–60.
Rackley RJ. In vitro-in vivo correlation for controlled-release dosage forms. Dissolut Technol, 1996; 3(1):12–7. CrossRef
Roman BI, Guedes RC, Stevens CV, García-Sosa AT. Recovering actives in multi-antitarget and target design of analogs of the myosin II inhibitor blebbistatin. Front Chem, 2018; 6:179. CrossRef
Schenone M, Dan?ík V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol, 2013; 9(4):232–40. CrossRef
Schmidt J, Marques MR, Botti S, Marques MA. Recent advances and applications of machine learning in solid-state materials science. NPJ Comput Mater, 2019; 5(1):1–36. CrossRef
Schmidt T, Bergner A, Schwede T. Modelling three-dimensional protein structures for applications in drug design. Drug Discov Today, 2014; 19(7):890–7. CrossRef
Schneider G, Coassolo P, Lavé T. Combining in vitro and in vivo pharmacokinetic data for prediction of hepatic drug clearance in humans by artificial neural networks and multivariate statistical techniques. J Med Chem, 1999; 42(25):5072–6. CrossRef
Street ME, Grossi E, Volta C, Faleschini E, Bernasconi S. Placental determinants of fetal growth: identification of key factors in the insulin-like growth factor and cytokine systems using artificial neural networks. BMC Pediatr, 2008; 7:8–24. CrossRef
Sullivan TJ, Sakmar E, Wagner JG. Comparative bioavailability: a new type of in vitro-in vivo correlation exemplified by prednisone. J Pharmacokinet Biopharm, 1976; 4(2):173–81. CrossRef
Tang YT, Marshall GR. Virtual screening for lead discovery. Methods Mol Biol, 2011; 716:1–22. CrossRef
Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, Li B, Madabhushi A, Shah P, Spitzer M, Zhao S. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov, 2019; 18(6):463–77. CrossRef
Wood RW, Martis L, Gillum AW, Roseman TJ, Lin L, Bernardo P. In vitro dissolution and in vivo bioavailability of commercial levothyroxine sodium tablets in the hypothyroid dog model. J Pharm Sci, 1990; 79(2):124–7. CrossRef
Yang H, Lou C, Sun L, Li J, Cai Y, Wang Z, Li W, Liu G, Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics, 2019a; 35(6):1067–9. CrossRef
Yang X, Wang Y, Byrne R, Schneider G, Yang S. Concepts of artificial intelligence for computer-assisted drug discovery. Chem Rev, 2019b; 119(18):10520–94. CrossRef
Yang X. Artificial neural networks. In: Hassan A. Karimi, Hand book of research on geoinformatics. IGI Global, USA, pp 122–8, 2009. CrossRef
Zador AM. A critique of pure learning and what artificial neural networks can learn from animal brains. Nat Commun, 2019; 10(1):1–7. CrossRef