INTRODUCTION
Human serum albumin (HSA) is a globular protein with a molecular weight of 66.5 kDa, which consists of 585 amino acids and 17 disulfide bonds in a single polypeptide. HSA is a primary protein found in blood plasma. It functions as a carrier for endogenous and exogenous hydrophobic molecules such as fatty acids, bilirubin, and hormones. Furthermore, it also acts as a flexible carrier protein for small lipophilic drug molecules (Charter and Ho, 1994; Raoufinia et al., 2016). HSA can act as a surfactant and maintain active protein conformation during storage (Mead et al., 2007; Shahin et al., 2020; Taguchi et al., 2012).
The application of albumin in therapy and diagnosis has developed into nanosized drug carriers over the last few years. Albumin has been used as a nanodelivery drug for insulin molecules, tumor drugs, cancer therapy, rheumatoid arthritis, and infectious diseases (Chai et al., 2006; Elsadek and Kratz, 2012; Karimi et al., 2016; Kratz, 2008; Pilati and Howard, 2020). One technique for early detection of cancer is the magnetic resonance imaging (MRI) tool. Detection using MRI usually uses gadolinium-diethylenetriamine pentaacetate (Gd-DTPA) contrast agents to obtain a cross-section of the body using magnetic fields (Meade, 2004; Na et al., 2009; Pierre et al., 2014; Zou and Lu, 2013). HSA reversibly can bind to Gd-DTPA, resulting in significantly higher relaxivity and intravascular retention than the existing contrast agent used in magnetic resonance (Wu et al., 2007; Goyen, 2008; Zhen et al., 2012). The HSA-Gd complex has been widely studied. One of the contrast agents containing HSA, gadofosveset, is approved for clinical use (Caravan et al., 2002).
Conventional production of plasma HSA (pHSA) has been done through fractionation of human plasma, which is limited by the availability of blood donations and may carry a risk of viral infections, e.g., hepatitis and human immunodeficiency virus; however, pHSA manufacturing has increased rapidly in recent years (Caravan et al., 2002; Raoufinia et al., 2016; Zhen et al., 2012). The issue of availability of blood donations and the safety of using pHSA underline the importance of producing recombinant HSA (rHSA) to replace pHSA (Erstad, 1996; MacLennan and Barbara, 2006). rHSA is an alternative to obtain authentic HSA in large quantities (Chen et al., 2013; Mead et al., 2007; Nguyen et al., 2020). rHSA has been expressed successfully in various host expression systems and appears to be indistinguishable from pHSA. The physicochemical and immunochemical properties of rHSA demonstrate the same properties as pHSA (Chen et al., 2013; Ohnishi et al., 2008; Ohtani et al., 1998). rHSA has been expressed using the Escherichia coli expression system (Latta et al., 1987; Lawn et al., 1981; Nguyen et al., 2020; Sharma et al., 2017), Saccharomyces cerevisiae (Kalman et al., 1990; Okabayashi et al., 1991; Sleep et al., 1990), Pichia pastoris (Belew et al., 2008; Kobayashi et al., 2000; Prevatt and Sreekrishna, 1994; Zhu et al., 2018), and transgenic plants or animals (He et al., 2011; Moghaddassi et al., 2014; Zhang et al., 2013). Yeast appears to be an efficient host to produce rHSA with the same property as pHSA proven in preclinical and clinical trials (Kobayashi, 2006). Meanwhile, rHSA produced in a rice seed transgenic plant demonstrates great potency for more cost-effective production (Chen et al., 2013).
The expression of rHSA in E. coli produced an insoluble fraction, which may be due to the high molecular weight of HSA and the lack of a protein folding factor in E. coli to facilitate folding of HSA, which has many disulfide bonds; therefore, it requires a time-consuming and expensive renaturation process (Latta et al., 1987; Lawn et al., 1981). However, recently Nguyen et al. (2020) showed that the E. coli Origami host significantly improved the expression and solubility of rHSA fused to the maltose-binding protein tag. Around 9.46 mg rHSA with 97% purity was obtained from a 500 ml cell culture. Meanwhile, the expression of rHSA in S. cerevisiae hosts results in a low expression level, possibly due to the considerable molecular weight of rHSA. Some of the rHSA is retained in the periplasmic cavity (Okabayashi et al., 1991). The P. pastoris expression systems seem to be most suitable for the expression of rHSA that has 17 disulfide bonds, which require posttranslational modification (Zhu et al., 2018).
Pichia pastoris demonstrates a remarkably attractive host for HSA production due to several benefits of this expression system. Pichia pastoris is a widely used host for recombinant protein expression due to the fact that it is easy to manipulate, its genetic structure is stable, it has a high cell density when growing in an inexpensive basal salt medium, it has strong inducible alcohol oxidase 1 (AOX1) promoter, it has the ability to perform posttranslational modification, and it has a high capacity of secretory expression and low endogenous protein secretion (Ahmad et al., 2014; Baghban et al., 2018; Gaffar et al., 2019). Recently, Zhu et al. (2018) have reported the production of rHSA in P. pastoris GS115 hosts in a shake flask and fermenter culture yielded 1.6 and 8.86 g/l, respectively.
Secreted expression of rHSA by either P. pastoris or S. cerevisiae has always resulted in proteolytic degradation of rHSA, particularly in continuous cultures with periodic feeding (Kang et al., 2000; Kobayashi et al., 2000). Although manipulations of medium were undertaken to control the protease’s expression, the protease activity still existed in the culture broth. During fermentation, the leakage of the protease can occur in lysed cells. The protein degradation was reported caused by an endogenous acidic protease that acts at an acidic pH. Kobayashi et al. (2000) observed a degradation rate of rHSA equal to 660 mg of rHSA/l/hour at pH 4.3, which caused a significant decrease in rHSA production in yeast. Another strategy to reduce HSA degradation has been achieved by interrupting the gene encoding with an aspartyl protease [Yeast Aspartic Protease3 (YAP3)]. However, the suspicion of proteases that cause such degradation of rHSA in yeast is still unclear (Kerry-Williams et al., 1998).
In this study, we report the production of rHSA in P. pastoris protease-deficient SMD1168 extracellularly using the α-factor signal peptide. rHSA production using this host has never been reported previously. SMD1168 is a host that lacks vacuole peptidase A (pep4) which is responsible for the activation of carboxypeptidase Y and protease B1 (Gleeson et al., 1998). Protease-deficient P. pastoris is an alternative host to avoid the effects of proteolysis of target proteins by the host system. Protease-deficient strains have been shown to be effective in reducing the degradation of several foreign proteins, particularly in secreted recombinant protein in fermenter cultures, because of the combination of high cell density and lysis of a small number of cells results in relatively high concentrations of vacuolar proteases. However, these protease-deficient cells were not as strong as the wild-type strain with respect to PEP4. These strains have lower viability and slower growth rates, are more difficult to transform, and are more difficult to handle in shake flasks and fermenter cultures (White et al., 1994). The rHSA produced furthermore was conjugated to Gd-DTPA which can later be applied as a contrast agent. This study will provide information on the expression level of HSA in the P. pastoris protease-deficient host and its conjugation with Gd-DTPA that has not been previously reported.
MATERIALS AND METHODS
Strains, plasmids, and growth medium
Escherichia coli TOP10F’ was used for DNA manipulation, cloning, and sequencing procedures. Pichia pastoris SMD1168 [Histidinol dehydrogenase4 (his4), pep4] was purchased from Invitrogen Co. (Carlsbad, Germany). Plasmid pD912 was obtained from DNA2.0 (Hong Kong). The E. coli recombinants were grown at 37°C in low salt Luria-Bertani medium plates (0.5% yeast extract, 1% tryptone, 0.5% NaCl, and 1.5% agar) containing 10 μg/ml tetracycline and 25 μg/ml ZeocinTM. Pichia pastoris SMD1168 strain was cultured in minimal dextrose (MD) medium plates [20 g/l dextroses, 10 g/l ammonium sulfate, 3.4 g/l yeast nitrogen base (YNB) without amino acids and ammonium sulfate, 400 μg/l biotins, and 15 g/l agar] and yeast extract peptone dextrose (YPD) medium plates (10 g/l yeast extract, 20 g/l tryptone, 20 g/l dextroses, and 15 g/l agar) containing 100–1,000 μg/ml. The yeast recombinants were screened based on resistance to the Zeocin antibiotic. The medium used for rHSA expression was buffered minimal glycerol medium (BMGY) (10 g/l yeast extract, 20 g/l tryptone, 10 g/l ammonium sulfate, 3.4 g/l YNB without amino acids and ammonium sulfates, 100 mM potassium phosphate buffer pH 6.0, 400 μg/l biotins, and 10 g/l glycerol) and buffered minimal methanol histidine medium (BMMH) (3.4 g/l YNB, 400 μg/l biotins, 40 mg/l histidines, 100 mM potassium phosphate pH 6, and 0.75% methanol).
Reagent
Diethylaminoethyl (DEAE)-cellulose, diethylenetriamine pentaacetate (DTPA), gadolinium trichloride hexahydrate, trifluoroacetic acid (TFA 1%), DTPA anhydride, (PBS) Phosphate Buffer Saline buffer, sodium hydroxide, gadolinium chloride hexahydrate in 2 ml of sodium acetate buffer (pH 5.5), and 1% sodium chloride were purchased from Sigma Aldrich. The grade of all of the reagents was pro analysis.
Apparatus
The equipment used was as follows: High Performance Liquid Chromatography (HPLC) Shimadzu LC 10 ADVP with C-18 column, Sephadex G-25 gel filtration column (1 m × 2 cm), autoclave sterilizer (Hirayama Autoclave HVE-50), SDS-PAGE Mini-Sub Cell (Bio-Rad) and SDS-PAGE (Bio-Rad PowerPacTM Basic), magnetic stirrer (HMS-79 Magnetic Heated), freeze dryer, ice bath, conductivity meter, Millipore, MiniSpin (Eppendorf), Balance (Mettler Toledo AL204), UV-lamp λ312 nm (Viber Lourmat TCP-20M), pH meter (Mettler Toledo InLab pH combination polymer electrodes), automatic micropipette (Thermo Fisher), centrifuge (Tomy MX-305), shaking incubator (DAIHAN Scientific), UV-Vis spectrophotometer (Ultrospec 3000 Pro UV/Vis), vortex (Boeco V-1 Plus), and water bath (Poly Science).
Construction of expression vector containing HSA gene
The sequence of the native HSA gene was obtained from GenBank (ACC: AF190168.1). The sequence was optimized according to the P. pastoris preference codon, using the gcua program (www.gcua.org). The HSA gene was then synthesized by DNA2.0 (Hong Kong) and inserted after the α-factor signal sequence in the pD912 expression vector to produce a recombinant plasmid pD912-rHSA. The recombinant plasmid was propagated in the E. coli TOP10F’ and subsequently isolated by TIANprep Mini Plasmid Kit following the procedure.
Pichia pastoris transformation and screening of transformants with high expression
Briefly, 80 μl of the electrocompetent P. pastoris SMD1168 was transformed with 500 ng SacI-linearized pD912-rHSA (10 μl) by electroporation (25 μF, 200 Ω, and 1.0 kV), using a MicroPulser Electroporator (Bio-Rad, CA). The cells were mixed with 1 ml prechilled sorbitol (1 M) and incubated at 28°C for 2 hours. The transformants were subsequently selected on the MD plates after growing the cells at 28°C for 48–72 hour and followed by screening on the YPD plates containing various concentrations of Zeocin (100–1,000 μg/ml) at 28°C, up to the point of colony formation (Zhu et al., 2018). The transformants were analyzed by colony Polymerase Chain Reaction (PCR) to confirm the HSA gene’s presence in the P. pastoris genome, using universal primers for pD912. The nucleotide sequence was confirmed by the Sanger dideoxy method by using primer pairs: pD912F (5?-ACGTCGCTGTTTTGCC-3?) and pD912R (5?-CTGCATCTCTCAG GCAAAT-3?).
Expression of rHSA by P. pastoris
Pichia pastoris transformants were grown in 20 ml of BMGY at 28°C and 230 rpm, through a period before the culture reached an OD600 of 15–20 (approximately 18 hours). The cells were harvested by centrifugation at 3,000 g and 25°C for 20 minutes. These were subsequently resuspended in a 100 ml flask containing 20 ml Buffered Methanol- Complex Medium (BMMY) and allowed to grow under similar conditions as before. Furthermore, methanol p.a. was added to the flasks every 24 hours to achieve a final concentration of 0.75% (v/v), required to induce the AOX1 promoter. The P. pastoris transformed with empty vector were exposed to the same treatment and used as a negative control. Also, samples were taken every 24 hours, and then the cultures were centrifuged at 12,000 g for 5 minutes after 144 hours. The presence of rHSA in the supernatant was confirmed using SDS-PAGE carried under moderated conditions using 10% running gel and 4% stacking gel. Then 25 μl of supernatant was analyzed. The protein bands present on the gel were stained with Coomassie Brilliant Blue R250.
Optimization of rHSA expression with various inducer methanol concentrations
Pichia pastoris transformants were grown in four flasks containing 20 ml BMGY and subsequently transferred to BMMH, following the procedure stated above. The rHSA expression was optimized using variations of methanol concentrations (0.75%, 1.0%, 1.25%, and 1.5%), and samples were collected every 24 hours up to the 144th hour. These cultures were then centrifuged at 12,000 g for 5 minutes, followed by detection of rHSA in the supernatant using SDS-PAGE.
Purification of rHSA with Sephadex G-25 gel filtration chromatography
The supernatants obtained were respectively placed into the Sephadex G-25 gel filtration column (1 m × 2 cm), previously equilibrated with a 1% sodium chloride solution. Furthermore, 5 ml of each eluate was taken for up to 57 fractions. The absorptions were measured using a UV-Vis spectrophotometer at 280 nm. The conductivity was also measured. A curve of absorption and conductivity value of the fractions was prepared. The fraction with a maximum peak was characterized by SDS-PAGE.
Characterization of rHSA by RP-HPLC
About 20 μl of HSA standard was injected into the C-18 column. The column was eluted using water-acetonitrile containing 0.1% TFA as a mobile phase. The flow rate was set to 1 ml/minutes for 20 minutes, and the peak was detected using a UV detector at 215 nm. The chromatogram showed the retention time of the HSA standard. The column was washed with the mobile phase to flush the remaining impurities. After cleaning the column, the rHSA sample was injected using the same protocol as a standard injection. This was used to determine sample purity based on the chromatogram generated. The protein concentration was determined using the Lowry method (Lowry et al., 1951) by using bovine serum albumin as a standard.
Synthesis and characterization of Gd-DTPA-rHSA
Gd-DTPA-rHSA was synthesized using the protocol described by Kundu et al. (2011). About 10 mg of rHSA was dissolved in 1 ml of 0.01 M PBS (pH 7.4), followed by the direct incorporation of 10 mg DTPA anhydride powder (mol ratio of rHSA:DTPA = 1:200). The pH was maintained at 7.0 using 0.1 M NaOH, and the mixture was then stirred at 4°C for 2 hours. Subsequently, 10 mg of GdCl2.6H2O in 2 ml Acetic acid/Sodium acetic (HAc/NaAc) (pH. 5.5) was added to DTPA-rHSA and stirred slowly for 4 hours at room temperature. The purity of the Gd-DTPA-rHSA generated was determined using Reverse Phase- High Performance Liquid Chromatography (RP-HPLC). The HPLC was run using a C-18 column, and water-acetonitrile containing 0.1% TFA served as the mobile phase. The flow rate was set to 0.5 ml/minute, and the peaks were monitored using a UV detector at 215 nm.
RESULTS
Generation of recombinant P. pastoris harboring rHSA
The sequence of the native HSA (GenBank Acc: AF190168.1) was optimized according to the P. pastoris preference codon using the Gene-GPS technology from DNA2.0 (Supplement material 1). The amino acid sequence of HSA was confirmed using the database on Protein Data Bank (Acc 1AO6 or 6JE7). The HSA gene was constructed in the pD912 plasmid (DNA2.0) to form recombinant plasmid pD912-rHSA under AOX1 promoter control with an α-factor signal sequence and subsequently cloned in E. coli TOP10F’. The clone was characterized by electrophoresis agarose as shown in Figure 1.
Plasmid pD912-rHSA was used to transform P. pastoris SMD1168 by electroporation. Colony transformants were analyzed by colony PCR to confirm the HSA gene’s presence in the P. pastoris genome, using universal primers for pD912 (data not shown). Confirmation of rHSA sequence by using the dideoxy Sanger method using universal primers for pD912 shows 100% homology to HSA sequence (Supplement material 2). The expression of HSA by P. pastoris was carried out extracellularly by using methanol as an inducer (Gaffar et al., 2019).
Optimization and production of rHSA in P. pastoris SMD1168
The expression of rHSA was induced by variation of methanol concentration (0.75%, 1.0%, 1.25%, and 1.5%). The result showed that 1.5% of methanol provided the best result (Fig. 2). Furthermore, rHSA was characterized on SDS-PAGE with a consistent band (66.5 kDa) starting 24 hours until 144 hours after induction. The expression level of rHSA in P. pastoris SMD1168 seems to be lower compared to a previous study. However, no proteolysis products were detected.
Figure 3 shows that the increase in methanol concentration does not correlate directly with protein levels. The highest protein concentration was obtained in a 1.5% methanol inducer at 96 hours after induction. However, the level is not very high compared to 0.75% methanol. Determination of protein concentration using the Lowry method calculates all the total protein in the sample. Indeed, only an rHSA band appears on the gel, and there is no other protein on the SDS-PAGE in Figure 2. The production of rHSA in 100 ml expression media induced by methanol every 24 hours until the 96th hour shows that the level of rHSA expression in P. pastoris SMD1168 was 0.44 g/l.
Purification of rHSA by Sephadex G-25 gel filtration chromatography
Figure 4 shows the result of purification of rHSA using gel chromatography filtration, with a Sephadex G-25 column (1 m × 2 cm), with sodium chloride 0.1% as the eluent. There are two major peaks. The first peak (fractions 14–19) was combined into fraction A, while the second peak (fractions 21–25) was combined into B. Figure 3 indicates a relatively higher absorbance of the last peak. At the same time, conductivity was increased after the protein was eluted, as shown in Figure 4. Both samples (fractions A and B) were concentrated by freeze-drying, removing water, and redissolution in 10 ml double-distilled water. SDS-PAGE analysis shows that the A fraction contains the HSA, whereas the B fraction does not show the protein band. The concentration of rHSA purified by gel filtration chromatography was 0.249 g/l, significantly dropping compared to the amount of HSA present in the culture supernatant (0.44 g/l). This was probably due to the presence of fraction B, which did not contain rHSA.
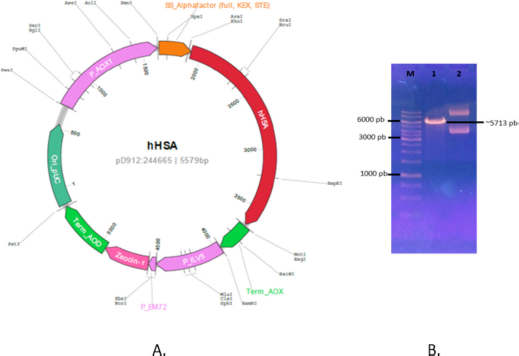 | Figure 1. Map of recombinant plasmid and characterization. (A) Map of pD912-HSA. (B) Characterization of pD912-HSA. Line (1) DNA marker, line (2) linear pD912-HSA/Sac1, and line (3) pD912-HSA circular. [Click here to view] |
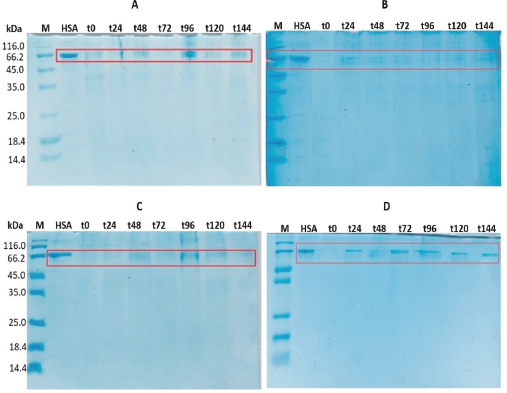 | Figure 2. SDS-PAGE of P. pastoris SMD1168 transformant culture broth supernatant, with methanol inducer concentration: (A) 0.75%, (B) 1.0%, (C) 1.25%, and (D) 1.5%. M: marker; HSA: HSA standard. The sample was taken every 24 hours, from t-0 (0 hour) to t-144 (144 hours). 20 μl of supernatant was loaded to each well. [Click here to view] |
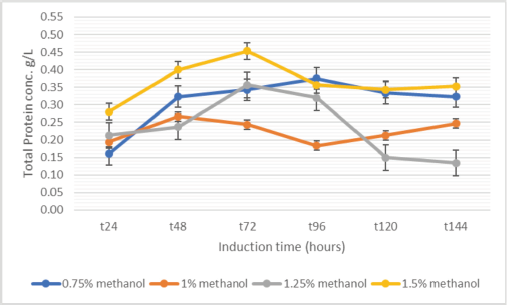 | Figure 3. [Click here to view] |
Characterization of the purity of rHSA by RP-HPLC
Characterization of rHSA using RP-HPLC (with a C-18 column) showed the peak of standard HSA appearing at 1.984 minutes, while the sample displayed a high peak at 2.122 minutes, with an area of 81.84% that indicates the purity of rHSA (Supplement material 3). The 0.138 minute difference in retention time was likely due to the effect of the solvent/medium used in this experiment, which might affect the solution’s polarity during separation. SDS-PAGE characterization confirms that the peak contains a 66.5 kDa band.
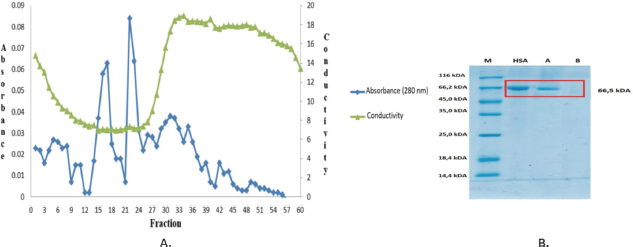 | Figure 4. (A) Chromatogram of gel filtration. The metrics used were Sephadex G-25 (1 m × 2 cm), using 1% sodium chloride as an elution buffer, at 5 ml/5 minute flow rate. (B). SDS-PAGE analysis gel filtration chromatography fractions results. Lane (a) fractions of 14–19 and (b) fractions of 21–25. 20 μl of the fraction was loaded, with pure HSA as the standard. [Click here to view] |
Synthesis of Gd-DTPA-rHSA
The Gd-DTPA was covalently linked to rHSA molecules via amide formation. The formation of DTPA-rHSA requires a reaction between the carboxyl groups in DTPA-dihydride and lysine residue of rHSA molecules to form an amide. Subsequently, the gadolinium (III) complexation proceeded by reaction Gd+3 ion of gadolinium trichloride hexahydrate with DTPA-rHSA (Kundu et al., 2011). Characterization using RP-HPLC (in a C-18 column; water-acetonitrile containing 0.1% TFA as the mobile phase) shows three single peaks: an unconjugated rHSA, rHSA-DTPA, and rHSA-DTPA-Gd with retention times of 7.155, 9.951, and 10.493 minutes, respectively (Fig. 5).
DISCUSSION
The P. pastoris SMD1168 (his4, pep4) is a protease-deficient strain that can grow in methanol at a similar rate to the wild type (Mut+). The Mut+ strain requires a higher concentration of methanol inducer than the Muts and Mut- strain. In general, methanol with a concentration of 0.5%–1.5% is used for recombinant protein expression. SMD1168 (his4, pep4) is widely used on fermenter cultures expression of heterologous proteins because the high cell density fermentation causes lysis of a small percentage of cells resulting in the secretion of a relatively high concentration of vacuolar proteases such as proteinase A (pep4) and proteinase B (prb1) (Ahmad et al., 2014; Baghban et al., 2018; Gleeson et al., 1998). However, the protease-deficient strains commonly exhibit slower growth rates subsequently producing low yields. The accumulation of methanol can negatively affect cell growth and later reduce expression levels. Hence, the optimum methanol inducer concentration needs to be evaluated for each expressed protein (White et al., 1994).
The results obtained in this study are one-third lower compared with the results reported by Zhu et al. (2018), which expressed rHSA in P. pastoris GS115. This was probably because the strain used in this study was a protease-deficient strain. The SMD1168 strain does not have vacuole peptidase A (pep4). This enzyme is responsible for the activation of carboxypeptidase Y and protease B1 (White et al., 1994). The shortcoming of the protease deficiency strain is that it has lower viability, possesses a slower growth rate, and is more difficult to handle in a shake flask and fermenter cultures. This is likely the cause of the low level of rHSA production in the P. pastoris SMD1168 strain. The protease-deficient P. pastoris strain produces intact HSA but a low expression level.
DTPA molecule binds covalently to the amine group present in the albumin and Gd (III) molecules through chelate ligands forming Gd-DTPA-HSA complexes (Liu and Chen, 2016). It is most likely that only one DTPA molecule can link to HSA through Lys199. According to Wang et al. (2020), this Lys residue can serve as a drug linking site. The partial conjugation possibly happens because there are multiple lysine residues that are available for linkage formation between the HSA and DTPA. The HPLC method that we used should be able to separate different degrees of conjugation since a different degree of conjugation will possess a different degree of polarity and the column used to separate samples based on the polarity of the sample. The albumin-(Gd-DTPA) conjugate was also synthesized by the reaction of the cyclic anhydride of DTPA with albumin and subsequent addition of an excess of Gd3+ ions before purification by extensive dialysis (Schmiedl et al., 1987). The DTPA was covalently bound to the amine group of the albumin, followed by chelating the Gd3+ to the DTPA ligand (Rinck et al., 2010; Wallnöfer et al., 2020).
We succeeded in conjugating the rHSA from P. pastoris SMD1168 with Gd-DTPA to form Gd-DTPA-rHSA with high purity. This serves as a contrast agent and is required in high purity since free DTPA molecules and Gd3+ ions are dangerous and toxic (Fauzia et al., 2015; Gallo et al., 2020; Rinck, 2018; Schmiedl et al., 1987; Wallnöfer et al., 2020). The results of this study indicate that the deficient protease host P. pastoris SMD1168 can be used to express rHSA, and conjugation with Gd-DTPA produced a complex with high purity. However, the low expression level of rHSA in the host P. pastoris SMD1168 needs to be increased. All HPLC chromatograms showed a single peak indicating single species in the sample analyzed (Fig. 5 A–C). Only a small amount of impurities was detected before the main peak of rHSA (Fig. 5A), which most likely is a minor protein still present in the sample. However, the impurities did not interfere with the formation of the DTPA-HSA conjugate since only one peak appeared in the chromatogram of the DTPA-HSA sample. No other peak was observed in the chromatogram (Fig. 5B). Finally, the final product Gd-DTPA-HSA (Fig. 5C) was detected as a single peak indicating a product with high purity (~99%). Since a high-purity product was obtained in our study, the product from our current study can be used as a contrast agent in clinical research to determine the influence of rHSA on Gd-DTPA’s performance as a contrast agent.
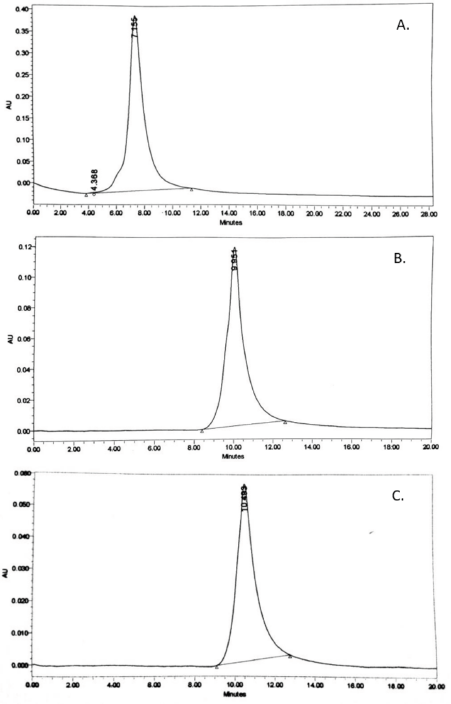 | Figure 5. Result of RP-HPLC analysis. (A) rHSA. (B). DTPA-rHSA. (C). Gd-DTPA-rHSA. RP-HPLC has performed at λ215 nm. Flow rate of 1 ml/minute. Column C-18, a mobile phase of water-acetonitrile containing 0.1% TFA. Flow rate 0.5 ml/minute. Peak was monitored using a UV detector at 215 nm. <AQ> [Click here to view] |
The application of Gd-DTPA-rHSA is to enhance imaging diagnosis with an MRI apparatus. The advantage of using serum protein is based on its biocompatibility, stability, and extended intravascular retention. The application of contrast agents, including the complex compound Gd-DTPA, has been adopted to improve image quality (Goyen, 2008). The contrast agents enhance internal body structures’ visibility in MRI. The most commonly used contrast agent enhancement is a gadolinium-based compound. Such contrast agents shorten the relaxation?times of nuclei within body tissues following oral or intravenous?administration in MRI (Bumb et al., 2010; Ouyang et al., 2020). After the intravenous administration, Gd-DTPA is reportedly distributed from the intravascular (inside a blood vessel) to the extracellular (outside a blood vessel) areas and is quickly removed from the body through renal excretion. This phenomenon reduces user preference, especially during blood analysis (Gallo et al., 2020). However, the conjugation with a control compound, e.g., serum protein, HSA, as seen with the Gd-DTPA-HSA complex, provides better imaging results (Bumb et al., 2010; Rinck et al., 2010; Wallnöfer et al., 2020). This complex compound can be synthesized through a reaction between DTPA and albumin molecules, followed by a reaction with excess Gd (III) ions. However, MR imaging contrast applications may extend beyond these typical radiographic applications (Ouyang et al., 2020; Rinck et al., 2010). Combining monoclonal antibodies and MR imaging contrast media technologies may improve the detection and characterization of malignant lesions and other antigenically distinct pathologic lesions, which is an exciting possibility for MR imaging contrast investigation (Ouyung et al., 2020).
CONCLUSION
rHSA was successfully expressed in protease-deficient P. pastoris SMD1168. The optimum yield of 0.44 g/l was obtained using a 1.5% methanol inducer at 96 hours after induction. Purification by gel filtration chromatography resulted in 81.84% purity. RP-HPLC with a C-18 column showed that the conjugation of rHSA with Gd-DTPA was successfully performed, evidenced by the presence of a single peak on the chromatogram.
ACKNOWLEDGMENTS
An Academic Leadership Grant supported this work (No. 3823/UN6.D/LT/2018) to Ukun MS Soedjanaatmadja from the Directorate of Research, Public Services, and Innovations, Universitas Padjadjaran, Indonesia.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHORS’ CONTRIBUTION
Shabarni Gaffar conceived, designed, and performed the experiments, analyzed and interpreted the data, and wrote and revised the manuscript. Rina Anggraeni conceived and designed the experiments and analyzed, interpreted, and conducted analysis of the data. Mia Tria Novianti analyzed and interpreted the data. Safri Ishmayana analyzed and interpreted the data and wrote and carried out a review of the manuscript. Ukun MS Soedjanaatmadja conceived and designed the experiments, analyzed and interpreted the data, and wrote, compiled, and revised the manuscript.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Ahmad M, Hirz M, Pichler H, Schwab H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol, 2014; 98(12):5301–17. CrossRef
Baghban R, Farajnia S, Ghasemi Y, Mortazavi M, Zarghami N, Samadi N. New developments in Pichia pastoris expression system, review and update. Curr Pharm Biotechnol, 2018; 19:451–67. CrossRef
Belew M, Li Yan M, Zhang, Wei, Caldwell K. Purification of recombinant human serum albumin (rHSA) produced by genetically modified Pichia Pastoris. Sep Sci Technol, 2008; 43:3134–53. CrossRef
Bumb A, Brechbiel MW, Choyke P. Macromolecular and dendrimer based magnetic resonance contrast agents. Acta Radiol, 2010; 51:751–67. CrossRef
Caravan P, Cloutier NJ, Greenfield MT, McDermid SA, Dunham SU, Bulte JWM, Amedio JC, Looby RJ, Subkowski RM, Horrocs W, de W, McMurry TJ, Lauffer RB. The interaction of MS-325 with human serum albumin and its effect on proton relaxation rates. J Am Chem Soc, 2002; 124:3152–62. CrossRef
Chai C, Zhou K, Wu Y, Wu L. Enhanced liver targeting of 5-fluorouracil using galactosylated human serum albumin as a carrier molecule. J Drug Target, 2006; 14:55–61. CrossRef
Charter DC, Ho JX. Structure of serum albumin. Adv Protein Chem, 1994; 45:153–203. CrossRef
Chen Z, He Y, Shi B, Yang D. Human serum albumin from recombinant DNA technology: challenges and strategies. Biochim Biophys Acta General Subject, 2013; 1830:5515–25. CrossRef
Elsadek B, Kratz F. Impact of albumin on drug delivery—new applications on the horizon. J Controlled Release, 2012; 157:4–28. CrossRef
Erstad BL. Viral infectivity of albumin and plasma protein fraction. Pharmacotherapy, 1996; 16:996–1001.
Fauzia RP, Mutalib A, Soedjanaatmadja UMS, Bahti HH, Anggraeni A, Gunawan AH, Pujiastuti H, Hudayati Y. Synthesis and characterization of gadolinium diethylenetriamine pentaacetate-folate. Proc Chem, 2015; 17:139–46. CrossRef
Gaffar S, Natalia D, Subroto T, Suprijana O, Soemitro S. Combination of genetic manipulation improved Saccharomycopsis fibuligera α-amylase secretion by Pichia pastoris. Indones J Chem, 2019; 19:305–18. CrossRef
Gallo E, Rosa E, Diaferia C, Rossi F, Tesauro D, Accardo A. Systematic overview of soft materials as a novel frontier for MRI contrast agents. RSC Adv, 2020; 10:27064. CrossRef
Gleeson MAG, White CE, Meininger DP, Komives EA. Generation of protease-deficient strains and their use in heterologous protein expression. In: Higgins DR, Cregg JM (eds.). Pichia protocols. Methods in molecular biology™, Humana Press, New Jersey, USA, p 103, 1998.
Goyen M. Gadofosveset-enhanced magnetic resonance angiography. Vasc Health Risk Manage, 2008; 4:1–9. CrossRef
He Y, Ning T, Xie T, Qiu Q, Zhang L, Sun Y, Jiang D, Fu K, Yin F, Zhang W, Shen L, Wang H, Li J, Lin Q, Sun Y, Li H, Zhu Y, Yang D. Large-scale production of functional human serum albumin from transgenic rice seeds. Proc Natl Acad Sci, 2011; 108:19078–83. CrossRef
Kalman M, Cserpan I, Bajszar G, Dobi A, Horvath E, Pazman C, Simoncsits A. Synthesis of a gene for human serum albumin and its expression in Saccharomyces cerevisiae. Nucl Acids Res, 1990; 18:6075–82. CrossRef
Kang HA, Choi ES, Hong WK, Kim JY, Ko SM, Sohn JH, Rhee SK. Proteolytic stability of recombinant human serum albumin secreted in the yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol, 2000; 53:575–82. CrossRef
Karimi M, Bahrami S, Ravari SB, Zangabad PS, Mirshekari H, Bozorgomid M, Shahreza S, Sori M, Hamblin MR. Albumin nanostructures as advanced drug delivery systems. Expert Opin Drug Deliv, 2016; 13:1609–23. CrossRef
Kerry-Williams SM, Gilbert SC, Evans LR, Ballance DJ. Disruption of the Saccharomyces cerevisiae YAP3 gene reduces the proteolytic degradation of secreted recombinant human albumin. Yeast, 1998; 14:161–9. CrossRef
Kobayashi K, Kuwae S, Ohya T, Ohda T, Ohyama M, Ohi H, Tomomitsu K, Ohmura T. High-level expression of recombinant human serum albumin from the methylotrophic yeast Pichia pastoris with minimal protease production and activation. J Biosci Bioeng, 2000; 89:55–61. CrossRef
Kobayashi K. Summary of recombinant human serum albumin development. Biologicals, 2006; 34:55–9. CrossRef
Kratz F. Albumin as a drug carrier: dessign of prodrugs, drug conjugates and nanoparticle. J Controlled Release, 2008; 132:171–83. CrossRef
Kundu A, Peterlik H, Krssak M, Bytzek AK, Pashkunova-Martic I, Arion VB, Helbich TH, Keppler BK. Strategies for the covalent conjugation of a bifunctional chelating agent to albumin: synthesis and characterization of potential MRI contrast agents. J Inorg Biochem, 2011; 105:250–5. CrossRef
Latta M, Knapp M, Sarmientos P, Bréfort G, Becquart J, Guerrier L, Jung G. Mayaux JF. Synthesis and purification of mature human serum albumin from E. Coli. Bio/Technol, 1987; 5:1309–14. CrossRef
Lawn RM, Adelman J, Bock SC, Franke AE, Houck CM, Najarian RC, Seeburg PH, Wion KL. The sequence of human serum albumin cDNA and its expression in E. coli. Nucleic Acids, 1981; 9:103–14. CrossRef
Liu Z, Chen X. Simple bioconjugate chemistry serves great clinical advances: albumin as a versatile platform for diagnosis and precision therapy. Chem Soc Rev, 2016; 45:1432–56. CrossRef
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem, 1951; 193:265–75. CrossRef
MacLennan S, Barbara MA. Risks and side effects of therapy with plasma and plasma fractions. Best Pract Res Clin Haematol, 2006; 19:169–89. CrossRef
Mead D, Dermot P, Marine D. Recombinant human albumin: applications as a biopharmaceutical excipient. Novozymes Delta Ltd., Bagsveard, Denmark, vol. 22, pp 42–3, 2007.
Meade JT. 2004. Magnetic resonance imaging agents for the delivery of therapeutic agents, United States US Patent, 6,713,046.
Moghaddassi S, Eyestone W, Bishop CE. The large-scale production of recombinant human serum albumin in the milk of transgenic cattle: strategy and implications. J Sci Appl, 2014; 02:6–19.
Na HB, Song IC, Hyeon T. Inorganic nanoparticles for MRI contrast agents. Adv Mater, 2009; 21:2133–48. CrossRef
Nguyen MT, Heo Y, Do BH, Baek S, Kim CJ, Jang YJ, Lee W, Choe H. Bacterial overexpression and purification of soluble recombinant human serum albumin using maltose-binding protein and protein disulfide isomerase. Protein Expr Purif, 2020; 167:105530. CrossRef
Ohnishi K, Kawaguchi A, Nakajima S, Mori H, Ueshima T. A comparative pharmacokinetic study of recombinant human serum albumin with plasma-derived human serum albumin in patients with liver cirrhosis. J Clin Pharmacol, 2008; 48:203–8. CrossRef
Ohtani W, Nawa Y, Takeshima K, Kamuro H, Kobayashi K, Ohmura T. Physicochemical and immunochemical properties of recombinant human serum albumin from Pichia pastoris. Anal Biochem, 1998; 256:56–62. CrossRef
Okabayashi K, Nakagawa Y, Hayasuke N, Ohi H, Miura M, Ishida Shimizu M, Mukarami K, Hirabayashi K, Minamino H, Horii H, Masaki A, Sumi A, Ohmura T, Kawabe H. Secretory expression of the human serum albumin gene in the yeast, Saccharomyces cerevisiae. J Biochem, 1991; 110:103–10. CrossRef
Ouyang Z, Li D, Shen M, Shi X. Dendrimer-based tumor-targeted systems. In: Huang R, Wang Y (eds.). New nanomaterials and techniques for tumor-targeted systems, Springer, The Gateway, Singapore, pp 337–69, 2020. CrossRef
Pierre CV, Allen MJ, Caravan P. Contras agents for MRI: 30+ years and where are we going? J Biol Inorg Chem, 2014; 19:127–31. CrossRef
Pilati D, Howard KA. Albumin-based drug designs for pharmacokinetic modulation. Expert Opin Drug Metab Toxicol, 2020; 16:783–5. CrossRef
Prevatt WD, Sreekrishna K. 1994. Expression of human serum albumin in Pichia pastoris, US Patent, 5,330,901; 107,435-450.
Raoufinia R, Mota A, Keyhanvar K, Safari F, Shamekhi S, Abdolalizadeh J. Overview of albumin and its purification methods. Adv Pharm Bull, 2016; 6:495–507. CrossRef
Rinck PA. Magnetic resonance in medicine a critical introduction. In: The basic textbook of the european magnetic resonance forum. 12th edition, BoD, Hamburg, Germany, pp 25–35, 2018.
Schmiedl U, Ogan M, Paajanen H, Marotti M, Crooks LE, Brito AC, Brasch RC. Albumin labeled with Gd-DTPA as an intravascular, blood pool-enhancing agent for MR imaging: biodistribution and imaging studies. Radiology, 1987; 162:205–10. CrossRef
Shahin H, Elmasry M, Steinvall I, Markland, K, Blomberg P, Sjoberg F, El-Serafi AT. Human serum albumin as a clinically accepted cell carrier solution for skin regenerative application. Sci Rep, 2020; 10:14486. CrossRef
Sharma A, Chaudhuri TK. Revisiting Escherichia coli as a microbial factory for enhanced production of human serum albumin. Microb Cell Fact, 2017; 16:173. CrossRef
Sleep D, Belfield GP, Goodey AR. The secretion of human serum albumin from the yeast Saccharomyces cerevisiae using five different leader sequences. J Biotech, 1990; 8:42–6. CrossRef
Taguchi K, Chuang VTG, Maruyam T, Otagiri M. Pharmaceutical aspects of the recombinant human serum albumin dimer: structural characteristics, biological properties, and medical applications. J Pharm Sci, 2012; 101:3033–46. CrossRef
Wallnöfer EA, Thurner GC, Kremser C, Talasz H, Stollenwerk MM, Helbok A, Klammsteiner N, Albrecht-Schgoer K, Dietrich H, Jaschke W. Albumin-based nanoparticles as contrast medium for MRI: vascular imaging, tissue and cell interactions, and pharmacokinetics of second-generation nanoparticles. Histochem Cell Biol, 2020; 10:235–43. CrossRef
Wang S, Liu S, Zhang Y, He J, Coy DH, Sun L. Human serum albumin (HSA) and its applications as a drug delivery vehicle. Health Sci J, 2020; 14:698.
White CE, Kempi NM, Komives EA. Expression of highly disulfide-bonded proteins in Pichia pastoris. Structure, 1994; 2:1003–5. CrossRef
Wu YK, Fang TS, Chen DC, Liu GQ, Xu YK. A comparative study of Gd-DTPA and HSA-Gd-DTPA in magnetic resonance lymphography. J South Med Univ, 2007; 27:1597–600.
Zhang Q, Yu H, Zhang FZ, Shen ZC. Expression and purification of recombinant human serum albumin from selectively terminable transgenic rice. J Zhejiang Univ Sci B, 2013; 14:867–74. CrossRef
Zhen M, Zheng J, Ye L, Li S, Jin C, Li K, Qiu D, Han H, Shu C, Yang Y, Wang C. Maximizing the relaxivity of Gd-complex by synergistic effect of HSA and Carboxylfullerene. ACS Appl Mat Interfaces, 2012; 4:3724−9. CrossRef
Zou Z, Lu ZR. Gadolinium-based contrast agents for magnetic resonance cancer imaging. WIREs Nanomed Nanobiotechnol, 2013; 5:1–18. CrossRef
Zhu W, Gong G, Pan J, Han S, Zhang W, Hu Y, Xie L. High level expression and purification of recombinant human serum albumin in Pichia pastoris. Prot Expr Purif, 2018; 147:61–8. CrossRef
SUPPLEMENT MATERIALS
 | Supplement material 1. The nucleotide sequence of HSA optimized by gene GPS (DNA 2.0) using P. pastoris codon table. [Click here to view] |
 | Supplement material 2. Alignment of rHSA (HSA_1) with HSA sequence. [Click here to view] |
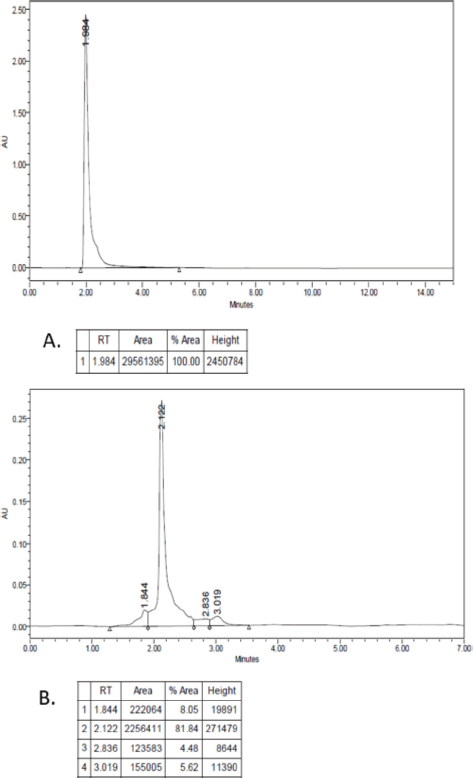 | Supplement material 3. Result of RP-HPLC. (A) HSA standard; (B) rHSA. Column; C-18, mobile phase; water-acetonitrile containing 0.1% TFA. Flow rate 0.5 ml/minute. Absorption is measured by a UV detector at 215 nm. [Click here to view] |