INTRODUCTION
Nowadays, antimicrobial peptides (AMPs) are of interest as new agents of food additives and preservatives due to less toxicity and good bioactivity (Ben Said et al., 2019; Wang et al., 2016a). Due to the increased development of antibacterial resistance, antibiotic alternative research has been of interest. For example, polymyxin B is a group of cyclic nonribosomal polypeptide that is isolated from the bacterium Bacillus polymyxa. It has been used as an antibiotic against Gram-negative bacteria (Michalopoulos and Falagas, 2008). Although many researchers have reported on designed antibacterial peptides, the amino acid sequences in this paper have not been reported before. Commercially, most AMPs are chemically synthesized because their production is easy, it takes a short time, and a large quantity is produced. Normally, AMPs have amphiphilic properties, 2–7 net charges, and approximately 50% hydrophobic percentage (Mishra et al., 2018). Hydrophobic boundary of a designed peptide is also an important factor in antibacterial peptide design (Chen et al., 2007; Guo et al., 2013; Lee et al., 2019).
AMPs can be used to improve the shelf life of cosmeceutical or food products such as in the case of Nisin, a well-known preservative peptide. Nisin was approved by food and drug administration (FDA) in 1998 as a preservative (FDA, 1998; Hsu et al., 2004). Moreover, lysozyme peptides (LzP) are well known as natural food preservatives (Abdoub et al., 2007). Also, MDpep9 which is obtained from housefly larvae is an antibacterial peptide which is of interest to further develop as a preservative (Tang et al., 2009).
Many peptides with antioxidant activity have been isolated from food hydrolysates, plant proteins, and chemical synthesis. The short peptide (YWYPGL) obtained from casein hydrolysates had good antioxidant activity in superoxide, hydroxyl, and 2,2-diphenyl-1-picrylhydrazyl (DPPH) assay (Suetsuna et al., 2000). The short peptide, WYSLAMAASAI, obtained from lactalbumin, had better antioxidant activity than that of butylated hydroxyanisole (BHA) (Hernández-Ledesma et al., 2005). BHA is both an antioxidant agent and a preservative which is widely used in food, drug, and cosmeceutical applications. The chemically synthetic peptide, GWWW, showed good antioxidant activity against peroxyl radicals and peroxynitrite (Matsui et al., 2018). They are interesting as food additives because some antioxidant peptides could not be destroyed in the human digestive system (Chabance et al., 1998; Webb et al., 1990).
The aim of this study is to design ultra-short peptides with antibacterial and antioxidant activities and low toxicity by varying hydrophobic and hydrophilic facets. The various hydrophobic and hydrophilic facet sizes of the designed peptides resulting in net charge, length, and % hydrophobic ratio differences were examined for hemolytic, antibacterial, and antioxidant activities.
MATERIALS AND METHODS
Materials
All peptides were obtained from GL Biochem (Shanghai, China). Other reagents were reagent grade.
Peptide synthesis
Ultra-short peptides were obtained from GL Biochem (Shanghai, China). They were synthesized by Fmoc solid phase. High performance liquid chromatography (HPLC) (stationary phase: C-18, mobile phase: 5%–20% acetone in water, time: 0–20 minutes) was performed to obtain peptides with a 95% purity level or greater. Electrospray ionization mass spectrometry (ESI-MS) chromatogram confirmed the synthesis efficiency by comparing the molecular weight of the synthesized peptides with the theoretical molecular weight of each peptide.
Minimum inhibitory concentrations (MICs) and bactericidal activities
Briefly, minimum inhibitory concentrations (MICs) and bactericidal activities were determined by using a methodology adapted from a previous report (Andrews, 2001). All tested-bacterial strains were obtained from the department of medical science, Thailand. Each bacterial strain was inoculated to enter its log growth phase. The bacterial suspension was adjusted to an optical density (OD) of 0.001 at 600 nm. The 90 ¼l of each microorganism in Luria-Bertani broth (Himedia, India) was added to 96-well plates. The 10 ¼l of various peptide concentrations (twofold dilution) in 10 mM Tris–HCl buffer (pH 7.4) was treated. The 10 mM Tris–HCl buffer (pH 7.4) was used as a control representing 100% growth ability. After incubation at 37°C for 16–18 hours, the OD600 absorbance of each well was calculated for the 90% inhibition (MIC90) by using the following formula:
where Abs 0 is the absorbance of a blank well, Abs 1 is the absorbance of a negative control (100% bacterial growth), and Abs 2 is the absorbance of a peptide treated sample. The clear suspensions of each well (MIC90) were further analyzed, on the 100% minimum bactericidal concentration (MBC) value, by using the drop plate technique. The concentrations without any bacterial colony were reported as 100% MBC.
Antioxidant activity
The antioxidant activity of all the designed peptides was determined by using the 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) assay. Briefly, the mixture of ABTS and potassium persulfate was prepared by dissolving ABTS and potassium persulfate in distilled water to obtain a final concentration of 7.2 and 2.6 mM, respectively. The mixture was left in the dark at room temperature for about 12 hours. The mixture was then measured at 734 nm and was adjusted to an absorbance of about 0.72 at 734 nm. The 10 ¼l of each peptide was incubated with 90 ¼l of ABTS free radical suspension in a 96-well plate for 10 minutes in the darkness. Glutathione was used as a positive control and distilled water was used as a negative control. The samples in 96-well plates were measured at 734 nm. All values were subtracted from the average value of the blank wells. The percent of ABTS radical scavenging activity was calculated according to the following formula:
where Abs 0 is the absorbance at 734 nm of the blank well, Abs 1 is the absorbance at 734 nm of the negative control (100% free radicals), and Abs 2 is the absorbance at 734 nm of the sample.
Hemolysis
The toxicity of all designed peptides was determined against human red blood cells. The red blood cells of a healthy human were collected and washed with phosphate-buffered saline (PBS) and then centrifuged three times at 5,000 rpm for 5 minutes each time. After that, the red blood cell suspension was adjusted to 2% red blood cells in PBS. Each peptide (0.781–1,000 ¼g/ml, 10 ¼l) was incubated with the suspension for 1 hour at 37°C. One percent Triton X100 was treated as a positive control and PBS was treated as a negative control. The samples were then centrifuged at 5,000 rpm for 5 minutes. The 100 ¼l of suspensions was collected and measured at 570 nm.
where Abs 0 is the absorbance of the negative control, Abs 1 is the absorbance at 570 nm of the positive control, and Abs 2 is the absorbance at 570 nm of the sample.
Circular dichroism
The secondary structure of P8 and P9 which had good antibacterial activity was then studied in different conditions by using circular dichroism. Shortly, each peptide was dissolved in phosphate buffer or 50% TFE in distilled water at the concentration of 0.1 mg/ml. A 1 mm path length cell was used. The Circular dichroism (CD) spectrum data were obtained by using the Jasco J-815 CD Spectrometer.
Scanning electron microscope
The effects of the peptides P8 and P9 on bacterial envelope change were studied by using a scanning electron microscope. Briefly, the bacterial suspension in mid-exponential growth phase was washed in PBS three times and diluted to approximately 108 CFU/ml in PBS. After that, 90 ¼l of the bacterial suspension was incubated with 10 ¼l of each peptide in the concentration of 4-time MIC for 1 hour, 37°C. The PBS was used as a negative control. The 20 ¼l of each sample was deposited on 0.1 micrometer polycarbonate membrane and was fixed with 5% glutaraldehyde for 4 hours. The samples were gradually dehydrated with ethanol series (30%, 50%, 70%, 90%, and 100% ethanol) for 15 minutes of each ethanol concentration. After sample drying in a vacuum for 6 hours, all samples were coated with gold. Scanning electron microscope (SEM) images were taken at low electron energies.
RESULTS AND DISCUSSIONS
Peptide design
In an ultra-short peptide design containing only two different amino acids, tryptophan represents a hydrophobic amino acid and lysine represents a positive charge amino acid. Tryptophan is found in many antibacterial peptides (Deslouches et al., 2013; Sitaram et al., 2003); therefore, it was selected to represent a hydrophobic amino acid. The NH of the aromatic side chain of tryptophan can form hydrogen bonds, which has a dipole moment, with the interfacial region of a membrane resulting in the peptide anchoring to the bilayer surface (Mishra et al., 2018). The reason why lysine was selected to represent a positive charge amino acid in this study is due to its selectivity as can be seen in a previous report (Anunthawan et al., 2013). Their facets were depicted by helical wheel patterns as can be seen in Figure 1 (Mól et al., 2019). Each peptide had different hydrophobic and hydrophilic facet sizes as can be seen in the helical wheel pattern (Fig. 1). Three pairs of designed peptides which were of equal lengths and various facet sizes (P4 and P7, P5 and P6, and P8 and P9) and the others which were of unequal lengths and various facet sizes (P1, P2, and P3) were examined for antibacterial, antioxidant, and hemolytic activities. The chemical and physical properties of all nine peptides, named P1–P9, can be seen in Table 1. The % hydrophobic ratio and protein-binding potential (Boman index) of all designed peptides were calculated by using an online program (APD3, 2019). All designed peptides were modified by C-terminal amidation (Strandberg et al., 2007). Their lengths were 3–15 amino acids. The net-positive charge including NH2 at both termini was from +4 to +11. Protein-binding potential (Boman index) was 2.39 for P3, P4, and P5 and was 1.61 for P8 and P9. The hydrophobic ratio of P1 was the lowest (33%) and the hydrophobic ratio of P2–P7 was 40%. P8 and P9 had the highest hydrophobic ratio at 50%. The designed peptides were compared to other peptide sequences stored in the antimicrobial peptide database (APD3 database) to ensure that their peptide sequences were not previously reported (Wang et al., 2016b).
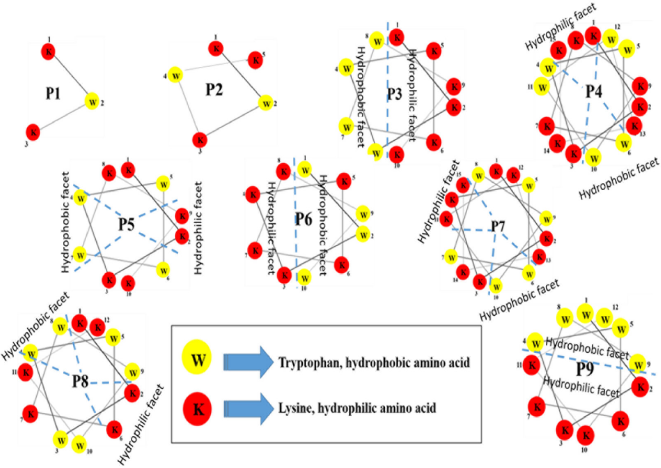 | Figure 1. Helical wheel patterns of all designed peptides. [Click here to view] |
Minimum inhibitory concentrations (MICs) and bactericidal activities
Antibacterial activity of all designed peptides was determined against seven bacterial strains (Escherichia coli DMST 15537, Escherichia coli O157:H7, Staphylococcus epidermidis DMST 5868, Pseudomonas aeruginosa DMST 5870, Staphylococcus aureus DMST 8013, Staphylococcus aureus (MRSA) DMST 4738, and Klebsiella pneumoniae DMST 37389) as can be seen in Table 2. The highest concentration of P8 and P9 (100 ¼g/ml) could not kill the largest number of bacteria. This was possibly caused by the defense activation of bacteria against the peptides or peptide aggregation (Azad et al., 2011). The peptides P8 and P9 had better antibacterial activity than the other designed peptides. P8 had MIC90 at 6.3 ¼g/ml, whereas P9 had MIC90 at 3.1 ¼g/ml against S. epidermidis DMST 5868. Normally, S. epidermidis can form biofilms that grow on implants resulting in infections. For S. aureus DMST 8013, which causes foodborne illnesses, P8 had MIC90 at 50 ¼g/ml, whereas P9 had MIC90 at 6.3 ¼g/ml. Only P9 could inhibit the bacterial growth of E. coli O157:H7, which is a cause of foodborne illnesses, at 12.5 ¼g/ml. However, other designed peptides needed a higher concentration for MIC90 against all bacterial strains.
P8 and P9 had the same number of amino acid residues which was 12. They had equal hydrophobic residues (six tryptophan residues) and hydrophilic residues (six lysine residues) resulting in 50% hydrophobic ratio, 8 positive charges, and 1.61 kcal/mol Boman index. The Boman index of P8 and P9 was lower than that of P3–P7 (2.39 KJ/mol) indicating that a high Boman index does not always result in good antibacterial activity (Boman, 2003). Comparing the sequences of P8 and P9 with other reported peptides in APD database, P8 and P9 were similar to the peptides named Inverso-CysHHC10 50% which has antibacterial activity against S. aureus and S. epidermidis and has low toxicity toward human red blood cells (Buckholtza et al., 2016). P8 and P9 could inhibit S. epidermidis at 6.3 and 3.1 ¼g/ml, respectively. The hydrophobic ratio of P8 and P9 was the highest among the designed peptides in this study. This may be one of the factors that increased their antibacterial activity. These results corresponded to the previous studies which reported that higher hydrophobicity or hydrophobic ratio of peptides results in higher antibacterial activity (Chen et al., 2007; Rosenfeld et al., 2010).
The difference between P8 and P9 is the amino acid sequence as can be seen in Table 1. P8 had two times an amino acid repeat pattern, KKWWW. But P9 had three times an amino acid repeat pattern, WKKW. This difference resulted in the difference in their secondary structure, hydrophobic and hydrophilic facets. P9 had a bigger hydrophobic facet than that of P8 resulting in its better antibacterial activity. Comparing the activities of P9 and P3, even though they had the same sizes of hydrophilic facet, P3 had no antibacterial activity indicating that its hydrophobic facet size was not suitable. The hydrophobic facet of P3 contained only four residues; this lower hydrophobic ratio has been hypothesized to be the reason for the lack of antibacterial activity (Chen et al., 2007; Rosenfeld et al., 2010). However, P4 and P7, containing 15 amino acid residues (more than the amino acid residue number of P8 and P9), had poor antibacterial activity against all bacterial strains indicating that a longer peptide chain and a more positive charge do not always improve antibacterial activity.
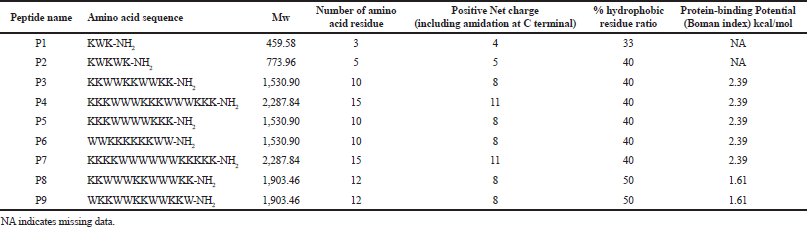 | Table 1. Biological and biophysical properties of designed peptides. [Click here to view] |
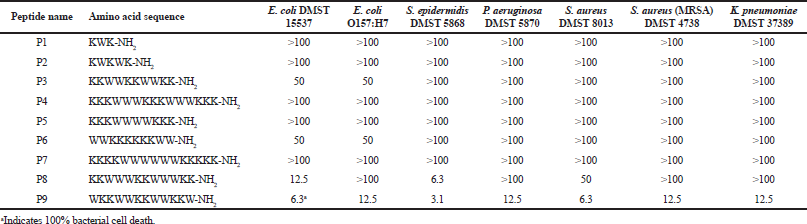 | Table 2. Antibacterial activity of designed peptides. MICs are deï¬ned as the concentration (¼g/ml) that inhibited bacterial growth at least by 90%. [Click here to view] |
Antioxidant activity
All designed peptides had antioxidant activity higher than approximately 80% free radical scavenging (< 20% remaining free radicals) at 0.1 mg/ml. IC50 of all peptides are shown in Table 3. The reason why all designed peptides have antioxidant activity is due to the tryptophan property, which has an indole ring for free radical scavenging (Zou et al., 2016). P3 demonstrated the best antioxidant activity among the designed peptides, with IC50 of 3.2 ¼g/ml or 2.07 ¼M. A comparison between P9 and P3 which had the same hydrophilic facet size and smaller hydrophobic facet showed that P3 had a better antioxidant activity. This indicates that the activity may not depend on hydrophobic facet size. P1, containing three amino acids, displayed IC50 almost equal to glutathione which is the positive control (6.1 ¼g/ml, 13.3 ¼M). P2, the slightly longer peptide (with five residues), had an antioxidant activity slightly better than that of P1. A comparison of the antioxidant activities of the three pairs of designed peptides which had the same length (P4 and P7, P5 and P6, and P8 and P9) showed that only the pair of P8 and P9 had different antioxidant activity. P8 had IC50 at 4.5 ¼g/ml or 2.35 ¼M, which was lower than that of P9 (10.8 ¼g/ml, 5.67 ¼M). Even though P8 and P9 had the same chemical properties, their hydrophobic, hydrophilic facets, and antioxidant activities were different. The larger hydrophobic facet sizes as visualized by the helical wheel had no direct effect on a higher antioxidant activity as can be seen in the comparison of the activities and the facets of the P3 and P9 and P8 and P9 pairs.
Hemolysis
In in vitro selectivity index, an antibiotic candidate should be nontoxic against human cells at the concentration required to achieve a therapeutic effect. All designed peptides except P9 had hemolytic activity lower than 5% at the concentration of 100 ¼g/ml as can be seen in Figure 2. P9 had a bigger hydrophobic region than that of P8; its toxicity against human red blood cells was higher. These results are consistent with the previous study which reported that higher hydrophobicity of the nonpolar face of amphipathic peptides results in higher hemolytic activity (Chen et al., 2007). Moreover, P9 had toxicity against human red blood cells of approximately 20% at 100 ¼g/ml and had no hemolytic activity at its MIC90 against S. epidermidis (3.1 ¼g/ml). For antioxidant activity, P2, P3, and P8 in particular had low toxicity toward human red blood cells, less than 5% at the concentration (100 ¼g/ml), and greater than their IC50, approximately more than 20–30 times.
Circular dichroism
The conformational change of antibacterial peptides is reported in a previous study (Andrushchenko et al., 2006). A secondary conformational analysis was performed to consider whether there was a conformational change when expressed in a hydrophobic environment compared to a conformation in hydrophilic environment. CD experiments of P8 and P9 were performed in phosphate buffer (pH 7.4) indicating no defined secondary structure. Fifty percent of TFE, which is a hydrophobic solution, induced maximum α helix structure in peptides. For P8, an α helix conformation was observed in 50% TFE, indicating a coil-to-α-helix transition upon interaction with hydrophobic environments as can be seen in Figure 3A. Also, the secondary structure of P9 was more ordered as a helix structure as can be seen in Figure 3B, observed in 50% TFE. It corresponds to the helical wheel pattern of P9 that its hydrophobic and hydrophilic facets are equal. The results suggest that their secondary structures may change when interacting with an antibacterial membrane.
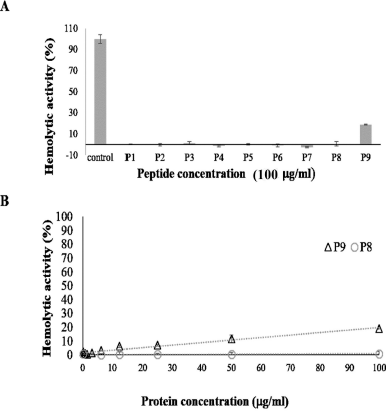 | Figure 2. Hemolytic activities of de novo short peptides. Hemolytic activities of all designed peptides at 100 ¼g/ml are shown in 2A. Only P9 had toxicity toward human red blood cells. 2B reveals comparing hemolytic activities of P8 and P9. While P8 had no hemolytic activities at all tested concentrations, P9 had toxicity less than 20% at 50 ¼g/ml. [Click here to view] |
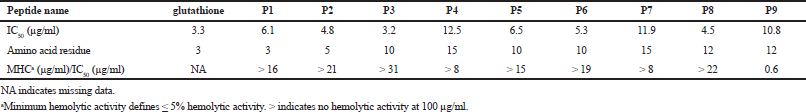 | Table 3. Antioxidant activity in IC50 (¼g/ml) of each peptide. [Click here to view] |
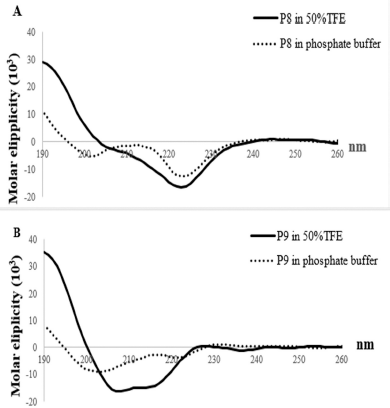 | Figure 3. Structure analysis of P8 and P9. The spectra (a black line) in 50% TFE were obtained at a peptide concentration of 0.1 mg/ml. The spectra (a black dot line) in water were obtained at 0.1 mg/ml. P8 (A) has unstructured conformation in water (a hydrophilic environment), whereas it presents α-helix structure in 50% TFE (a hydrophobic environment). P9 (B) has unstructured conformation in water (a hydrophilic environment), whereas it presents α-helix structure in 50% TFE (a hydrophobic environment). [Click here to view] |
 | Figure 4. SEM images show the effects of P8 and P9 on S. epidermidis. The negative control which is PBS buffer without peptide is displayed in the A panel. The B and C panels visualized the bacterial cells incubating with P8 and P9, respectively. The bacterial cells were treated with each peptide at a final concentration of 0.1 mg/ml. [Click here to view] |
Scanning electron microscope
The effects of P8 and P9 on the envelope were observed. As P8 and P9 had the best MIC against S. epidermidis, they were selected to be studied and it was found that even though they had different facet sizes and different bacterial activities, they could make cell wall blebs as can be seen in Figure 4. These blebs may result in bacterial membrane pores and finally cell deaths (Chileveru et al., 2015).
CONCLUSION
In conclusion, not only the net charge and % hydrophobic ratio but also hydrophobic facet size is important factors for antibacterial peptide design. P9 had less than 5% toxicity toward human red blood cells at 6.3 ¼g/ml which was higher than its MIC against S. epidermidis DMST 5868, approximately 2 times. All designed peptides exhibited antioxidant activity. P2, P3, and P8 in particular had low toxicity toward human red blood cells, less than 5% at the concentration (100 ¼g/ml), and higher than their IC50, approximately more than 20–30 times. All designed peptides are interesting to further develop in antioxidant applications. Also, P8 and P9 are interesting to further develop as antibiotic agents.
ACKNOWLEDGMENTS
The authors would like to thank National Research Council of Thailand and Rajamangala University of Technology Isan for financial assistance. Also, they would like to thank Nakhon Ratchasima Rajabhat University for providing the microplate reader. Finally, they would like to thank Assoc. Prof. Dr. Chartchai Krittanai, Institute of Molecular Biosciences, Mahidol University, for providing some chemical reagents of CD.
CONFLICT OF INTEREST
The authors declare that they have no known competing financial interests or personal relationships that could appear to influence the work reported in this paper.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Abdoub AM, Higashiguchia S, Aboueleinin AM, Kim M, Ibrahim HR. Antimicrobial peptides derived from hen egg lysozyme with inhibitory effect against Bacillus species. Food Control, 2007; 18(2):173–8. CrossRef
Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother, 2001; 48:5–16. CrossRef
Andrushchenko VV, Vogel HJ, Prenner EJ. Solvent-dependent structure of two tryptophan-rich antimicrobial peptides and their analogs studied by FTIR and CD spectroscopy. Biochim Biophys Acta, 2006; 1758:1596–608. CrossRef
Anunthawan T, Yaraksa N, Phosri S, Theansungnoen T, Daduang S, Dhiravisit A, Klaynongsruang, S. Improving the antibacterial activity and selectivity of an ultra short peptide by hydrophobic and hydrophilic amino acid stretches. Bioorg Med Chem Lett, 2013; 23(16):4657–4662. CrossRef
APD3. Antimicrobial peptide calculator and predictor. 2019. Available via http://aps.unmc.edu/AP/prediction/prediction_main.php (Accessed 01 January 2019).
Azad MA, Huttunen-Hennelly HEK, Friedman CR. Bioactivity and the first transmission electron microscopy immunogold studies of short de novo-designed antimicrobial peptides. Antimicrob Agents Chemother, 2011; 55:2137–45. CrossRef
Ben Said L, Fliss I, Offret C, Beaulieu L. Antimicrobial peptides: the new generation of food additives. Encyclopedia Food Chem, 2019; 3:576–82. CrossRef
Boman H. Antibacterial peptides: basic facts and emerging concepts. Intern Med J, 2003; 254:197–215. CrossRef
Buckholtza GA, Regera NA, Anderton WD, Schimoler PJ, Roudebush SL, Meng WS, Miller MC, Gawalt ES. Reducing Escherichia coli growth on a composite biomaterial by a surface immobilized antimicrobial peptide. Mater Sci Eng C, 2016; 65:126–34. CrossRef
Chabance B, Marteau P, Rambaud JC, Migliore-Samour D, Boynard M, Perrotin P, Guillet R, Jollès P, Fiat AM. Casein peptide release and passage to the blood in humans during digestion of milk or yogurt. Biochimie, 1998; 80(2):155–65. CrossRef
Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob Agents Chemother, 2007; 51(4):1398–406. CrossRef
Chileveru HR, Lim SA, Chairatana C, Wommack AJ, Chiang I-L, Nolan EM. Visualizing attack of Escherichia coli by the antimicrobial peptide human Defensin 5. Biochemistry, 2015; 54(9):1767–77. CrossRef
Deslouches B, Steckbeck JD, Craigo JK, Doi Y, Mietzner TA, Montelaroa RC. Rational design of engineered cationic antimicrobial peptides consisting exclusively of arginine and tryptophan, and their activity against multidrug-resistant pathogens. Antimicrob Agents Chemother, 2013; 57:2511–21. CrossRef
FDA. Nisin preparation: afï¬rmation of GRAS status as a direct human food ingredient. Fed Regist, 1998; 53:12247.
Guo X, Ma C, Du Q, Wei R, Wang L, Zhou M, Chen T, Shaw C. Two peptides, TsAP-1 and TsAP-2, from the venom of the Brazilian yellow scorpion, Tityus serrulatus: evaluation of their antimicrobial and anticancer activities. Biochimie, 2013; 95(9):1784–94. CrossRef
Hernández-Ledesma B, Dávalos A, Bartolomé B, Amigo L. Preparation of antioxidant enzymatic hydrolysates from alpha-lactalbumin and beta-lactoglobulin. Identification of active peptides by HPLC-MS/MS. J Agric Food Chem, 2005; 53(3):588–93. CrossRef
Hsu ST, Breukink E, Tischenko E, Lutters MA, de Kruijff B, Kaptein R, Bonvin AMJJ, Nuland NAJ. The nisin–lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol, 2004; 11:963–7. CrossRef
Lee PC, Chu CC, Tsai YJ, Chuang YC, Lung FD. Design, synthesis, and antimicrobial activities of novel functional peptides against Gram-positive and Gram-negative bacteria. Chem Biol Drug Des, 2019; 94(2):1537–44. CrossRef
Matsui R, Honda R, Kanome M, Hagiwara A, Matsuda Y, Togitani T, Ikemoto N, Terashimaet M. Designing antioxidant peptides based on the antioxidant properties of the amino acid side-chains. Food Chem, 2018; 245:750–5. CrossRef
Michalopoulos A, Falagas ME. Colistin and polymyxin B in critical care. Crit Care Clin, 2008; 24:377–91. CrossRef
Mishra AK, Choi J, Moon E, Baek KH. Tryptophan-rich and proline-rich antimicrobial peptides. Molecules, 2018; 23:815. CrossRef
Mól AR, Fontes W, Castro MS. NetWheels: Peptides helical wheel and net projections maker. 2019. Available via http://lbqp.unb.br/NetWheels/ (Accessed 01 January 2019).
Rosenfeld Y, Lev N, Shai Y. Effect of the hydrophobicity to net positive charge ratio on antibacterial and anti-endotoxin activities of structurally similar antimicrobial peptides. Biochemistry, 2010; 49:853–61. CrossRef
Sitaram N, Subbalakshmi C, Nagaraj R. Indolicidin, a 13-residue basic antimicrobial peptide rich in tryptophan and proline, interacts with Ca(2+)-calmodulin. Biochem Biophys Res Commun, 2003; 309:879–84. CrossRef
Strandberg E, Tiltak D, Ieronimo M, Kanithasen N, Wadhwani P, Ulrich SA. Influence of C-terminal amidation on the antimicrobial and hemolytic activities of cationic α-helical peptides. Pure Appl Chem, 2007; 79(4):717–28. CrossRef
Suetsuna K, Ukeda H, Ochi H. Isolation and characterization of free radical scavenging activities peptides derived from casein. J Nutr Biochem, 2000; 11(3):128–31. CrossRef
Tang YL, Shi YH, Zhao W, Hao G, Le GW. Discovery of a novel antimicrobial peptide using membrane binding-based approach. Food Control, 2009; 20(2):149–56. CrossRef
Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res, 2016b; 44(D1):D1087–93. CrossRef
Wang S, Zeng X, Yang Q, Qiao S. Antimicrobial peptides as potential alternatives to antibiotics in food animal industry. Int J Mol Sci, 2016a; 17(603):1–12. CrossRef
Webb KE Jr. Intestinal absorption of protein hydrolysis products: a review. J Anim Sci, 1990; 68(9):3011–22. CrossRef
Zou TB, He TP, Li HB, Tang HW, Xia EQ. The structure-activity relationship of the antioxidant peptides from natural proteins. Molecules, 2016; 21:72. CrossRef