INTRODUCTION
The major drawbacks of conventional cancer treatments are increased systemic toxicity and low efficiency to eradicate the disease. Therefore, single-drug therapy is the most commonly used treatment modality and is considered to be less effective due to its side effects and susceptibility towards drug resistance (Khdair et al., 2009; Partridge et al., 2001).
Combining two or more chemotherapeutic drugs with a different mechanism of action on cancer cells is more effective than mono-drug therapy. In addition, the synergistic effect of two or more drugs reduces the therapeutic dosage of individual drugs (Albain et al., 2008). However, the clinical application of chemotherapeutics encounters a major problem. The anti-cancer drugs are administered to the body with organic solvents or surfactants due to high hydrophobicity. Usage of two or more drugs can lead to aggregation, precipitation, and drugs losing their original pharmaceutical activity (Bae et al., 2007). Thus, there is a need for a novel drug delivery system that can simultaneously deliver the drugs and overcome the drawbacks of conventional chemotherapy.
Paclitaxel (Fig. 1A), with molecular weight 853.91 g/mol, (pKa: 10.36; logP: 3) is the potent anti-cancer drug obtained from the bark of Taxus brevifolia. It is used in the treatment of various types of cancers by hindering neoplastic growth in malignant tumors like breast cancer, ovarian cancer, head and neck cancers (Fu et al., 2009; Okano and Rustgi, 2001). Paclitaxel acts on cancer cells by interfering with the mitotic phase of cell division where it stabilizes tubulin polymerization, thereby it makes the dynamics of the microtubules stable and dysfunctional and consequently arresting the cell cycle at the mitotic phase (Panchagnula, 1998; Wang et al., 2000; Wani et al., 1971). Prolonged exposure to Paclitaxel in cancer therapy makes cancer cells acquire resistance over a period, leading to the failure of treatment (Ambudkar et al., 1999; Blum et al., 2001). There are many mechanisms for drug resistance in cancer therapy. The most commonly addressed in most of the cancer cells are the increased efflux of anti-cancer drugs mediated through Adenosine triphosphate (ATP)-binding cassette transporter and P-glycoprotein [P-gp also known as Multidrug resistance (MDR1)] induced multidrug-resistant (Szakacs et al., 2006) P-gp shows resistance to a wide range of structurally, chemically, and functionally diverse compounds. However, the precise location of P-gp drug binding sites where the specific substrate binds within the transmembrane segments is still unclear (Ecker et al., 2002). Sparreboom et al. (1997) studied the efflux orally, and IV administered Paclitaxel on wild type and mdr1a(-/-) mice. He showed that P-gp resists the uptake of administered Paclitaxel in wild type mice, whereas a higher amount of Paclitaxel was found in the blood plasma of mdr1a knocked out mice.
Studies have also revealed that Paclitaxel activates the nuclear factor kappa B (NF?B) in mouse peritoneal macrophages (Hwang and Ding, 1995). The transcription factor NF?B is composed of 5 REL proteins. However, the NF?B is concealed within the cytoplasm bound to its regulatory protein called inhibitor of kappa B (I?B), which helps inhibit the activation of NF?B. Upon phosphorylation, the NF?B: I?B complex dissociates, leading to the activation of NF?B (Ghosh et al., 1998). The activation of NF?B results in cellular growth, and it plays a crucial role in all hallmarks of cancer. Most cancers like lung cancer, leukemia, and breast cancer have shown the activation of NF?B (Chua et al., 2007; Tew et al., 2008; Vilimas et al., 2007). Increased levels of NF?B result in poor prognosis in cancer therapy (Annunziata et al., 2010). Studies on rat hepatoma cell line containing NF?B binding site at mdr1b promoter, when exposed to a hepato carcinogen (2-acetylaminofluorene), showed upregulation of mdr1b through the activation of reactive oxygen species and dissociation of NF?B: I?B complex, which leads to the activation of NF?B (Kuo et al., 2002). Drug resistance in cancer cells arises and causes failure in cancer treatment due to the activation of NF?B and up-regulation of efflux pump proteins during treatment with Paclitaxel (Jin et al., 2009).
Studies have shown that Paclitaxel with Curcumin (Molecular weight: 368.4 g/mol; it has a pKa1, pKa2, and pKa3 value of 7.8, 8.5, and 9.0, respectively, for three acidic protons; logP: 3.2) will help downregulate NF?B and will improve the therapeutic efficiency of Paclitaxel (Bava et al., 2011). In addition, the phytochemical extracted from Curcuma longa perennial herb called Curcumin (Fig. 1B) inhibits the NF?B and interrupts cancer cell growth-inducing apoptosis (Singh and Aggarwal, 1995; Tang et al., 2002; Yallapu et al., 2012).
The usage of dual drugs with different mechanisms of action will improve the therapeutic efficiency of the treatment (Sreekanth et al., 2011). In addition, the usage of drug combinations reduces the therapeutic dosage of individual drugs, thereby reducing the severity of side effects caused due to chemotherapy.
Though Paclitaxel and Curcumin are efficient anti-cancer drugs, their therapeutic applications are constrained by poor bioavailability, slow dissolution rate, and high hydrophobicity. Therefore, a promising strategy to overcome these drawbacks is needed for efficient cancer treatment. Recent research has shown that nanoparticles (NPs), micelles, nanocapsules, liposomes, and polymeric microspheres can overcome the problems associated with Paclitaxel and Curcumin’s bio-availability (Madusanka et al., 2015; Verderio et al., 2013).
 | Figure 1. (A) Structure of Paclitaxel; (B) Structure of Curcumin; and (C) Structure of PLGA. [Click here to view] |
Physicochemical properties of the anticancer drugs are considered an important factor in developing new formulations for cancer therapy. The highly hydrophobic Paclitaxel/Curcumin’s physicochemical properties have been altered for improved solubility and bioavailability through triazine conjugated cyclodextrin polymer complexation. Furthermore, the encapsulation of Paclitaxel/Curcumin (separately) with an oligosaccharide cyclodextrin altered the drugs’ physiochemical properties and showed drugs’ synergistic effect in inhibiting cell growth (Boztas et al., 2013). A T7 mediated polylactide-co-glycolic acid (PLGA) NPs entrapped with oleic acid magnetic NPs and Paclitaxel with Curcumin together as a dual targeting system has shown synergistic cytotoxicity on glioma cells. In addition, the formulation gives improved chemotherapeutic efficacy compared to the single use of each drug (Cui et al., 2016).
The use of graphene oxide as a cargo for cancer drug delivery has gained substantial interest. Reduced graphene oxide (G) cargo, functionalized with amphiphilic triblock copolymer PF-127 (P) and loaded with Paclitaxel and Curcumin together showed a synergistic effect on A549 and MDA-MB-231 cells (Muthoosamy et al., 2016). In addition to the inorganic nano-carriers, protein nano-carriers are also extensively studied due to their biocompatibility and the presence of various functional groups that can be utilized for ligand attachments. Albumin (Alb) NPs co-bounded with Paclitaxel and Curcumin have been shown to have enhanced cytotoxicity and intracellular uptake on mia-paca-2 human pancreatic carcinoma cells (Kim et al., 2016).
PEGylation is considered as the most widely used technique for imparting stealth properties to nano-carriers and enhance their uptake. Co-administration of Paclitaxel and Curcumin, encapsulated separately in flaxseed oil nanoemulsion [with or without PEG Poly (ethylene glycol) modification]significantly induced apoptosis and enhanced cytotoxic properties on SKVO3 and SKVO3TR cells (Ganta et al., 2009).
The above mentioned previously published articles have encapsulated dual drugs together in a single drug delivery vehicle or simultaneously delivered dual drugs using polymer complexes or nanoemulsions (Boztas et al., 2013; Cui et al., 2016; Ganta et al., 2009; Kim et al., 2016; Muthoosamy et al., 2016). This study’s main concern is increasing therapeutic efficacy without increasing the dosage of the drugs by using Food and Drug Administration (FDA) approved polymer poly(lactic-co-glycolic acid) (PLGA) (Fig. 1C) as a drug delivery vehicle. Paclitaxel/Curcumin was encapsulated in PLGA separately and delivered simultaneously during therapy to find how simultaneous delivery helps in increasing the therapeutic efficacy. Furthermore, the Enzyme Linked Immunosorbent Assay (ELISA) kit tested the inhibition of NF?B activity with the nano-formulation of individual drugs and a combination of drugs.
The present study focuses on preparing biodegradable and biocompatible polymeric NPs made of PLGA encapsulated with Curcumin/Paclitaxel drugs. The efficacy of formulated NPs was tested as a mono drug/combination of the drug for a sustained drug delivery system. It also evaluated whether Paclitaxel and Curcumin in nano-formulation or Curcumin with Paclitaxel reduced the activity of NF?B on cell lines of U-87 (glioblastoma cells) and A549 (lung cancer cells).
MATERIALS AND METHODS
Materials
PLGA (lactide to glycolide ratio 50:50; average molecular weight (MW) 1,00,000-1,20,000 Da) was purchased from DURECT Corporation, AL; Polyvinyl alcohol (PVA; MW 1,25,000) was obtained from Thomas Baker Mumbai, India; high-performance liquid chromatography (HPLC) grade organic solvent acetonitrile was obtained from Merck Life Science Private Limited, Mumbai, India; Paclitaxel (PTX; MW 853.91) was obtained from Xi’an Natural Field Bio-Technique Co., Ltd, China; Curcumin (CUR; MW 368.38) was procured from Alfa Aesar, Mumbai, India; Trifluoroacetic acid was obtained from Lobo Chemie, Mumbai, India; U87 (glioblastoma) cells and A549 lung carcinoma cells were procured from The National Centre for Cell Science (NCCS), Pune, India. The cell culture growth media, namely minimum essential medium eagle, Nutrient Mixture F-12 Ham, Kaighn’s modification media, fetal bovine serum (FBS), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent were obtained from HiMedia, Mumbai, India. NF?B p65 (Total) Human InstantOneTM ELISA kit was purchased from Invitrogen Bioservices India Pvt. Ltd, Thermo Fisher Scientific, India.
Methods
Preparation of Paclitaxel/Curcumin/Coumarin-6 loaded PLGA NPs
A single emulsion solvent evaporation technique was used (Hakkimane et al., 2018) to formulate PTX loaded PLGA NPs, and CUR loaded PLGA NPs. Briefly, PLGA 50:50 (90 mg-amount of polymer used), PTX (45 mg), or CUR (45 mg) 2:1 (Hakkimane and Guru, 2017) ratio of polymer. The drug (PTX and CUR were dissolved separately to formulate PTX loaded PLGA NPs and CUR loaded PLGA NPs) was first dissolved in 1.8 ml of chloroform (organic solvent). 12 ml of 2% (w/v) aqueous PVA solution was added and emulsified using a probe sonicator to form an oil-in-water emulsion (QSonica; Sonicator, CL-334) for 6 minutes with 40% amplitude over an ice bath. Coumarin-6 (300 μg) was used instead of anti-cancer drugs to prepare the fluorescent probed NPs, and the same experimental procedure was followed to get Coumarin-6 loaded NPs. The organic solvent was removed by evaporation with constant stirring overnight using a magnetic stirrer (SCHOTT magnetic stirrer). Figure 2 depicts the schematic representation of the formulation of NPs. The formed NPs were washed four times by suspending in pure water followed by centrifugation at 13,000 rpm for 45 minutes at 4°C. NPs were resuspended in deionized water, frozen at −80°C (ultra-low temperature freezer, Model-U410 premium, New Brunswick, UK), and then freeze-dried (LaboGene ScanVac cool safe) at −110°C and 0.1 mbar. The NPs obtained were characterized using different characterization techniques.
Characterization of NPs
Dynamic light scattering (DLS)
Hydrodynamic diameter, polydispersity, size distribution, and zeta potential of NPs were determined using a particle size analyzer (Zetasizer nano series, Model-Nano ZS ZEN 3600, Malvern Instruments, Malvern, UK) by suspending the freeze-dried NPs in deionized water. The analysis was performed at room temperature at a scattering angle of 90°.
Determination of drug loading
The amount of drug successfully loaded in the formulated PTX/CUR loaded PLGA NPs was determined using reverse-phase-HPLC (RP-HPLC) by dispersing the NPs (1 mg/ml) in methanol followed by constant shaking for two days using a tilting rocker shaker (Rotek instruments) (Hakkimane et al., 2018). Methanol bulges PLGA NPs and helps to loosen its construction to free the encapsulated drug to get dissolved in methanol. After centrifugation for 20 minutes (Eppendorf centrifuge), the supernatant containing the dissolved drug was removed and evaporated to remove the methanol. The evaporated sample was dissolved in 1 ml of acetonitrile and analyzed for the concentration of the PTX/CUR by RP-HPLC (Shimadzu LC-20AD, SPD-20A, SIL-20AC HT) using C18 150*4.6 mm and 250*4.6 mm column of 5-micron pore size. HPLC method for PTX was developed using acetonitrile and water (65:35) as mobile phase and C18 reverse phase column of 150 mm. The retention time of pure drug was found to be 3.7 minutes (Fig. 3). The HPLC method for CUR was developed using acetonitrile and water with 0.05% TFA (70:30) as mobile phase and was eluted at 3.9 minutes (Fig. 4). The RP-HPLC method developed was validated in accordance with the guidelines of the International Conference on Harmonization (ICH, 2005). The drug loading was expressed as the amount of drug (in μg) present in one mg of the formulated nanoparticle. The unknown amount of drug in the NPs was calculated using the calibration chart prepared for the drug.
 | Figure 2. Schematic representation of formulation of NPs. [Click here to view] |
Surface morphology of the NPs
The uniformity, shape, and size were analyzed by a scanning electron microscopy. The samples were prepared by dispersing them in deionized water and coating it on the carbon tape placed on the surface of the sample stub and was dried overnight. The next day the sample was sputter-coated with a thin layer of gold before examining using a scanning electron microscope (SEM) (Make JEOL JSM-6380LA).
Drug stability analysis
RP-HPLC studied the pure drug’s stability at different pH conditions (4.5, 5.4, and 7.4). A specific quantity of the drug PTX was weighed and dissolved in a known amount of phosphate buffer saline (PBS) at different pH conditions. This was followed by centrifugation at 10,000 rpm to remove the undissolved drug. The supernatant was collected and analyzed for the initial drug stability through HPLC, then incubated at 37°C and analyzed at different time intervals. All the samples were prepared in triplicates and analyzed for the drug stability studies.
In vitro release studies
The formulated PLGA-Paclitaxel nanoparticles (PTX NPs) were evaluated for their In-vitro drug release at different pH conditions at different times using RP-HPLC. The PBS of different pH (4.5, 5.4, and7.4) was used for the PTX NPs. Three milligrams of the drug-loaded NPs were suspended in 1 ml of PBS in triplicates and incubated at 37°C on a rocker shaker. The samples were collected at designated time intervals and centrifuged at 10,000 rpm for 20 minutes. The supernatant was removed and analyzed for the drug content using HPLC. The samples were re-dispersed in 1 ml of buffer, and the procedure was continued till the last sample collection. The drug release was calculated as a cumulative release of the drug to time.
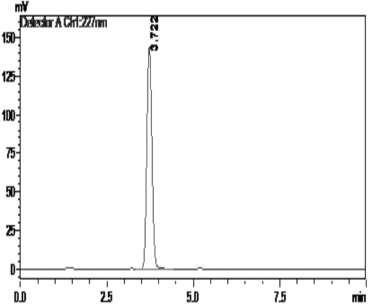 | Figure 3. Retention time of Paclitaxel. [Click here to view] |
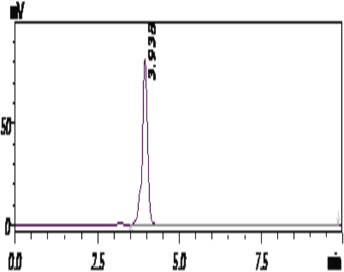 | Figure 4. Retention time of Curcumin. [Click here to view] |
Cell culture
U87 (glioblastoma) cell line and A549 lung carcinoma cell lines were procured from the National Centre for Cell Science (NCCS, Pune, India). The cell lines were retained in MEM and Nutrient Mixture F-12 Ham Kaighn’s Modification media, respectively, supplemented with 10% FBS and 1% antibiotic solution and incubated at 37°C with 5% of CO2.
In vitro cellular uptake of NPs
U87 and A549 cells were seeded 50,000 cells/well in 24 well plates and incubated overnight. After incubation, free Coumarin of concentration 1 μg/ml and Coumarin-6 equivalent to 1 μg in NPs were added to the wells and incubated for 1.5, 3, and 6 hours, respectively, at 37oC. After the treatment period, the wells were washed thrice with ice-cold PBS and lysed with lysis buffer (0.5% Triton X-100, 10 mM tris HCl, and 150 mM of NaCl). The lysed cells were centrifuged at 13,000 rpm for 20 minutes. One part of the cell lysate was used for analyzing the total protein concentration through Bradford assay, and for the other part of cell lysate, methanol was added to extract the Coumarin-6 in methanol. This was then analyzed for its fluorescence intensity using Thermo Fisher Varioskan LUX-(3020) multi-plate reader.
MTT cytotoxic assay
5,000 cells per well were seeded in 96 well plates and incubated at 37°C for 24 hours with 5% CO2. The stock solutions of 1 mM Paclitaxel and Curcumin were first prepared by dissolving them in dimethyl sulfoxide and serially diluted with a culture medium. After 24 hours of incubation, the cells were treated with different concentrations (25–100 nM) of free drug Paclitaxel (PTX FD), free drug Curcumin (CUR FD), the combination of free drugs (COMBI FD), equivalent drug encapsulated PTX NP, Curcumin nanoparticles (CUR NP), a combination of drug encapsulated nanoparticles (COMBI NP) and incubated for 24, 72, 120, and 144 hours. After the incubation period, MTT (3-(4, 5-dimethyl thiazol-2yl)-2, 5-diphenyl tetrazolium bromide) solution is added to all the wells and incubated for 4 hours for the formation of formazan crystals. The plates are then analyzed in the microplate reader to find the absorbance. GraphPad Prism software and CompuSyn software calculated IC50 values and Combination Index (CI) values.
NF?B analysis
The NF?B p65 activity on A549 and U87 cell lines was conducted using an InstantOneTM ELISA kit and the manufacturer’s instructions were followed. Briefly, 105 cells per well were seeded and incubated overnight. The cells were treated with 50 nM concentration of PTX FD, CUR FD, the COMBI FD, equivalent of 50 nM drug encapsulated PTX NP, CUR NP, and COMBI NP and incubated for 24 and 72 hours. After the incubation period, the cells were gently washed with PBS and lysed with cell lysis buffer mix for 10 minutes. 50 μL of cell lysate and 50 μL of antibody cocktail mixture (capture antibody reagent: detection antibody reagent) per well were added and incubated for 1 hour at room temperature. The lyophilized cell lysate supplied with the ELISA kit reconstituted in double distilled water was used as a control. The solutions were aspirated and washed three times with (1×) wash buffer. 100 μL of detection reagent was added to each well and incubated for 10–30 minutes for the development of color, followed by the addition of 100 μl of stop solution, which turns the sample solution’s color from blue to yellow. The absorbance was taken at 450 nm. The NF?B p65 activity (in percentage) was determined by calculating it with the absorbance of the control.
Data analysis and statistics
Statistical analysis was conducted with GraphPad Prism 8.0 trial version software. Data were expressed as mean ± SD (n = 6). A criterion for statistical significance for all tests was set at p < 0.05.
RESULTS AND DISCUSSIONS
Preparation and characterization of PTX/CUR loaded PLGA NPs
The PTX/CUR loaded PLGA NPs were prepared by using the single emulsion solvent evaporation method. The copolymer PLGA helps hydrophobic anti-cancer drugs PTX/CUR to load in the nanoparticle’s core. The size of the NPS and their surface charge (zeta potential) were determined through the Dynamic Light Scattering technique. The Z-average for PTX NPs was 183.2 ±4 nm with a polydispersity index (PDI) of 0.34 ± 0.002 (Fig. 5A). ~ 97% of the PTX NPs found to be in the range mentioned above. The Z-average of CUR NPs was 216.8 ± 11 nm with a PDI of 0.4 ± 0.022 (Fig. 6A). The zeta potential of PTX and CUR NPs was—42 ± 7.6 mV (Fig. 5B) and −34 ± 6.51 mV (Fig. 6B), respectively. Zeta potential value is the measure of the nanoparticle’s surface charge, which is crucial in determining the stability of the NPs when dispersed in a medium. Higher zeta values stabilize the NPs in suspension due to electrostatic repulsion between like-charged particles (Ravikumar et al., 2004). Table 1 shows the size, PDI, and zeta potential of formulated Paclitaxel and Curcumin NPs. The particles with low zeta potential values tend to agglomerate more than particles with high zeta potential values (Akhavan et al., 2020). The strong zeta potential of NPs also reveals that the drug has been loaded in the core and not on the surface (Du et al., 2020). Figure 7 represents the SEM picture of NPs. Dual drug loading of Paclitaxel and Curcumin together in a single drug delivery vehicle was not chosen due to the reduced stability of Paclitaxel and Curcumin drugs when put together (Praveena and Guru, 2021). Furthermore, it will be more complicated to find out the molar concentration of each drug when dually loaded into the PLGA nanoparticle.
Calibration graph and drug loading
1mg of PTX/CUR dissolved in 1 ml of acetonitrile was diluted serially into different concentrations and analyzed using HPLC with the developed HPLC method. The R2 value for Paclitaxel and Curcumin was 0.9986, and 0.9988, respectively, revealing that the graph is linear with a good relationship between concentration range and area under the curve (AUC). The method developed was validated, and the results are shown in Supplementary Data [S1(a–d), S2 (a–d) and ST1–ST6)]. Drug loading was measured as the amount of drug in μg present in 1 mg of NPs. The drug loading of PTX NPS was 150 ± 11 μg/mg, and the drug loading of CUR NPs was 300 ± 7 μg/mg (Table 2). The entrapment efficiency (EE%) was checked by a direct method of calculating the total mass of the drug in the nanoparticle and the initial amount of drug used. The EE% of PTX NPs was 20.3%, with a loading capacity of 15%. CUR NPs EE% was 48% with a loading capacity of 30%.
 | Figure 5. (A) Size of Paclitaxel NPs and (B) zeta potential of Paclitaxel NPs. [Click here to view] |
Stability studies of PTX in different buffers
The PTX was analyzed for its stability at different pH 4.5, 5.4, and 7.4 to mimic the body conditions. When the administration route of NPs is intravenous, they enter the bloodstream, where the pH is 7.4. NPs are taken up by endocytosis, where the endosomal pH is approximately around 5.4 and then NPs move to lysosomes where the pH is approximately around 4.5. Hence, the stability of the pure drug was checked in all three (4.5, 5.4, and 7.4) pH conditions. The stability of the drug Paclitaxel was analyzed for 96 hours. Almost 71% of PTX degraded within 24 hours of exposure at 7.4 pH. Around 36% and 16% of PTX degraded at 4.5 and 5.4 pH within 24 hours. PTX degrades rapidly at 7.4 pH compared to 4.5 and 5.4 pH. The drug was 66% stable at the end of 96 hours for pH 4.5, around 45% stable for pH 5.4, and around 13% stable at pH 7.4 after 96 hours. Figure 8 represents the stability analysis graph of Paclitaxel.
In vitro drug release
The rate at which the drug releases from the NPs is one of the crucial factors that control the therapy’s efficiency. A quick or insignificant amount of drug release might result in a similar or reduced therapeutic effect of the free drug (Allen and Cullies, 2004). The drug release behavior of PTX loaded NPs was studied at different pH environments. The pH 4.5 (lysosomal pH), 5.4 (endosomal pH), and 7.4 (blood pH) were chosen to simulate different physiological conditions. Figure 9 represents the drug release studies of PTX NPs at different pH conditions. pH-dependent drug release of PTX was studied in PBS. The release of PTX was slower at 4.5 pH compared to 5.4 pH and 7.4 pH. There was a small initial burst release in all three pHs, and then it releases slowly for a substantial time. The drug release studies for PTX NPs were carried out for 28 days. Almost 36% PTX was released at 4.5 pH. PTX NPs 5.4 pH showed the highest release of PTX compared to all other pHs; ~84% of PTX was released by the end of 28 days. At 7.4 pH, almost 68% of the drug was released in 28 days.
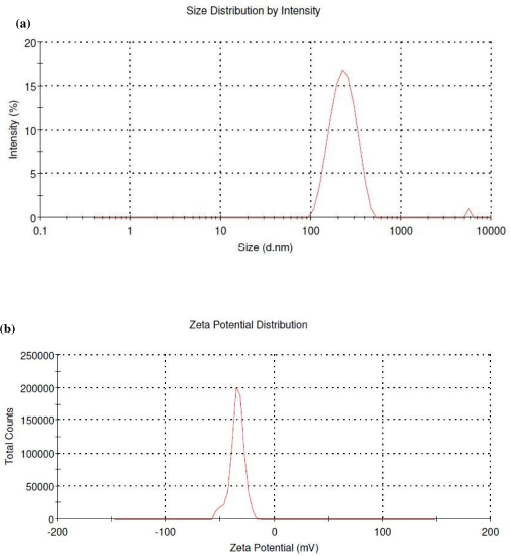 | Figure 6. (A) Size of curcumin NPs and (B) zeta potential of Curcumin NPs. [Click here to view] |
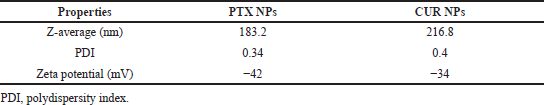 | Table 1. Particle size distribution, PDI, and zeta potential of PTX/CUR NPs. [Click here to view] |
Release studies of CUR NPs were not conducted due to the high hydrophobicity of the CUR as it has very low solubility in the release medium (PBS). Chen et al. (2013) have shown the drug release profile of CUR from solid lipid NPs using 0.5% of Tween-80 in the release medium (PBS 7.4 pH) that resulted in slow-release at the initial stage of 7.8% in 24 hours. This gradually increased up to ~90% of cumulative release within 84 hours. However, the non-ionic tween-80 acts as a surfactant to disrupt the cancer cell membrane. Therefore, it may disrupt the stability of the CUR. Therefore, the in vitro release study of the CUR NPs was not performed in the present study.
In vitro cellular uptake of NPs—quantification
The internalization of NPs through endocytosis is the key to cellular uptake of NPs without targeting moieties (Salatin and Yari Khosroushahi, 2017). The quantification of cellular uptake of NPs was used to analyze the internalization of free Coumarin-6, or Coumarin-6 loaded NPs on U87 and A549 cell lines. Figures 10 and 11 represent the internalization results of free Coumarin-6, and Coumarin-6 loaded NPs on U87 and A549 cell lines after 1.5, 3, and 6 hours of incubation. The Coumarin-6 content in the cells increased significantly with prolonged incubation time, proving time-dependent internalization. The results through high fluorescence intensity reveal that Coumarin-6 loaded NPs were taken up by cancer cells. The cellular uptake efficiency was calculated by normalizing it with total protein concentration. These studies revealed that Coumarin-6 loaded NPs took up ~>2fold (p < 0.0001) increase in internalization compared to the free Coumarin-6 treatment.
 | Figure 7. SEM of NPs. [Click here to view] |
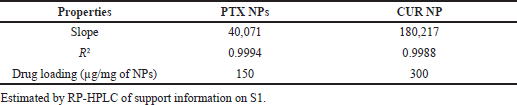 | Table 2. Linear regression coefficient and drug loading of PTX/CUR NPs. [Click here to view] |
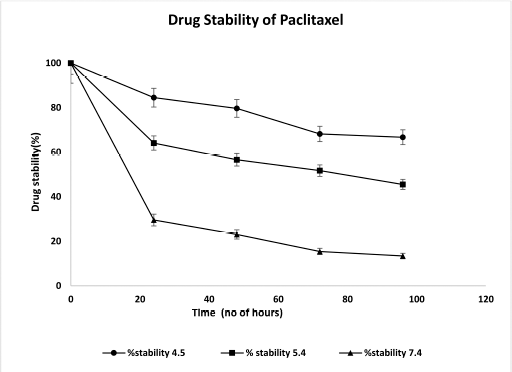 | Figure 8. Stability analysis graph of Paclitaxel. [Click here to view] |
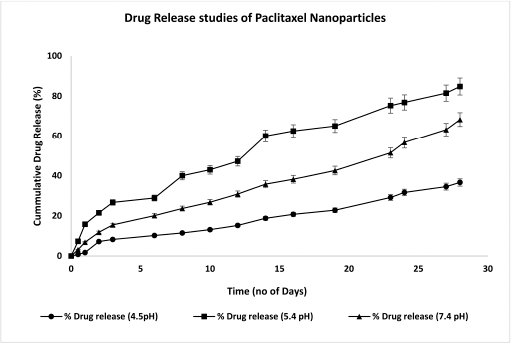 | Figure 9. Drug release profile graph of PTX NPs. [Click here to view] |
 | Figure 10. Quantitation of Free Coumarin/Coumarin NPs uptake in U87 cell lines. [Click here to view] |
In vitro cytotoxic assay
The cytotoxicity of the drug (PTX/CUR) loaded PLGA NPs, the combination of PTX & CUR NPs, and PTX FD/CUR FD were evaluated using the MTT assay technique on U87 and A549 cell lines. The assay was conducted for 24–144 hours. A total of six different treatments were used. Free Paclitaxel (PTX FD), free Curcumin (CUR FD), the combination of free COMBI FD, PTX NPs, CUR NPs, and a combination of COMBI NPs with optimized concentrations of 25, 50, 75, &100 nM were used. All NPs treatments showed increased cell death for an increase in drug concentration and the incubation time.
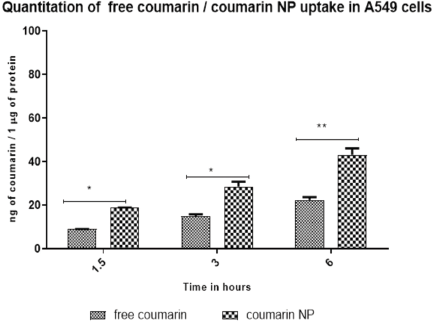 | Figure 11. Quantitation of Free Coumarin/Coumarin NPs uptake in A549 cell lines. [Click here to view] |
As shown in Figures 12A and B and 13A and B, the cytotoxic effects on different incubation periods with PTX/CUR as a free drug or as NPs either as single-drug therapy or combination therapy inhibited cell growth in a dose-dependent manner against U87 cell lines.
Comparison of Figure 12A and B reveals PTX FD/CUR FD reduced the cell viability with increased incubation time up to 72 hours. The decrease in IC50 values for PTX FD, CUR FD upon increased incubation time of more than 72 hours is shown in Table 3. Treatment with the combination of free drugs PTX and CUR (equimolar) with the concentration (25–100 nM) was conducted, and the results showed an increase in cytotoxicity on U87 cells. IC50 values for COMBI FD (Table 3) when incubated up to 72 hours resulted in significantly lesser IC50 values than PTX FD/CUR FD. A comparative analysis of Figure 12B with Figure 13A and B reveals that prolonged exposure (120 and 144 hours) of all the free drug treatments increased the viability of U87cells inferring the reduced potential of free drugs after 72 hours.
Treating U87 cell lines with the formulated nanoparticles either as PTX NPs/CUR NPs alone or as COMBI NPs showed increased cytotoxicity. Unlike the free drug, the treatment with NPs was effective against U87 cell lines even at an increased incubation period (Fig. 13A and B) of 120 and 144 hours. The results also show that COMBI NPs treatment is more effective than PTX NPs/CUR NPs alone. Treatment with the COMBI NPs has reduced the drug dosage compared to individual drugs. The amount of drug required to show approximately 70% of viable cells in single drug NPs treatment was 50 nM, but the COMBI NPs show a similar effect at 25 nM concentration. The IC50 values of the formulated NPs decreased with an increase in the incubation period. Combination Index (CI) values were analyzed by CompuSyn v.1 software and the corresponding effects were demonstrated by Fa. The Fa is the fraction of cells inhibited after drug exposure. According to CompuSyn software, it is defined between 0 and 1. For example, Fa = 0.5 shows cell growth inhibited by 50%. According to the Chou-Talalay method, CI > 1, CI = 1, and CI < 1 were, respectively, defined as an antagonistic, additive, and synergistic effect in drug combination (Chou, 2010). Based on the cytotoxicity results, CI values were calculated and found to be below 1. This infers a synergistic anti-cancer effect of PTX and CUR against U87 cells. The calculated IC50 (Table 3) of the treatment in combination showed a significantly lower IC50 value than in single-drug therapy.
The cytotoxic efficiency of PTX NPs, CUR NPs, and COMBI NPs was compared with PTX FD/CUR FD & COMBI FD on A549 cells. The applied concentrations ranged from 25nM to 100nM with an incubation period of 24, 72, 120, and 144 hours. The obtained results infer that cell viability is concentration dependent. As shown in Figures 14 and 15, treatment with NPs either alone or in combination reduced the viability of cells compared to all the treatments with free drugs. A549 cells show a trend similar to the one obtained in U87 cells. The potency of the free drugs reduced after 72 hours resulting in increased viability at 120 and 144 hours (Fig. 15A and B). Comparison of Figure 14A and B with Figure 15A and B shows that the potency of free drugs reduces after 72 hours. This was supported by the IC50 values shown in Table 4, which reveals that all free drug treatments, including the combination therapy, resulted in increased IC50 values upon prolonged incubation.
A549 cells were found to be more sensitive towards NPs treatment. Even after 144 hours of treatment, the cytotoxicity effect reduced the cell viability to a meager count. This explains that NPs serve as a protection to the drug, which helps retain the drug for an extended period and release the drug sustainably. The viability of the cell difference seen between 120 and 144 hours at lower concentrations (25 and 50 nM) was diminished by half, while higher concentrations 75 and100 nM showed a non-significant difference in cell viability for all NP treatments. Low IC50 values shown for the COMBI NP treatment when compared to other treatments in all treatment periods reveals that the combination of NPs treatment is much more efficient in killing cancer cells. Dose responsive IC50 graphs are shown in supplementary data S3 and S4. Statistical analysis (ANOVA followed by Post Hoc test) was performed for cytotoxic assay and was found to be significant (p < 0.05). The results generated using Prism Software are shown in supplementary data S5-S12. CI values using the cytotoxic assay results reveal (CI < 1) a synergistic anti-cancer effect of PTX and CUR against A549 cells combination index plots are provided in the Supplementary Information S13–S20.
 | Figure 12. (A) U87 cells cytotoxic assay on 24 hours of treatment and (B) U87 cells Cytotoxic assay on 72 hours of treatment. [Click here to view] |
 | Table 3. IC50 values of PTX & CUR as free drug/NPs in single or in combination against U87 cells. [Click here to view] |
 | Figure 13. (A) U87 cells cytotoxic assay on 120 hours of treatment and (B) U87 cells Cytotoxic assay on 144 hours of treatment. [Click here to view] |
NF?B activity
The A549 and U87 cells after the treatment with PTX FD, CUR FD, COMBI FD, PTX NP, CUR NP, and COMBI NP were lysed, and the cell lysate was used to check the NF?B activity. 24-hours treatment on A549 cells resulted in reduced levels of NF?B for all NPs treatments. CUR FD, which is supposedly expected to suppress NF?B, showed similar activity when treated with PTX FD (94%). A combination of PTX FD & CUR FD (25 nM + 25 nM) showed no significant difference in NF?B activity (97%) when compared to the mono drug treatment with free PTX/CUR. However, PTX/CUR drugs encapsulated in PLGA NPs showed a reduction in NF?B activity compared to the free drug treatment. PTX loaded PLGA NPs showed 76% of NF?B activity, while when treated with CUR loaded NPs the activity of NF?B was much more reduced (61%). Combination treatment yielded an intermediate result (72%). This could be due to the reduced dosage of the individual drugs (25 nM + 25 nM). Results of NF?B activity after 72 hours of treatment on A549 cells showed considerable reduction for PTX FD (76%) but not for CUR FD (97%). The combination of free PTX and CUR resulted in a significant reduction in NF?B activity (75%) compared to the 24-hour treatment. The drug-loaded PTX/CUR NPs after 72 hours of treatment showed considerable attenuation in NF?B activity compared to 24-hour treatment. The COMBI NPs yielded much more reduced NF?B activity (43%) on 72-hour treatment, proving our hypothesis that using drug combinations with NPs can reduce the NF?B activity on A549 cells with lower individual doses. Figure 16 represents the NF?B analysis results on A549 cell lines.
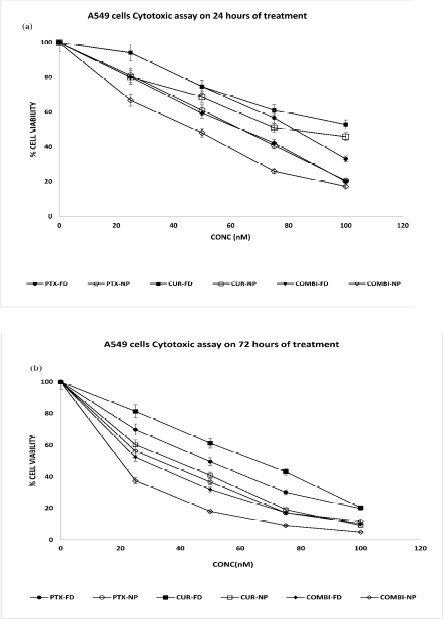 | Figure 14. (A) A549 cells cytotoxic assay on 24 hours of treatment and (B) A549 cells Cytotoxic assay on 72 hours of treatment. [Click here to view] |
The NF?B activity of U87 glioblastoma cells after 24 hours of treatment with CUR FD was found to be 94%, similar to the 24 hours results of CUR FD on A549 cells. PTX FD resulted in 85% of NF?B activity, while the combination of PTX FD and CUR FD showed no statistical difference (84%) in NF?B reduction compared to the mono drug treatment. PTX/CUR loaded PLGA NPs showed a similar reduction of NF?B activity (27% and 26%) but showed a significant difference in the reduction of NF?B activity compared to the free drug treatment. The treatment with the combination of PTX and CUR NPs resulted in 21% of NF?B activity. 72 hours of treatment period on U87 cells showed an ambiguous result; all treatments, including NPs treatment, resulted in higher NF?B activity when compared with 24-hour treatments. There was no significant difference in NF?B activity level seen for free drug treatments between 24 and 72 hours. However, the NF?B activity was seen doubled for mono drug NPs treatment on 72 hours, and no considerable increase of NF?B activity was seen for the COMBI NP treatment between 24 and 72 hours. This leads us to a discussion of different possibilities for the increased activity of NF?B since NF?B activity also depends on cytokine production like TNF-α during chemotherapy. Studies have shown increased NF?B activity on glial cells when exposed to TNF-α (Badr et al., 2011).
Further studies are required to clear this ambiguity. Other possibilities are the insufficient concentration of the treatments used, which resulted in higher NF?B activity at 72 hours of treatment. Though studies have shown that free Curcumin reduces the NF?B activity on cancer cells, the present study reveals that NPs are more efficient in attenuating the NF?B activity. Further studies would help with understanding the working of NF?B activity. Figure 17 represents the NF?B analysis results on U87 cells.
 | Figure 15. (A) A549 cells cytotoxic assay on 120 hours of treatment and (B) A549 cells cytotoxic assay on 144 hours of treatment. [Click here to view] |
 | Table 4. IC50 values of PTX & CUR as Free drug /Nanoparticles in single or in combination against A549 cells. [Click here to view] |
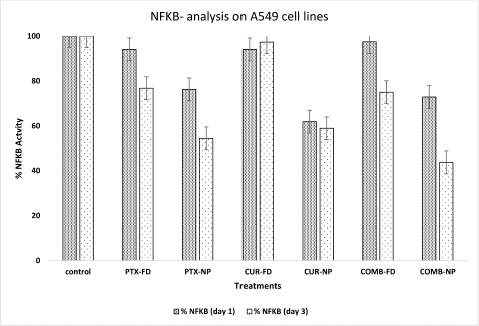 | Figure 16. NFkB analysis on A549 cells (day 1 & day 3 treatment). [Click here to view] |
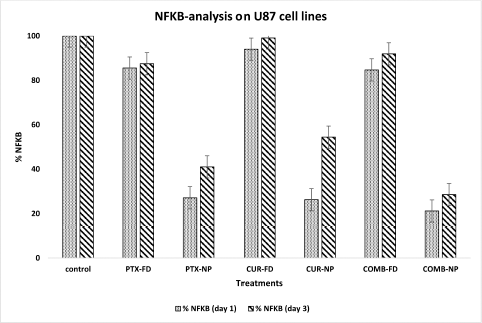 | Figure 17. NFkB analysis on A549 cells (day 1 & day 3 treatment). [Click here to view] |
CONCLUSION
We formulated Paclitaxel and Curcumin-loaded PLGA NPs and evaluated their effects as mono drug NPs formulation and combination of drugs NPs formulation on A549 and U87 cell lines. The characterization of NPs confirmed that they are negatively charged, spherical shaped with smooth surface and stable NPs. In vitro drug release studies showed that Paclitaxel releases from the NPs slowly and sustainably with some initial burst release. The cytotoxic effect of Paclitaxel and Curcumin-loaded PLGA NPs as single-drug therapy and combination therapy is more efficient on U87 and A549 cell lines compared to free drug or combination of drugs in free form. Treatment with the combination of drugs in nanoparticles is more efficient in killing cancer cells than individual drugs in nanoparticles. The cytotoxicity of formulated NPs was seen even after the fifth and sixth day of treatment, while free drug treatments lose efficacy after three days. In vitro cellular uptake studies showed increased fluorescent dye uptake, Coumarin-6 loaded NPs on A549 and U87 cells compared to free Coumarin-6. This result shows that NPs are taken up by a different mechanism called endocytosis. The transcription factor NF?B activity was evaluated on the formulated NPs. Paclitaxel and Curcumin-loaded PLGA NPs reduced the NF?B activity in both single drug treatments and a combination of drug treatments on A549 cell lines. NPs treatment on U87 cells also reduces NFKB activity substantially compared to the free drug but gave an ambiguous result of increased NF?B activity upon 72 hours of treatment both in free drug and in NPs compared to after 24 hours of treatment. Further studies are required to understand the activity of NF?B when treating with a combination of PTX and CUR in free drug form or NPs formulations form. The results obtained show that the combination of drugs in NPs formulation is more efficient for cancer therapy.
AUTHOR’S CONTRIBUTION
The concept and design of this study were done by Dr. Bharath Raja Guru and Joyceline Praveena. Acquisition of data was made by Joyceline Praveena and Dr. Sushruta S. Hakkimane. Statistical analysis and manuscript writing were done by Joyceline Praveena. The acquired data analysis and the written manuscript were supervised and approved by Dr. Bharath Raja Guru.
ACKNOWLEDGMENTS
The authors would like to thank the Department of Biotechnology, Manipal Institute of Technology, Manipal, for conducting this research.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
FUNDING
This work was partially funded by Vision Group of Science and Technology, Karnataka. The grant and the number are K-FIST LEVEL 1 GRD 267 was awarded to Dr. Bharath Raja Guru.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
REFERENCES
Akhavan Farid E, Davachi SM, Pezeshki-Modaress M, Taranejoo S, Seyfi J, Hejazi I, Tabatabaei Hakim M, Najafi F, D’Amico C, Abbaspourrad A. Preparation and characterisation of polylactic-co-glycolic acid/insulin nanoparticles encapsulated in methacrylate coated gelatin with sustained release for specific medical applications. J Biomater Sci Polym Ed, 2020; 31(7):910–37. CrossRef
Albain KS, Nag SM, Calderillo-Ruiz G, Jordaan JP, Llombart AC, Pluzanska A, Rolski J, Melemed AS, Reyes-Vidal JM, Sekhon JS, Simms L, O’Shaughnessy J. Gemcitabine plus Paclitaxel versus Paclitaxel monotherapy in patients with metastatic breast cancer and prior anthracycline treatment. J Clin Oncol, 2008; 26(24):3950–57. CrossRef
Allen TM, Cullis PR. Drug delivery systems: entering the mainstream. Science, 2004; 303(5665):1818–22. CrossRef
Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol, 1999; 39:361–98. CrossRef
Annunziata CM, Stavnes HT, Kleinberg L, Berner A, Hernandez LF, Birrer MJ, Steinberg SM, Davidson B, Kohn EC. Nuclear factor kappaB transcription factors are coexpressed and convey a poor outcome in ovarian cancer. Cancer, 2010; 116(13):3276–84. CrossRef
Badr CE, Niers JM, Morse D, Koelen JA, Vandertop P, Noske D, Wurdinger T, Zalloua PA, Tannous BA. Suicidal gene therapy in an NF-κB-controlled tumor environment as monitored by a secreted blood reporter. Gene Ther, 2011; 18(5):445–51. CrossRef
Bae Y, Diezi TA, Zhao A, Kwon GS. Mixed polymeric micelles for combination cancer chemotherapy through the concurrent delivery of multiple chemotherapeutic agents. J Control Release, 2007; 122(3):324–30. CrossRef
Bava SV, Sreekanth CN, Thulasidasan AK, Anto NP, Cheriyan VT, Puliyappadamba VT, Menon SG, Ravichandran SD, Anto RJ. Akt is upstream and MAPKs are downstream of NF-κB in paclitaxel-induced survival signaling events, which are down-regulated by Curcumin contributing to their synergism. Int J Biochem Cell Biol, 2011; 43(3):331–41. CrossRef
Blum JL, Dieras V, Lo Russo PM, Horton J, Rutman O, Buzdar A, Osterwalder B. Multicenter, Phase II study of capecitabine in taxane-pretreated metastatic breast carcinoma patients. Cancer. 2001; 92(7):1759–68. CrossRef
Boztas AO, Karakuzu O, Galante G, Ugur Z, Kocabas F, Altuntas CZ, Yazaydin AO. Synergistic interaction of Paclitaxel and Curcumin with cyclodextrin polymer complexation in human cancer cells. Mol Pharm, 2013; 10(7):2676–83. CrossRef
Chen J, Dai WT, He ZM, Gao L, Huang X, Gong JM, Xing HY, Chen WD. Fabrication and evaluation of curcumin-loaded nanoparticles based on solid lipid as a new type of colloidal drug delivery system. Indian J Pharm Sci, 2013; 75(2):178–84.
Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res, 2010; 70(2):440–6. CrossRef
Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H. NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene, 2007; 26(5):711–24. CrossRef
Cui Y, Zhang M, Zeng F, Jin H, Xu Q, Huang Y. Dual-targeting magnetic PLGA nanoparticles for codelivery of Paclitaxel and Curcumin for brain tumor therapy. ACS Appl Mater Interfaces, 2016; 8(47):32159–69. CrossRef
Du J, Wu Q, Li Y, Liu P, Han X, Wang L, Yuan J, Meng X, Xiao Y. Preparation and characterisation of Keratin-PEG conjugate-based micelles as a tumor microenvironment-responsive drug delivery system. J Biomater Sci Polym Ed, 2020; 31(9):1163–78. CrossRef
Ecker GF, Csaszar E, Kopp S, Plagens B, Holzer W, Ernst W, Chiba P. Identification of ligand-binding regions of P-glycoprotein by activated-pharmacophore photoaffinity labeling and matrix-assisted laser desorption/ionisation-time-of-flight mass spectrometry. Mol Pharmacol, 2002; 61(3):637–48. CrossRef
Fu Y, Li S, Zu Y, Yang G, Yang Z, Luo M, Jiang S, Wink M, Efferth T. Medicinal chemistry of Paclitaxel and its analogues. Curr Med Chem, 2009; 16(30):3966–85. CrossRef
Ganta S, Amiji M. Coadministration of Paclitaxel and Curcumin in nanoemulsion formulations to overcome multidrug resistance in tumor cells. Mol Pharm, 2009; 6(3):928–39. CrossRef
Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998; 16:225–60. CrossRef
Hakkimane SS, Guru BR. Biodegradable polymer nanoparticles of anti-tubercular drug: formulation and in vitro release studies. Adv Sci Lett, 2017; 23(3):1718–23. CrossRef
Hakkimane SS, Shenoy VP, Gaonkar SL, Bairy I, Guru BR. Antimycobacterial susceptibility evaluation of rifampicin and isoniazid benz-hydrazone in biodegradable polymeric nanoparticles against Mycobacterium tuberculosis H37Rv strain. Int J Nanomed, 2018; 13:4303–18. CrossRef
Hwang S, Ding A. Activation of NF-kappa B in murine macrophages by taxol. Cancer Biochem Biophys, 1995; 14(4):265–72.
ICH, Q2R1, “Validation of analytical procedures: text and methodology”, ICH Harmonised Tripartite Guideline. International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use, Chicago, USA, 2005.
Jin X, Qiu L, Zhang D, Zhang M, Wang Z, Guo Z, Deng C, Guo C. Chemosensitization in non-small cell lung cancer cells by IKK inhibitor occurs via NF-kappaB and mitochondrial cytochrome c cascade. J Cell Mol Med, 2009;13(11–12):4596–607. CrossRef
Khdair A, Handa H, Mao G, Panyam J. Nanoparticle-mediated combination chemotherapy and photodynamic therapy overcomes tumor drug resistance in vitro. Eur J Pharm Biopharm, 2009; 71(2):214–22. CrossRef
Kim B, Lee C, Lee ES, Shin BS, Youn YS. Paclitaxel and curcumin co-bound albumin nanoparticles having antitumor potential to pancreatic cancer. Asian J Pharm Sci, 2016; 11(6):708–14. CrossRef
Kuo MT, Liu Z, Wei Y, Lin-Lee YC, Tatebe S, Mills GB, Unate H. Induction of human MDR1 gene expression by 2-acetylaminofluorene is mediated by effectors of the phosphoinositide 3-kinase pathway that activate NF-kappaB signaling. Oncogene, 2002; 21(13):1945–54. CrossRef
Madusanka N, de Silva KM, Amaratunga G. A curcumin activated carboxymethyl cellulose-montmorillonite clay nanocomposite having enhanced curcumin release in aqueous media. Carbohydr Polym, 2015; 134:695–99. CrossRef
Muthoosamy K, Abubakar I, Bai R, Loh HS, Manickam S. Exceedingly higher co-loading of Curcumin and Paclitaxel onto polymer-functionalized reduced graphene oxide for highly potent synergistic anticancer treatment. Sci Rep, 2016; 6:32808. CrossRef
Okano J, Rustgi AK. Paclitaxel induces prolonged activation of the Ras/MEK/ERK pathway independently of activating the programmed cell death machinery. J Biol Chem, 2001; 276(22):19555–64. CrossRef
Panchagnula, R. Pharmaceutical aspects of Paclitaxel. Int J Pharm, 1998; 172(1–2):1–15. CrossRef
Partridge AH, Burstein HJ, Winer EP. Side effects of chemotherapy and combined chemohormonal therapy in women with early-stage breast cancer. J Natl Cancer Inst Monogr, 2001; (30):135–42. CrossRef
Praveena J, Guru BR. Simultaneous estimation of Paclitaxel and Curcumin in nano-formulation: Stability analysis of drugs, optimisation and validation of HPLC method. J Appl Pharm Sci, 2021; 11(03):71–83.
Ravi Kumar MN, Bakowsky U, Lehr CM. Preparation and characterisation of cationic PLGA nanospheres as DNA carriers. Biomaterials, 2004; 25(10):1771–7. CrossRef
Salatin S, Yari Khosroushahi A. Overviews on the cellular uptake mechanism of polysaccharide colloidal nanoparticles. J Cell Mol Med, 2017; 21(9):1668–86.
Singh S, Aggarwal BB. Activation of transcription factor NF-kappa B is suppressed by Curcumin (diferuloylmethane). J Biol Chem, 1995; 270(42):24995–5000. CrossRef
Sparreboom A, van Asperen J, Mayer U, Schinkel AH, Smit JW, Meijer DK, Borst P, Nooijen WJ, Beijnen JH, van Tellingen O. Limited oral bioavailability and active epithelial excretion of Paclitaxel (Taxol) caused by P-glycoprotein in the intestine. Proc Natl Acad Sci USA, 1997; 94(5):2031–5. CrossRef
Sreekanth CN, Bava SV, Sreekumar E, Anto RJ. Molecular evidences for the chemosensitising efficacy of liposomal Curcumin in paclitaxel chemotherapy in mouse models of cervical cancer. Oncogene, 2011; 30(28):3139–52. CrossRef
Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov, 2006; 5(3):219–34. CrossRef
Tang B, Ma L, Wang HY, Zhang GY. Study on the supramolecular interaction of Curcumin and beta-cyclodextrin by spectrophotometry and its analytical application. J Agric Food Chem, 2002; 50(6):1355–61. CrossRef
Tew GW, Lorimer EL, Berg TJ, Zhi H, Li R, Williams CL. SmgGDS regulates cell proliferation, migration, and NF-kappaB transcriptional activity in non-small cell lung carcinoma. J Biol Chem, 2008; 283(2):963–76. CrossRef
Verderio P, Bonetti P, Colombo M, Pandolfi L, Prosperi D. Intracellular drug release from curcumin-loaded PLGA nanoparticles induces G2/M block in breast cancer cells. Biomacromolecules, 2013; 14(3):672–82. CrossRef
Vilimas T, Mascarenhas J, Palomero T, Mandal M, Buonamici S, Meng F, Thompson B, Spaulding C, Macaroun S, Alegre ML, Kee BL, Ferrando A, Miele L, Aifantis I. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med, 2007; 13(1):70–7. CrossRef
Wang TH, Wang HS, Soong YK. Paclitaxel-induced cell death: where the cell cycle and apoptosis come together. Cancer, 2000; 88(11):2619–28. CrossRef
Wani MC, Taylor HL, Wall ME, Coggon P, McPhail AT. Plant antitumor agents. VI. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J Am Chem Soc, 1971; 93(9):2325–27. CrossRef
Yallapu MM, Jaggi M, Chauhan SC. Curcumin nanoformulations: a future nanomedicine for cancer. Drug Discov Today, 2012; 17(1–2):71–80. CrossRef
SUPPLEMENTARY MATERIALS
https://drive.google.com/file/d/1oTTMMONnDZdZfTKPVYRdYg5n5Lg-wikt/view