INTRODUCTION
An antibiotic is a natural or synthetic substance that kills bacteria or blocks their growth (Lewis, 2020). Antibiotics act on the bacterium by attacking either their cell wall (Kobras et al., 2020), their cytoplasmic membrane (Heesterbeek et al., 2021), their protein synthesis metabolism (Stokes et al., 2019), their nucleic acid (RNA and DNA), or even more than one of these elements (Grandclaudon et al., 2020). Since their discovery, many antibiotic molecules have emerged and their role in the treatment of various bacterial infections in both humans and animals is no longer to be demonstrated. In the United States, recent estimates show that 80% of antibiotics are used in livestock breeding as feed additives and growth promoters to improve the quality and yield of production (Ventola, 2015). Unfortunately, these antibiotics are excreted by animals in feces and then end up in the groundwater environment or are used as fertilizers (Ventola, 2015). As a result of this practice, beyond the direct environmental impacts, we are witnessing the emergence of increasingly resistant and multiresistant microorganisms (Ventola, 2015). This adaptation phenomenon is mainly due to the enzymatic degradation of antibiotics by bacteria (Reis et al., 2020), the modification of the target of the antibiotic (Schaenzer et al., 2020), the change in membrane permeability (Xu et al., 2020), and alternative metabolic pathways (Pollock et al., 2020). Interbacterial transmission of antibiotic resistance through horizontal gene transfer (conjugation, transduction, and transformation) has made the situation critical worldwide (Lima et al., 2020). Broadly speaking, antibiotic resistance is defined as the ability of a bacteria to resist the inhibitory or destructive activity of an antimicrobial to which it was not resistant (Palma et al., 2020). This problem concerns both veterinary medicine and human medicine, and every year numerous studies demonstrating the increasing resistance of bacterial strains are carried out (Mbarga et al., 2020; Rabello et al., 2020). In hospitals, the most resistant germs encountered are Staphylococcus aureus, Coagulase-negative Staphylococci, Pseudomonas aeruginosa, Klebsiella pneumoniae, Escherichia coli, Enterobacter spp., Enterococcus spp., and Acinetobacter (Antoinette and Dieudonne, 2020; Cleven et al., 2006). These germs are generally involved in various classic infections and some of them are sometimes implicated in nosocomial infections (Cleven et al., 2006). According to several researchers, the fight against antibiotic resistance requires a more measured use of antibiotics, a prescription exclusively reserved for professionals, systematic antibiograms before administration, and the search for new effective molecules on already resistant germs (Cleven et al., 2006; Lima et al., 2020; Palma et al., 2020). However, beyond the search for new antibiotics commonly called classic, several studies have been carried out to propose alternative techniques and methods having an effect on the bacteria. This review aims to discuss, without claiming to be exhaustive, most of the possible alternatives to conventional antibiotics.
REVIEW METHODOLOGY
This review article was carried out by exploiting numerous review articles, original articles, and related books from repuTable databases, such as Web of Science, PubMed, and Scopus. No open access published papers have been made available using the facilities provided by the People’s Friendship University of Russia, Moscow, Russia. The literature investigation process was conducted between June 2020 and June 2021 and the literature investigations were conducted in English and French. The keywords explored during literature searching included combinations of the words constituting the titles of each section, that is, “resistance to antibiotics” “phage therapy”, “antimicrobial peptides” (AMP), “combination of antibiotic and antimicrobial peptides”, “vaccines and antibiotics”, “herbal medicine”, “antimicrobial activity of plant materials”, “modulation of antibiotics with plant materials”, “antibacterial properties of nanoparticles”, “probiotics and antibiotics”; “phagothérapie”, “plantes médécinales”, “activité antibactérienne des nanoparticules’’, “peptides antimicrobiens”, and “synergie entre les extrait des plantes et les antibiotiques classiques”.
Resistance to Antibiotics and Current Issues
Antibiotic resistance poses one of the most serious threats to global health, food security, and development today (World Health Organization (WHO), 2019). An increasing number of infections, such as pneumonia, tuberculosis or gonorrhea, and salmonellosis, are becoming more difficult to treat as the antibiotics used to treat them lose their effectiveness (WHO, 2019). This situation is likely to affect people of all ages and genders in all corners of the globe and both the losses and the dangers it represents are considerable. Indeed, antibiotic resistance leads to prolonged hospitalizations, an increase in medical expenses, and, increasingly, an increase in mortality (Jit et al., 2020; WHO, 2019). Likewise, recent estimates have shown that antibiotic resistance is responsible for 700,000 annual deaths worldwide, 230,000 of which have resulted from multidrug-resistant tuberculosis (WHO, 2019). Furthermore, the WHO, 2019, estimates that if nothing is done to address this problem, drug-resistant diseases may cause 10 million deaths each year by 2050 and damage to the economy as catastrophic as the 2008–2009 global financial crises. In addition, economically (linked directly or not to agriculture and animal breeding), antimicrobial resistance could force up to 24 million people into extreme poverty by 2030 (WHO, 2019). However, in order to reduce the spread of this phenomenon, the researchers recommend more careful use of these substances both in animal breeding and in human medicine (Doidge et al., 2020; Morel et al., 2020). This consists mainly of stopping the use of antibiotics in breeding as growth factors and their use only when necessary to treat identified infections, under prescription, and with prior antibiograms (Mbarga et al., 2020; WHO, 2020). Moreover, it is also suggested that a major part of the solution to this problem lies in the establishment of new therapeutic protocols which include new biologically active compounds (Morel et al., 2020) including AMP, nanoparticles (NPs), antibodies, cytokines, phytochemicals, viral particles (bacteriophages), and microorganisms (probiotics).
Phage Therapy
The first use of phage therapy for the treatment of patients dates to 1919 when Félix d’Hérelle used them at the Institut Pasteur in Paris (Dublanchet and Schwartz, 2017). After this discovery, the practice quickly spread and the establishment of therapeutic protocols involving phages as a means of combating bacterial diseases was recognized by many scientists (Summers, 2005). However, it was only in 1925 that the first mass treatments took place in Brazil at the Oswaldo Cruz Institute and the results published by this institute were very promising. Among the first 10,000 cases treated, only 1 patient remained ill although his recovery was observed later (Biacchesi et al., 2017). Despite the good results obtained by this method, it has been abandoned in favor of the use of antibiotics because of their simplicity of handling, production, and use (Mireille et al., 2020). Today, due to the growing problem of multidrug resistance to antibiotics, phages are gradually regaining their place in the fight against bacterial infections although their use is still almost limited to highly equipped centers.
Mechanism of action of phages and application in medicine
Unlike conventional antibiotics whose mechanism of action is to attack one or more elements of the bacterial cell (wall, cytoplasmic membrane, metabolic pathways, or nucleic acids), phages have their own mechanisms and present a highly narrow specificity with the target bacteria (Koskella and Meaden, 2013). Without the host, it is impossible for them to multiply. As shown in Figure 1, the phage replication mechanism is like that of normal viruses. This figure shows that, after the attachment or adsorption (1) of the phage on the bacterial cell, the latter makes (2) its DNA penetrate the bacterium and rout all the enzymatic machinery of the latter to synthesize (3) the genome and viral proteins. After this step, the synthesized constituents will assemble (4) and cause the lysis of the bacterial cell which will lead to the release (5) of the new phages formed and the phenomenon will continue until the lysis of all the target bacteria (Jun, 2017). The time interval between attachment and cell lysis generally depends on several parameters (Van Belleghem et al., 2018). Indeed, modeling the pharmacokinetics of bacteriophages requires considering parameters related to their viral cycle, such as the adsorption constant of the viruses on the host bacteria, the number of virions released per cycle, the latency time, the initial number of bacteria values, and the bacteriophage/bacterium ratio (Van Belleghem et al., 2018). Likewise, the minimum bacterial density to ensure initiation of viral multiplication/transmission must be known. The modeling becomes even more complex when one considers the dynamics of the bacterial population over time (the uninfected cells continuing to replicate) as well as the phenomena of resistance to bacteriophages and their amplification over time treatment (Skurnik and Strauch, 2006). This pharmacokinetics is barely addressed in experimental animal studies, typically because of its complexity (determination of viral constants in vivo, bacterial inoculum, spontaneous viral elimination constant, etc.). It is, however, a fundamentally important approach and failure to take this into account could explain several previous failures of phage therapy (Payne and Jansen, 2001). Theoretical modeling of pharmacokinetics has thus made it possible to correlate the predicted results positively and significantly with what is observed in vitro (Cairns et al., 2009) but with difficulty in vivo (Weld et al., 2004), proving the need to continue studies in this area.
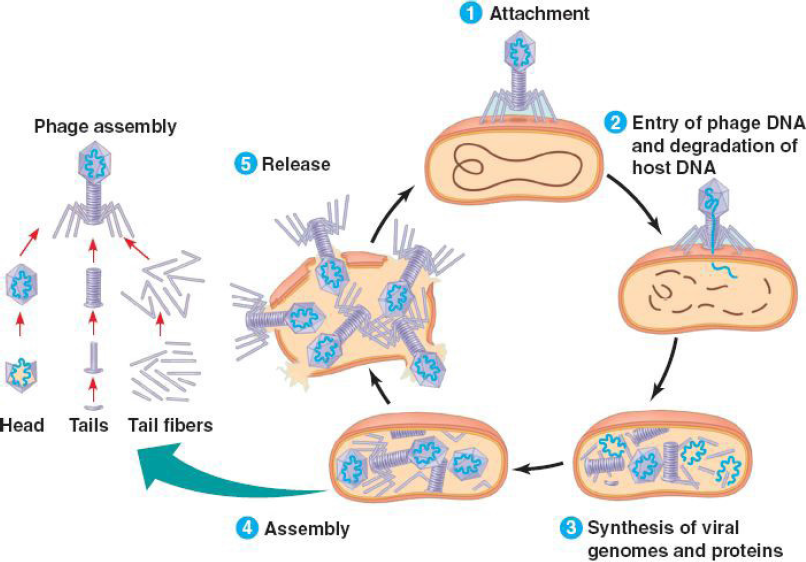 | Figure 1. Phage mechanisms of action (Jun, 2017 ). [Click here to view] |
However, despite the complexity of mastering phage pharmacodynamics in vivo, many recent studies have demonstrated their efficiency in human medicine. This is the case of a study conducted in the UK in 2018 which demonstrated the therapeutic potential of three bacteriophages (Escherichia phage ECP311, Klebsiella phage KPP235, and Enterobacter phage ELP140) against E. coli, K. pneumoniae, and Enterobacter cloacae (Prasanth et al., 2018). In addition, excellent results were obtained in terms of both safety and therapeutic efficiency in an Food and Drug Administration (FDA)-approved phase during the clinical trial (clinicaltrials.gov NCT00663091) on 42 patients with skin ulcers to evaluate the safety of 8 combinations of phages capable of lysing S. aureus, P. aeruginosa, and E. coli (Criscuolo et al., 2017). Three years later, still in Spain, another study (clinictrials.gov NCT00937274) was conducted on the safety, tolerability, and efficiency of oral administration of phage T4 in the treatment of toxigenic E. coli diarrhea in children; the results have been shown to be an alternative to antibiotic therapy, especially in developing countries, without modification of the patients’ intestinal flora (Criscuolo et al., 2017). Moreover, in a study carried out at the Ludwik Hirszfeld Institute in Poland on 157 patients (for 3 years) presenting various infections with multidrug-resistant bacteria, in particular urinary, bone, or respiratory, and treated with phage therapy for several weeks, beyond the observed efficiency, the biological parameters (blood count, liver function, renal function, and C reactive protein) demonstrated that no clinically relevant variation was observed, thus demonstrating the safety of bacteriophages under these conditions (Miedzybrodzki et al., 2012). Similarly, in another study involving 15 people who took three oral doses per day (for 2 days) of the bacteriophage T4 targeting E. coli, in addition to the efficiency, no adverse effects or an increase in markers of hepatic cytolysis or the development of antibodies against the T4 bacteriophage has been observed (Bruttin and Brussow, 2005). Unfortunately, in recent years, some researchers have gradually evoked the resistance mechanisms of certain bacterial groups against the phages that once infected them (Jorge and Eduardo, 2019; Sadhana et al., 2018).
Advantages and disadvantages of phage therapy
Table 1 presents the advantages and disadvantages of phages. One of the main drawbacks of phage therapy is the need to quickly establish the etiology of the bacteria causing the infection (Mireille et al., 2020) due to the excessive specificity of many phages. However, this problem could be solved with the help of phage mixtures and selection of potent lytic phages from a collection prior to bacterial characterization which could form the basis of empirical combination therapy (Nobrega et al., 2015). Otherwise, the possible involvement of humoral immunity with the production of neutralizing antibodies and the variable stability over time, influenced by storage conditions (pH, temperature, UV, etc.) as well as the potential capacity of phages to induce horizontal gene transfer by generalized transduction, is not negligible (Jun, 2017). During cell lysis, bacterial endotoxins and other released substances are a specific disadvantage of phages and other antimicrobial agents (Jun, 2017). However, the zero impact on nontarget bacteria and eukaryotic cells, rapid bactericidal activity independent of antibiotic resistance (Bruttin and Brussow, 2005), and antibiofilm activity (Sadhana et al., 2018) are certain assets that make this method a real alternative to conventional antibiotics.
Use of AMP as Alternatives to Antibiotics
AMP, also called host defense peptides, are essential components of the defense of multicellular beings and are part of the innate immune response (Andersson et al., 2016). They are amphiphilic and cationic molecules, mostly 12–50 amino acids, which exhibit a wide variety of structures. They have a broad spectrum of antimicrobial action as well as antibiofilm, anti-inflammatory, and immunomodulatory properties and constitute a class of molecules of choice in the face of the problems of antibiotic resistance (Vasilchenko and Rogozhin, 2019).
Antibacterial properties of known AMPs
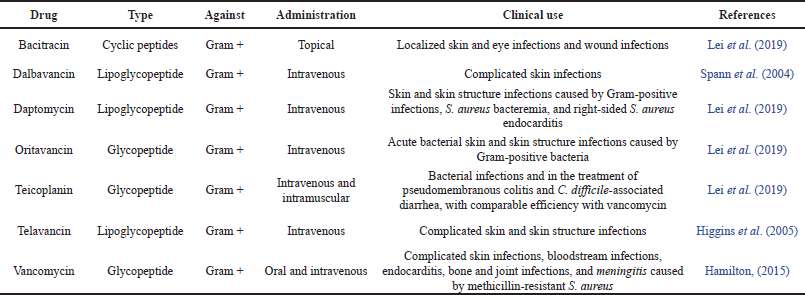
According to the antimicrobial peptide database (http://aps.unmc.edu/AP/main.php), 3,201 AMPs have been identified to date, of which 2,680 have antibacterial activity. Several AMPs have been shown to have a broad spectrum of activity against various microorganisms including Gram-positive and Gram-negative bacteria (Balter and Brown, 2011). In a recent study, the AMP PMAP-36PW and PMAP-36PK were found to have an expanded antibacterial spectrum, and in the evaluation of efficiency in vivo with mice infected with Salmonella choleraesuis C78-1 and Listeria monocytogenes CICC 21533, these peptide analogs have exhibited impressive therapeutic effect by reducing bacterial gene copies and decreasing inflammatory damage in mouse liver and lungs, resulting in reduction in mortality (Zhou et al., 2019). Each year, many similar studies are conducted and that provide baseline data for the design of clinically effective antibacterial peptides. In addition, unlike common antibiotics (Mahlapuu et al., 2016), several AMPs such as melittin, LL37, and alamethicin have demonstrated their ability (in vivo and in vitro) to induce membrane permeability in spheroplasts in E. coli (Faust et al., 2017). Furthermore, several antibacterial peptides act on other intracellular target processes, such as inhibition of DNA and RNA synthesis (buforin II, pleurocidin, and dermaseptin), protein synthesis (indolicidin and PR-39), and enzymatic activity (Jenssen et al., 2006). As shown in Table 1, most antibacterial peptides are associated either with carbohydrates (glycoproteins) or with lipids + carbohydrates (lipoglycopeptides) and their action is mainly directed toward Gram-positive bacteria. Some antimicrobial peptide drugs approved by the Food and Drug Administration (FDA) are presented in Table 2 AMPs are generally active on resistant bacteria and can be used for the treatment of complex infections involving resistant bacteria (Lei et al., 2019); such is the case with Telavancin and Teicoplanin which can be used either in the treatment of pseudomembranous colitis or treatment of diarrhea associated with Clostridium difficile; and the other in complicated skin infections, blood infections, endocarditis, bone and joint infections, and meningitis caused by methicillin-resistant S. aureus (Lei et al., 2019). The same could be said for daptomycin which was marketed in 2003 as an anionic AMP for the treatment of skin infections caused by Gram-positive bacteria and has shown inhibitory effects on Salmonellae highly resistant to classical antibiotics and on S. aureus (Lei et al., 2019).
 | Table 1. Summary of the advantages and disadvantages of the therapeutic use of bacteriophages from Mireille et al. (2020) with several modifications. [Click here to view] |
Combinations of AMP–AMP and AMP–Antibiotics
AMPs have shown their effectiveness on biofilms. This is the case of gH625, its analog gH625-GCGKKKK, and the synergy of cyclic polymyxin B and gramicidin S AMPs which are active on P. aeruginosa biofilms (Ron?evi? et al., 2019). In the same sense, the combination of different substances, Geitani et al. (2019) demonstrated an interesting antibacterial activity of the AMPs–antibiotic association. Indeed, human cathelicidin (LL-37) and cecropin (1–7)-melittin A (2–9) amide in combination with the antibiotics colistin and imipenem has proven to be effective against antibiotic-sensitive or resistant S. aureus and P. aeruginosa. Moreover, their combination considerably reduces the minimum inhibitory concentration (MIC) of imipenem and colistin and reduces their toxicity (Geitani et al., 2019).
AMP and brain infections
A highly desirable feature in the treatment of infectious agents is the ability of the antimicrobial to be able to cross the blood–brain barrier. To this end, several AMPs, such as oncocin, apidecin (137), and drosocin (Pro5Hyp), can cross the blood–brain barrier to selectively target brain cells. These AMPs could be potential therapies for brain infections (Li et al., 2014). Concisely, considering all the advantages they present, AMP constitute a major alternative to fight against antibiotic resistance. However, although they are a serious option to replace antibiotics, they also have many drawbacks that limit their production and use. This is linked to their MIC which is often too high and can constitute a factor of toxicity (Raheem and Straus, 2019). Thus, it would be more judicious to use them: on resistant bacteria on which they are active; for infections where their MIC is not very high; in combination with antibiotics to overcome resistance and limit the use of conventional antibiotics.
Use of Vaccines to Reduce the Use of Antibiotics
Since the discovery of the first vaccine in the 1790s, many other vaccines have been developed and have since played a major role in the prevention of various diseases. Vaccines are defined as preparations based on microorganisms with attenuated virulence or on their constituents, which the administration aims to protect the host from the disease caused by the pathogen involved (Rappuoli et al., 2014). The best-known antibacterial vaccines are those for diphtheria (caused by Corynebacterium diphtheriae), cholera (caused by Vibrio cholerae), tetanus (caused by Clostridium tetani), and pertussis (caused by Bordetella pertussis). In some countries, the oral Ty21a vaccine to prevent infections with Salmonella typhi (the causative agent of typhoid) is widely used. Certain vaccines can be combined, and this makes it possible to protect the host against several infectious agents at the same time; this is the case with the four-valent combination vaccines marketed under the names Infanrix Tetra®, Tetravac acellulaire®, Repevax®, and Boostrix Tetra®, which simultaneously prevent diphtheria, tetanus, pertussis, and poliomyelitis. The introduction of vaccines against resistant pathogenic bacteria could constitute a sufficient immunological barrier allowing the host to fight the incriminated bacteria without resorting to antibiotics (Terlizzi et al., 2017). However, setting up a vaccine against a specific bacterial germ requires the presence of one or more factors likely to trigger an immune response (Terlizzi et al., 2017). These factors typically include adhesins, iron receptors, secreted toxins, capsules (and their K antigens), capsule flagella (and their H antigens), outer membrane proteins, and lipopolysaccharides which may be transferred by plasmids, transposons, bacteriophages, and islets of pathogenicity (Terlizzi et al., 2017). Numerous studies have shown that the use of vaccines in certain pathologies of bacterial origin can help reduce the use of antibiotics (Bloom et al., 2018; Klugman and Black, 2018; Kobayashi et al., 2016; Rappuoli et al., 2014) and some have demonstrated the effectiveness of vaccines in the fight against antibiotic resistance (Klugman and Black, 2018; Ouldali et al., 2019). The most recent example is the introduction of pneumococcal conjugate vaccines, which has led to a significant drop in the proportion of resistant pneumococcal strains. Unfortunately, due to the excessive consumption of antibiotics, some of the nonvaccine serotypes have also become resistant to antibiotics, raising (still moderately) the level of resistance (Ouldali et al., 2019). In addition, a team of researchers from Johns Hopkins University have demonstrated the effectiveness in vitro (on macaques) of a vaccine against S. aureus (the major cause of nosocomial infections) whose treatment is generally complicated due to the resistance of this germ to antibiotics (Kuklin et al., 2006). Ultimately, despite the difficulties presented by this alternative, it can constitute significant protective means against the bacterial strains for which vaccines exist and consequently limit the use of antibiotics in a possible infection that can be caused by these pathogens.
Use of Cytokines and Antibodies as an Alternative to Antibiotics
Cytokines are cell signaling glycoproteins, expressed by cells of the immune system, and antibodies are glycoproteins capable of specifically detecting and neutralizing pathogens (Larochette et al., 2019). With the growth of antibiotic resistance, several researchers have proposed antimicrobial antibodies and cytokines as possible alternatives to reduce the use of conventional antibiotics (Holland, 2000). The ability of antibodies and cytokines to fight against various bacteria in vitro (Bebbington and Yarranton, 2009; Conti et al., 2000) and in vivo (Hancock et al., 2012; Morita et al., 2021; Rumfield et al., 2020) has been demonstrated. Some studies have proposed antibody–antibiotic combinations to overcome antibiotic resistance and to cure serious bacterial infections (Song et al., 2012; Sawa et al., 2014). Furthermore, certain monoclonal antibodies such as pagibaximab (BSYX-A110) (chimeric monoclonal antibody directed against lipoteichoic acid, a key component of the wall of Gram+ bacteria) have been set up to overcome certain disadvantages of antibiotics in vulnerable people and fragile individuals such as premature babies (Hancock et al., 2012). Finally, although antibodies cannot completely replace antibiotics, they can nevertheless help to reduce their use and fight against antibiotic resistance.
Phytochemicals and Plant Extracts as Alternatives to Antibiotics
Herbal medicine refers to the use of plants for therapeutic purposes. Depending on the case, the whole plant or parts of it are used, including the roots, stems, leaves, flowers, and fruits. To exploit their properties in various diseases, they generally proceed by extraction methods such as infusions, maceration, decoctions, or even extractions with solvents such as ethanol, methanol, and acetone. Plants with high antibacterial potential can contribute significantly to the diversification of antibacterials and fight against antibiotic resistance either by using them as they are or by using them in synergy with conventional antibiotics to thwart bacteria that are already resistant (Arsène et al., 2021; Mbarga et al., 2021; Nascimento et al., 2000).
Antibacterial properties of plant extracts and phytochemicals
Between 2015 and 2019, around 11,000 studies were published on the antibacterial properties of plants across the world (Source: PubMed). These studies, whether in vitro or in vivo, have demonstrated the antibacterial properties of many plants on various bacteria (Nascimento et al., 2000; Alrozeky and Nakahara, 2002; Arsène et al., 2021; Mbarga et al., 2021). Among these varied plants, certain spices and edible plants have also proved to be antibacterial (Alrozeky and Nakahara, 2002; Fankam et al., 2011). In a study conducted in China in 2002, several edible Asian plants demonstrated antibacterial activity on E. coli, Salmonella infantis, L. monocytogenes, S. aureus, and Bacillus cereus (Alrozeky and Nakahara, 2002). In Cameroon, a study involving spices used in cooking showed that 15 of the 20 plants used had an antibacterial effect on Mycobacterium tuberculosis H37Rv and H37Ra and extract of Echinops giganteus exhibited the most significant activity with a MIC value of 32 and 16 μg/ml (Fankam et al., 2011). The list of studies having obtained significant results with plant extracts cannot be exhaustive here but more detailed information on this subject can be found in our previous review (Mbarga et al., 2021). However, it is necessary to emphasize that most of the studies carried out on the antibacterial properties of plant extracts were carried out in vitro. Such results cannot be transposed directly to a possible in vivo application without additional studies. Notwithstanding the above, given the use of certain plant extracts in traditional medicine as a treatment of infections of the urinary tract (Ibrahim et al., 2015) and diarrheal infections (Ullah et al., 2016) among others, it clearly appears that plant extracts can play a major role in reducing the use of antibiotics.
Synergy between common antibiotics and phytochemical compounds to counteract antibiotic resistance
In most of the in vitro studies carried out on the antibacterial properties of plant extracts or phytochemical compounds, a part is reserved for the study of the effect of their combination with common antibiotics on multiresistant bacteria. This is, for example, the case with the study conducted by Nascimento et al. (2000) which demonstrated the synergistic effect of the combination of antibiotics with extracts of clove, jambolan, pomegranate, and thyme on P. aeruginosa (resistant to 19 antibiotics) and K. pneumoniae. Likewise, the combination of anacardic acid and totarol with methicillin has shown positive results in inhibiting methicillin-resistant strains of S. aureus (Nascimento et al., 2000). The mechanism of action of this synergistic action would involve multidrug efflux pumps (EPs) located on the cytoplasmic membrane of bacteria. Indeed, plants are an important reservoir of bioactive compounds which can serve as a source of potent EP inhibitors (EPIs) (Shriram et al., 2018). One study published in 2020 demonstrated that flavonoids could serve as EPIs and antimicrobials against both environmental and pathogenic intracellular mycobacterial species (Solnier et al., 2020). In the same vein, the results obtained by Tran et al. (2020) demonstrated that carvotacetones from Sphaeranthus africanus could have an antibacterial activity and play an EPI role against Mycobacteria. Briefly, the antibacterial properties of several plant extracts and isolated phytochemical compounds are undeniable but more in vivo studies should be carried out to determine the optimal conditions under which these preparations could be safely used as such or in combination with conventional antibiotics.
Use of NPs in Synergy with Conventional Antibiotics
NPs are a wide class of materials that include particulate substances, which have one dimension less than 100 nm at least (Laurent et al., 2010). NPs are considered a feasible alternative to antibiotics, especially with the emergence of bacterial multidrug resistance, since it has been discovered that they do not lead to bacterial resistance (Rai et al., 2012). As shown in Table 3, several recent investigations have used different methods to synthesize various NPs and these NPs have shown very encouraging antimicrobial properties with very low minimum inhibitory and bactericidal concentrations. NPs have a high surface area to volume ratio and unique chemical and physical properties, which makes them promising antimicrobial agents (Crisan et al., 2021; Yin et al., 2020). It has been observed that NPs target multiple cellular pathways at once (Crisan et al., 2021; Li et al., 2012), which makes it extremely difficult for bacteria to develop resistance against them (Rai et al., 2014). NPs with size less than 20 nm can kill bacteria by penetrating the cell wall and destruct cell organelles (Arakha et al., 2015; Crisan et al., 2021) and can hamper the synthesis of nucleic acids in several microorganisms (Fayaz et al., 2010). In recent years, silver nanoparticles (AgNPs) were considered as a possible new class of antimicrobial (Arsène et al., 2021; Crisan et al., 2021; Rai et al., 2014). Several studies have shown that AgNPs can enhance the effect of antibiotics against susceptible and resistant bacteria (Birla et al., 2009). Combinations between antibiotics and NPs have also shown efficiency against multiresistant bacteria. Indeed, in a study conducted by Mala et al. (2016) where they impregnated urinary catheters with a synergistic combination of antibiotics and AgNPs to evaluate antibiofilm activity in vitro and in vivo, the authors reported that the synergistic combination showed a 90% inhibition of bacterial adhesion and concluded that a synergistic combination of antibiotics and AgNPs is an efficient method for preventing biofilm formation (Mala et al., 2016). Besides AgNPs, copper nanoparticles (CuNPs) also demonstrated high antibacterial and antifungal effects (Amer and Awwad, 2021; Ramzan et al., 2021). Examples showing the antibacterial activity of other NPs are presented in Table 3. Although NPs have proven to be a great substitute for conventional antibiotics, they can be used not only to treat antibiotic-resistant bacterial infections but also to avoid the potential for resistance development. However, research should no longer stop at the in vitro aspect but should propose formulations likely to be applied to treat bacterial infections.
 | Table 2. Some antimicrobial peptide drugs approved by the FDA. [Click here to view] |
 | Table 3. Some recent research on the green synthesis of NPs and their antibacterial activity. [Click here to view] |
 | Figure 2. Mechanism of protection of probiotics against infections (Ghosh et al., 2019 ). [Click here to view] |
Probiotics as a Potential New Antibacterial Strategy
Probiotics are living microorganisms (bacteria or yeasts), which when consumed in adequate quantities produce a beneficial effect on the health of the host beyond the traditional nutritional effects (WHO and FAO, 2002). The main strains recognized as probiotics in humans are bacteria belonging to the genera Lactobacillus, Bifidobacterium, Enterococcus, and Streptococcus and yeasts of the genus Saccharomyces (Ayivi et al., 2020; Joseph et al., 2021). As described in Figure 2 (Ghosh et al., 2019; Markowiak and ?li?ewska, 2017), it has been shown that certain probiotics are able to inhibit the growth of pathogenic bacteria by specific and nonspecific competition for adhesion to epithelial cells (Bajaj et al., 2021), by the production of antimicrobial substances (Arsene et al., 2021; Silva et al., 2020), by competition in the use of nutrients (Abd El-Hack et al., 2020), and by the stimulation of immune defense mechanisms (Abd El-Hack et al., 2020). With the resurgence of the resistance to antibiotics issue, the antagonist and immunomodulatory properties of probiotics are today the most used (Arsene et al., 2021; Manga et al., 2019; Silva et al., 2020). Thanks to these properties, probiotics could constitute a credible alternative to antibiotics in the fight against specific bacterial infections for which their effectiveness has already been proven. Our previous review comprehensively presented the main probiotics that can effectively replace antibiotics in breeding (Arsène et al., 2021). Otherwise, several in vitro and in vivo studies, both in clinical medicine and in veterinary medicine have demonstrated the efficiency of probiotics against specific germs (Abd El-Hack et al., 2020; Bajaj et al., 2021; Huang et al., 2014; Homan and Orel 2015; Makras, 2006). For example, the study conducted by Makras (2006) demonstrated the antagonist activity of six strains of Lactobacilli (L. acidophilus, L. amylovorus, L. casei, L. johnsonii, L. plantarum, and L. rhamnosus) on diarrhea caused by Salmonella enterica. In addition, certain combinations of probiotics with each other or with antibiotics have also demonstrated their effectiveness (Homan and Orel, 2015). In the search for new antibacterial control strategies and the reduction of the use of antibiotics, probiotics have various advantages such as the possibility of being included in food such as yogurts, cheeses, dietary supplements, and fermented drinks (Mbarga et al., 2019), their immunomodulatory capacity (Silva et al., 2020), and their ability to cleanse and balance the intestinal flora (Wang et al., 2020). However, although they have positive effects on certain enteric infectious pathologies, probiotics have various drawbacks such as their ineffectiveness on major bacterial infections affecting other organs such as the skin, heart, or brain.
CONCLUSION
All the methods discussed in this review can help to reduce the use of antibiotics, but none alone can replace them. Our analysis of the situation shows that the development of vaccines can effectively contribute to reducing the use of antibiotics by strengthening the immune system in order to protect it against specific pathogens as is already the case with bacteria such as Neisseria meningitis, Haemophilus influenzae, Streptococcus pneumoniae, M. tuberculosis, C. diphtheriae, and C. tetani. In addition, probiotics, thanks to their multiple barrier effects on the intestinal flora, their antagonist effect, and their immunomodulation capacity of the immune system, can be a significant contribution in reducing the use of antibiotics. As for phages, cytokines, and antimicrobial antibodies, given their narrow specificity, they appear to be pragmatic solutions to drastically limit the use of antibiotics in the treatment of diseases caused by bacteria on which their effect is recognized. However, phages have some drawbacks such as the release of enterotoxin and other metabolites during lysis, the possible involvement of humoral immunity with the production of neutralizing antibodies, their variable stability over time influenced by storage conditions (pH, temperature, UV, etc.), and their potential capacity to induce horizontal transfer of genes by generalized transduction (virulence factor; resistance to antibiotics). Although the antibacterial efficiency of several plants has been demonstrated, there is an eternal dilemma as to the isolation of active molecules because the totum of preparations based on these plants would be more bioavailable and more effective than isolated phytochemicals and their potential ability to induce horizontal gene transfer by generalized transduction (virulence factor; antibiotic resistance). In addition, the green synthesis of NPs (silver, gold, zinc, copper, iron, etc.) is also an important option against the scourge of antibioresistance and the combination of common antibiotics with other antibacterial molecules (phytochemicals, NPs, and AMP) can give a second life or improve the effectiveness of certain antibiotics which are losing their effectiveness. Finally, research should continue to be carried out to effectively exploit all the alternatives presented in this study to face this major public health issue of antibiotic resistance.
ACKNOWLEDGMENT
This publication has been supported by the RUDN University Strategic Academic Leadership Program.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
Not Applicable
REFERENCES
Abd El-Hack ME, El-Saadony MT, Shafi ME, Qattan SY, Batiha GE, Khafaga AF, Abdel-Moneim AM, Alagawany M. Probiotics in poultry feed: a comprehensive review. J Anim Physiol Anim Nutr (Berl), 2020; 104(6):1835–50. CrossRef
Akintelu SA, Yao B, Folorunso AS. Green synthesis, characterization, and antibacterial investigation of synthesized gold nanoparticles (AuNPs) from Garcinia kola pulp extract. Plasmonics, 2021; 16(1): 157–65. CrossRef
Alzoreky NS, Nakahara K. Antibacterial activity of extracts from some edible plants commonly consumed in Asia. Int J Food Microbiol, 2002; 80:223–30. CrossRef
Andersson DI, Hughes D, Kubicek-Sutherland JZ. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist Updat, 2016; 26:43–57. CrossRef
Antoinette KD, Dieudonne A. Etiologic profile and sensitivity pattern of germs responsible for urinary tract infection among underfive in Douala: a hospital-based study. Int J Infect Dis, 2020; 101:358. CrossRef
Amer MW, Awwad AM. Green synthesis of copper nanoparticles by Citrus limon fruits extract, characterization and antibacterial activity. Chem Int, 2021; 7:1–8.
Arakha M, Pal S, Samantarrai D, Panigrahi TK, Mallick BC, Pramanik K, Mallick B, Jha S. Antimicrobial activity of iron oxide nanoparticle upon modulation of nanoparticle-bacteria interface. Sci Rep 2005; 5:14813 CrossRef
Arsène MM, Podoprigora IV, Davares AK, Razan M, Das MS, Senyagin AN. Antibacterial activity of grapefruit peel extracts and green-synthesized silver nanoparticles. Vet World, 2021; 14(5):1330–41. CrossRef
Arsene MM, Zangue DS, Ngoune TL, Nyasha K, Louis K. Antagonistic effects of raffia sap with probiotics against pathogenic microorganisms. Foods Raw Mater, 2021; 9(1):24–31. CrossRef
Ayivi RD, Gyawali R, Krastanov A, Aljaloud SO, Worku M, Tahergorabi R, Silva RC, Ibrahim SA. Lactic acid bacteria: food safety and human health applications. Dairy, 2020; 1(3):202–32. CrossRef
Bajaj BK, Claes IJ, Lebeer S. Functional mechanisms of probiotics. J Microbiol Biotechnol Food Sci, 2021; 2021:321–7. CrossRef
Baltzer SA, Brown MH. Antimicrobial peptides?: promising alternatives to conventional antibiotics. J Mol Microbiol Biotechnol, 2011; 20(4):228–35. CrossRef
Bebbington C, Yarranton G. Antibodies for the treatment of bacterial infections: current experience and future prospects. Curr Opin Biotechnol, 2008; 19:613–9. CrossRef
Bhuiyan MS, Miah MY, Paul SC, Aka TD, Saha O, Rahaman MM, Sharif MJ, Habiba O, Ashaduzzaman M. Green synthesis of iron oxide nanoparticle using Carica papaya leaf extract: application for photocatalytic degradation of remazol yellow RR dye and antibacterial activity. Heliyon, 2020; 6(8):e04603. CrossRef
Birla SS, Tiwari VV, Gade AK, Ingle AP, Yadav AP, Rai MK. Fabrication of silver nanoparticles by Phoma glomerata and its combined effect against Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus. Lett Appl Microbiol, 2009; 48:173–9. CrossRef
Bloom DE, Black S, Salisbury D, Rappuoli R. Antimicrobial resistance and the role of vaccines. Proc Natl Acad Sci USA, 2018; 115:12868–71. CrossRef
Biacchesi S, Christophe C, Marie G, Christelle L, Ronan LG, Michel B. Les virus : ennemis ou alliés ? Enjeux Sciences, Versailles (France), 112 p, 2017. CrossRef
Bruttin A, Brussow H. Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob Agents Che-mother, 2005; 49(7):2874–8. CrossRef
Cairns BJ, Timms AR, Jansen VA, Connerton IF, Payne RJ. Quantitative models of in vitro bacteriophage-host dynamics and their application to phage therapy. PLoS Pathog, 2009; 5(1):e1000253.25. CrossRef
Cittrarasu V, Kaliannan D, Dharman K, Maluventhen V, Easwaran M, Liu WC, Balasubramanian B, Arumugam M. Green synthesis of selenium nanoparticles mediated from Ceropegia bulbosa Roxb extract and its cytotoxicity, antimicrobial, mosquitocidal and photocatalytic activities. Sci Rep, 2021; 11(1):1–15. CrossRef
Cleven BE, Palka-Santini M, Gielen J, Meembor S, Kro?nke M, Krut O. Identification and characterization of bacterial pathogens causing bloodstream infections by DNA microarray. J Clin Microbiol, 2006; 44(7):2389. CrossRef
Crisan CM, Mocan T, Manolea M, Lasca LI, T?b?ran FA, Mocan L. Review on silver nanoparticles as a novel class of antibacterial solutions. Appl Sci, 2021; 11(3):1120. CrossRef
Criscuolo E, Spadini S, Lammanna J, Ferro M, Burioni R. Bacteriophages and their immunological applications against infectious threats. J Immunol, 2017; 2017:3780697. CrossRef
Conti S, Magliani W, Arseni S, Dieci E, Frazzi R, Salati A, Varaldo PE, Polonelli L. In vitro activity of monoclonal and recombinant yeast killer toxin-like antibodies against antibiotic-resistant gram-positive cocci. Mol Med, 2000; 6:613–9. CrossRef
Doidge C, Ruston A, Lovatt F, Hudson C, King L, Kaler J. Farmers’ perceptions of preventing antibiotic resistance on sheep and beef farms: risk, responsibility and action. Front Vet Sci, 2020; 7:524. CrossRef
Dublanchet A, Maxime S. Autobiographie de Fe?lix d’He?relle 1873-1949. Editions Me?dicales Internationales, Paris, France, 2017.
Fankam A, Kuete V, Voukeng I, Kuiate J. Antibacterial activities of selected Cameroonian spices and their synergistic effects with antibiotics against multidrug-resistant phenotypes. BMC Complement Altern Med, 2011; 11(1):104. CrossRef
Faust JE, Yang PY, Huang HW. Action of antimicrobial peptides on bacterial and lipid Membranes?: a direct comparison. Biophys J, 2017; 112(8):1663–72. CrossRef
Fayaz AM, Balaji K, Girilal M, Yadav R, Kalaichelvam PT, Venketesan R. Biogenic synthesis of silver nanoparticles and their synergistic effect with antibiotics: a study against gram-positive and gram-negative bacteria. Nanomed. Nanotechnol. Biol Med, 2010; 6(1):103–9. CrossRef
Geitani R, Moubareck CA, Touqui L, Sarkis DK. Cationic antimicrobial peptides?: alternatives and/or adjuvants to antibiotics active against methicillin-resistant Staphylococcus aureus and multidrug-resistant Pseudomonas aeruginosa. BMC Microbiol, 2019; 19(1):54. CrossRef
Ghosh C, Sarkar P, Issa R, Haldar J. Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol, 2019; 27(4):323–38. CrossRef
Grandclaudon C, Birudukota NS, Elgaher WA, Jumde RP, Yahiaoui S, Arisetti N, Hennessen F, Huettel S, Stadler M, Herrmann J, Miethke M. Semisynthesis and biological evaluation of amidochelocardin derivatives as broad-spectrum antibiotics. Eur J Med Chem, 2020; 188:112005. CrossRef
Gudimalla A, Jose J, Varghese RJ, Thomas S. Green synthesis of silver nanoparticles using Nymphae odorata extract incorporated films and antimicrobial activity. J Polym Environ, 2021; 29(5):1412–23. CrossRef
Hancock RE, Nijnik A, Philpott DJ. Modulating immunity as a therapy for bacterial infections. Nat Rev Microbiol, 2012; 10:243–54. CrossRef
Hamilton R. Tarascon pocket pharmacopoeia 2015 deluxe lab-coat edition. Jones & Bartlett Learning, Burlington, MA, 91 p, 2015. ISBN 9781284057560.
Heesterbeek DA, Muts RM, van Hensbergen VP, de Saint Aulaire P, Wennekes T, Bardoel BW, van Sorge NM, Rooijakkers SH. Outer membrane permeabilization by the membrane attack complex sensitizes Gram-negative bacteria to antimicrobial proteins in serum and phagocytes. PLoS Pathog, 2021; 17(1):e1009227. CrossRef
Higgins DL, Chang R, Debabov DV, Leung J, Wu T, Krause KM, Sandvik E, Hubbard JM, Kaniga K, Schmidt Jr DE, Gao Q. Telavancin, a multifunctional lipoglycopeptide, disrupts both cell wall synthesis and cell membrane integrity in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Ch, 2005; 49(3):1127–34. CrossRef
Holland SM. Cytokine therapy of mycobacterial infections. Adv Intern Med, 2000; 45:431–52.
Homan M, Orel R. Are probiotics useful in Helicobacter pylori eradication? World J Gastroenterol, 2015; 21(37):10644–53. CrossRef
Huang YF, Liu PY, Chen YY, Nong BR, Huang IF, Hsieh KS, Chen KT. Three-combination probiotics therapy in children with salmonella and rotavirus gastroenteritis. J Clin Gastroenterol, 2014; 48(1):37–42. CrossRef
Ibrahim OMS, Sarhan SR, Hameed AA. In vivo and in vitro antibacterial activities of cranberry extract against E. coli o157: H7 in urinary tract infected rats. Adv Anim Vet Sci, 2015; 3:233–44. CrossRef
Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev, 2006; 19(3):491. CrossRef
Jit M, Ng DH, Luangasanatip N, Sandmann F, Atkins KE, Robotham JV, Pouwels KB. Quantifying the economic cost of antibiotic resistance and the impact of related interventions: rapid methodological review, conceptual framework and recommendations for future studies. BMC Med, 2020; 18(1):1–14. CrossRef
Jorge AMS, Eduardo R. Environmental structure drives resistance to phages and antibiotics during phage therapy and to invading lysogens during colonisation. Sci Rep, 2019; 28;9(1):3149. CrossRef
Joseph AMM, Davares AK, Andreevna SL, Vladimirovich EA, Carime BZ, Marouf R, Khelifi I. The use of probiotics in animal feeding for safe production and as potential alternatives to antibiotics. Vet World, 2021; 14(2):319–28. CrossRef
Joshi NC, Siddiqui F, Salman M, Singh A. Antibacterial activity, characterizations, and biological synthesis of manganese oxide nanoparticles using the extract of Aloe vera. Asian Pac J Health Sci, 2020; 7(3):27–9. CrossRef
Jun K. Phage as a therapeutic agent, 2017. Available via https://thryve.medium.com/phage-as-a-therapeutic-agent-ed4c466302e5 (Accessed 22 December 2020).
Khan MI, Akhtar MN, Ashraf N, Najeeb J, Munir H, Awan TI, Tahir MB, Kabli MR. Green synthesis of magnesium oxide nanoparticles using Dalbergia sissoo extract for photocatalytic activity and antibacterial efficacy. Appl Nanosci, 2020; 10(7):2351–64. CrossRef
Klugman KP, Black S. Impact of existing vaccines in reducing antibiotic resistance: primary and secondary effects. Proc Natl Acad Sci USA, 2018; 115:12896–901. CrossRef
Kobayashi, M, Vekemans J, Baker CJ, Ratner AJ, Le Doare K, Schrag SJ. Group B Streptococcus vaccine development: present status and future considerations, with emphasis on perspectives for low and middle income countries. F1000 Res, 2016; 5:2355. CrossRef
Kobras CM, Piepenbreier H, Emenegger J, Sim A, Fritz G, Gebhard S. BceAB-type antibiotic resistance transporters appear to act by target protection of cell wall synthesis. Antimicrob Agents Ch, 2020; 64(3):e02241–19. CrossRef
Koskella B, Meaden S. Understanding bacteriophage specificity in natural microbial communities. Viruses, 2013; 5:806–23. CrossRef
Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, McNeely T, Noble L, Brown MJ, Zorman JK, Wang XM, Pancari G. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun, 2006; 74:2215–23. CrossRef
Larochette V, Miot C, Poli C, Beaumont E, Roingeard P, Fickenscher H, Jeannin P, Delneste Y. IL-26, a cytokine with roles in extracellular DNA-induced inflammation and microbial defense. Front Immunol, 2019; 10:204. CrossRef
Laurent S, Forge D, Port M, Roch A, Robic C, Vander Elst L, Muller RN. Magnetic iron oxide nanoparticles: synthesis, stabilization, vectorization, physicochemical characterizations, and biological applications. Chem Rev, 2010; 110:2574. CrossRef
Lei J, Sun L, Huang S, Zhu C, Li P, He J, Mackey V, Coy DH, He Q. The antimicrobial peptides and their potential clinical applications. Am J Transl Res, 2019; 11(7):3919–31.
Lewis K. The science of antibiotic discovery. Cell, 2020; 181(1):29–45. CrossRef
Li W, Tailhades J, O’Brien-Simpson NM, Separovic F, Otvos L, Hossain MA, Wade JD. Proline-rich antimicrobial peptides?: potential therapeutics against antibiotic-resistant bacteria. Amino Acids, 2014; 46(10):2287–94. CrossRef
Lima T, Domingues S, Da Silva GJ. Manure as a potential hotspot for antibiotic resistance dissemination by horizontal gene transfer events. Vet Sci, 2020; 7(3):110. CrossRef
Li Y, Zhang W, Niu J, Chen Y. Mechanism of photogenerated reactive oxygen species and correlation with the antibacterial properties of engineered metal-oxide nanoparticles. ACS Nano, 2012; 6:5164–73. CrossRef
Mahlapuu M, Håkansson J, Ringstad L, Björn C. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol, 2016; 6:194. CrossRef
Makras L, Triantafyllou V, Fayol-Messaoudi D, Adriany T, Zoumpopoulou G, Tsakalidou E, Servin A, De Vuyst L. Kinetic analysis of the antibacterial activity of probiotic Lactobacilli towards Salmonella enterica serovar Typhimurium reveals a role for lactic acid and other inhibitory compounds. Res Microbiol, 2006; 157:241–7. CrossRef
Markowiak P, ?li?ewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients, 2017; 9(9):1021. CrossRef
Mala R, Aglin AA, Celsia AS, Geerthika S, Kiruthika N, VazagaPriya C, Kumar KS. Foley catheters functionalised with a synergistic combination of antibiotics and silver nanoparticles resist biofilm formation. IET Nanobiotechnol, 2017, 11:612–20. CrossRef
Manga JA, Zangué SC, Zargar M, Albert E, Bayat M. Producing probiotic beverage based on raffia sap fermented by Lactobacillus fermentum and Bifidobacterium bifidum. Res Crops, 2019; 20(3):629–34. CrossRef
Mbarga MJA, Andreevna SL, Viktorovna PI. Evaluation of apparent microflora and study of antibiotic resistance of coliforms isolated from the shells of poultry eggs in Moscow-Russia. J Adv Microbiol, 2020; 20(4):0–77.
Mbarga MJA, Podoprigora IV, Anyutoulou KLD, Ngah Esther, Senyagin AN. Urinary tract infections (UTIs): virulence factors, resistance to antibiotics and management of uropathogenic (UP) bacteria with medicinal plants —a review. J Appl Pharm Sci, 2021; 11(07):001–12.
Mbarga MJA, Zangué SCD, Tatsadjieu LN, Zargar M, Albert E, Bayat M. Producing probiotic beverage based on raffia sap fermented by Lactobacillus fermentum and Bifidobacterium bifidum. Res Crop, 2019; 20(3):629–34. CrossRef
Mi?dzybrodzki R, Borysowski J, Weber-D?browska B, Fortuna W, Letkiewicz S, Szufnarowski K, Pawe?czyk Z, Rogó? P, K?ak M, Wojtasik E, Górski A. Clinical aspects of phage therapy. Adv Virus Res, 2012; 83:73–121. CrossRef
Mireille A, Pascale B, Charlotte B, Laurent D, Nicolas D, Rémy F, Sylvain G, Claire LH, Marie-Agnès P, Eduardo R, Clara Torres-B. Les applications antibactériennes des bactériophages. Virologie, 2020; 24(1):23–36.
Morel CM, Lindahl O, Harbarth S, de Kraker ME, Edwards S, Hollis A. Industry incentives and antibiotic resistance: an introduction to the antibiotic susceptibility bonus. J Antibiot, 2020; 73(7):421–8. CrossRef
Morita Y, Masters EA, Schwarz EM, Muthukrishnan G. Interleukin-27 and its diverse effects on bacterial infections. Front Immunol, 2021; 12:1752. CrossRef
Nascimento GGF, Locatelli J, Freitas PC, Silva Gl. Antibacterial activity of plant extracts and phytochemicals on antibiotic-resistant bacteria. Braz J Microbiol, 2000; 31:247–56. CrossRef
Naseer M, Ramadan R, Xing J, Samak NA. Facile green synthesis of copper oxide nanoparticles for the eradication of multidrug resistant Klebsiella pneumonia and Helicobacter pylori biofilms. Int Biodeter Biodegr, 2021; 159:105201. CrossRef
Nawaz M, Tahir MB, Iqbal T, Pervaiz M, Rafique M, Aziz F, Younas U, Alrobei H. Synthesis, characterization and antibacterial activity of NiO NPs against pathogen. Inorg Chem Commun, 2020; 122:108300. CrossRef
Nobrega FL, Costa AR, Kluskens LD, Azeredo J. Revisiting phage therapy: new applications for old resources. Trends Microbiol, 2015; 23:185–91. CrossRef
Ouldali N, Cohen R, Levy C, Gelbert-Baudino N, Seror E, Corrard F, Vie Le Sage F, Michot AS, Romain O, Bechet S, Bonacorsi S. Pneumococcal susceptibility to antibiotics in carriage: a 17 year time series analysis of the adaptive evolution of non-vaccine emerging serotypes to a new selective pressure environment. J Antimicrob Chemother, 2019; 74:3077–86. CrossRef
Pachaiappan R, Rajendran S, Ramalingam G, Vo DV, Priya PM, Soto-Moscoso M. Green synthesis of zinc oxide nanoparticles by Justicia Adhatoda Leaf and its antimicrobial activity. Chem Eng Technol, 2021; 44(3):551–8. CrossRef
Palma E, Tilocca B, Roncada P. Antimicrobial resistance in veterinary medicine: an overview. Int J Mol Sci, 2020; 21(6):1914. CrossRef
Panda MK, Dhal NK, Kumar M, Mishra PM, Behera RK. Green synthesis of silver nanoparticles and its potential effect on phytopathogens. Mater Today Proc, 2021; 35:233–8. CrossRef
Payne RJ, Jansen VA. Understanding bacteriophage therapy as adensity-dependent kinetic process. J Theor Biol, 2001; 208(1):37–48. CrossRef
Pollock J, Low AS, McHugh RE, Muwonge A, Stevens MP, Corbishley A, Gally DL. Alternatives to antibiotics in a one health context and the role genomics can play in reducing antimicrobial use. Clin Microbiol Infect, 2020; 26(12):1617–21. CrossRef
Prasanth M, Ramesh N, Bruno SL. The therapeutic potential of bacteriophages targeting gram-negative bacteria using Galleria mellonella infection model. BMC Microbiol, 2018; 18:97. CrossRef
Rabello RF, Bonelli RR, Penna BA, Albuquerque JP, Souza RM, Cerqueira AM. Antimicrobial resistance in farm animals in Brazil: an update overview. Animals, 2020; 10(4):552. CrossRef
Raheem N, Straus SK. Mechanisms of action for antimicrobial peptides with antibacterial and antibiofilm functions. Front Microbiol, 2019; 10:2866.
Rai MK, Deshmukh SD, Ingle AP, Gade AK. Silver nanoparticles: the powerful nanoweapon against multidrug-resistant bacteria. J Appl Microbiol, 2012; 112:841–52. CrossRef
Rai M, Kon K, Ingle A, Duran N, Galdiero S, Galdiero M. Broad-spectrum bioactivities of silver nanoparticles: the emerging trends and future prospects. Appl Microbiol Biotechnol, 2014; 98:1951–61. CrossRef
Ramzan M, Obodo RM, Mukhtar S, Ilyas SZ, Aziz F, Thovhogi N. Green synthesis of copper oxide nanoparticles using Cedrus deodara aqueous extract for antibacterial activity. Mater Today Proc, 2021; 36:576–81. CrossRef
Rappuoli R, Pizza M, Del Giudice G, De Gregorio E. Vaccines, new opportunities for a new society. Proc Natl Acad Sci, 2014; 111(34):12288–93. CrossRef
Reis AC, Kolvenbach BA, Nunes OC, Corvini PF. Biodegradation of antibiotics: the new resistance determinants–part I. New Biotechnol, 2020; 54:34–51. CrossRef
Ron?evi? T, Puizina J, Tossi A. Antimicrobial peptides as anti-infective agents in pre-post-antibiotic era? Int J Mol Sci, 2019; 20(22):5713. CrossRef
Rumfield C, Hyseni I, McBride JW, Walker DH, Fang R. Activation of ASC inflammasome driven by toll-like receptor 4 contributes to host immunity against rickettsial infection. Infect Immun, 2020; 88(4):e00886–19. CrossRef
Sadhana SS, Ram Na, Rajesh K, Shilpa DK. Reversal of antibiotic resistance by phage resistant Pseudomonas aeruginosa PA01. J Biosci Bioeng, 2018; 6(1):11–5. CrossRef
Sawa T, Ito E, Nguyen VH, Haight M. Anti-PcrV antibody strategies against virulent Pseudomonas aeruginosa. Hum Vaccin Immunother, 2014; 10(10):2843–52. CrossRef
Schaenzer AJ, Wright GD. Antibiotic resistance by enzymatic modification of antibiotic targets. Trends Mol Med, 2020; 26(8):768–82. CrossRef
Shriram V, Khare T, Bhagwat R, Shukla R, Kumar V. Inhibiting bacterial drug efflux pumps via phyto-therapeutics to combat threatening antimicrobial resistance. Front Microbiol, 2018; 9:2990. CrossRef
Silva DR, Sardi JD, de Souza Pitangui N, Roque SM, da Silva AC, Rosalen PL. Probiotics as an alternative antimicrobial therapy: current reality and future directions. J Funct Foods, 2020; 73:104080. CrossRef
Skurnik M, Strauch E. Phage therapy: facts and fiction. Int J Med Microbiol, 2006; 296(1):5–14. CrossRef
Solnier J, Martin L, Bhakta S, Bucar F. Flavonoids as novel efflux pump inhibitors and antimicrobials against both environmental and pathogenic intracellular mycobacterial species. Molecules, 2020; 25:734. CrossRef
Song Y, Baer M, Srinivasan R, Lima J, Yarranton G, Bebbington C, Lynch SV. PcrV antibody-antibiotic combination improves survival in Pseudomonas aeruginosa-infected mice. Eur J Clin Microbiol Infect Dis, 2012; 31(8):1837–45. CrossRef
Spann CT, Taylor SC, Weinberg JM. Topical antimicrobial agents in dermatology. Dis Mon, 2004; 50 (7):407–21. CrossRef
Stokes JM, Lopatkin AJ, Lobritz MA, Collins JJ. Bacterial metabolism and antibiotic efficacy. Cell Metab, 2019; 30(2):251–9. CrossRef
Summers WC. History of phage research and phage therapy in phages: their role in bacterial pathogenesis and biotechnology. ASM Press, Washington, DC, pp 3–17, 2005.
Terlizzi ME, Gribaudo G, Maffei ME. Uropathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front Microbiol, 2017; 8:1566. CrossRef
Tran HT, Solnier J, Pferschy-Wenzig EM, Kunert O, Martin L, Bhakta S, Huynh L, Le TM, Bauer R, Bucar F. Antimicrobial and efflux pump inhibitory activity of Carvotacetones from Sphaeranthus africanus against mycobacteria. Antibiotics, 2020; 9(7):390. CrossRef
Ullah N, Parveen A, Bano R, Zulfiqar I, Maryam M, Jabeen S, Liaqat A, Ahmad S. In vitro and in vivo protocols of antimicrobial bioassay of medicinal herbal extracts: a review. Asian Pac J Trop Dis, 2016; 6:660–7. CrossRef
Van Belleghem JD, Dabrowska K, Vanee M, Barr JJ, Bol-lyky PL. Interactions between bacteriophage, bacteria and the mammalian immune system. Viruses, 2018; 11(1):10. CrossRef
Vasilchenko AS, Rogozhin EA. Sub-inhibitory effects of antimicrobial peptides. Front Microbiol, 2019; 10:1160. CrossRef
Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther, 2015; 40 (4): 277–83.
Wang S, Hou Q, Guo Q, Zhang J, Sun Y, Wei H, Shen L. Isolation and characterization of a deoxynivalenol-degrading bacterium Bacillus licheniformis YB9 with the capability of modulating intestinal microbial flora of mice. Toxins, 2020; 12(3):184. CrossRef
Weld RJ, Butts C, Heinemann JA. Models of phage growth and their applicability to phage therapy. J Theor Biol, 2004; 227(1):1–11. CrossRef
WHO: World Health Organization. Antibiotic resistance. World Health Organization, Geneva, Switzerland. Available via https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance. 2020 (Accessed 31 May 2021).
WHO: World Health Organization. New report calls for urgent action to avert antimicrobial resistance crisis. World Health Organization, Geneva, Switzerland. Available via https://www.who.int/news/item/29-04-2019-new-reportcalls-for-urgent-action-to-avert-antimicrobial-resistancecrisis. 2019 (Accessed 28 January 2021).
WHO and FAO. Probiotics in food FOOD AND NUTRITION. Food and Nutrition Paper, 85, 2001.
Xu L, Zhou Z, Zhu L, Han Y, Lin Z, Feng W, Liu Y, Shuai X, Chen H. Antibiotic resistance genes and microcystins in a drinking water treatment plant. Environ Pollut, 2020; 258:113718. CrossRef
Yin IX, Zhang J, Zhao IS, Mei ML, Li Q, Chu CH. The antibacterial mechanism of silver nanoparticles and its application in dentistry. Int J Nanomedicine, 2020; 15:2555–62. CrossRef
Zhou J, Liu Y, Shen T, Chen L, Zhang C, Cai K, Liao C, Wang C. Antimicrobial activity of the antibacterial peptide PMAP-36 and itsanalogues. Microb Pathog, 2019; 136:103712. CrossRef