INTRODUCTION
Colorectal cancer (CRC) is one of the most common causes of death due to cancer in the world (Deng, 2017). The incidence and rate of death due to colon cancer have increased in recent decades due to lifestyle changes in modern-day society, including decreased dietary fiber intake and increased processed food consumption (O'Keefe, 2016). In colon cancer treatment, conventional chemotherapy is administered to reduce tumor size and eradicate residual tumor tissue after a curative resection (Polastro et al., 2018). Nevertheless, both intrinsic and acquired resistance can decrease the effectiveness of chemotherapeutics (Hu et al., 2016). Chemoresistance mechanisms arise due to the existence of a minor population called cancer stem cells (Kozovska et al., 2014). Taken together, these lines of evidence indicate the necessity of developing agents that can improve the effectiveness of colon cancer chemotherapy and overcome chemoresistance.
Sinensetin (Fig. 1A), a flavonoid compound found in citrus fruits, is known to have anticancer activity in various cancer cells. Sinensetin exerts cytotoxicity on MDA-MB-468 breast cancer cells (Androutsopoulos et al., 2009), adenocarcinoma gastric cell lines human gastric cancer cells (Dong et al., 2011), and K562 human chronic myeloid leukemia cells (Danışman et al., 2019). An increase in cytotoxicity of the tyrosine kinase inhibitor imatinib was observed in a combinatorial study of sinensetin with imatinib in K562 leukemic cells (Danışman et al., 2019). Recent studies have revealed the molecular mechanisms of sinensetin in cancer cells, including induction of cell death by modulating the p53-5' AMP-activated protein kinase/mTOR signaling pathway in HepG2 liver cancer cells (Kim et al., 2020) and inhibition of migration and invasion of the human gallbladder TJ-GBC2 cell line through inhibition of the phosphatase and tensin homolog (PTEN)/phosphatidylinositide 3-OH kinase (PI3K)/AKT serine/threonine kinase (AKT) signaling pathway (Huang et al., 2020). In addition, another recent study showed that sinensetin increased the effectiveness of 5-fluorouracil (5-FU) in spheroids derived from CRC cells (Pereira et al., 2019); however, the molecular mechanism remains elusive.
 | Figure 1. (A) Chemical structure of sinensetin. (B) Venn diagram of CRC resistance regulatory genes and sinensetin-predicted targets. (C) GO enrichment analysis of potential target genes of sinensetin in overcoming CRC resistance. [Click here to view] |
In this study, we used an integrated bioinformatics approach to explore the targets of sinensetin and the possible molecular mechanisms by which it acts to circumvent colon cancer resistance to chemotherapy. Sinensetin targets and colon cancer resistance regulatory genes were downloaded from free public databases. Protein–protein interaction (PPI) networks, gene ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses revealed the potential therapeutic targets of sinensetin against chemoresistance in colon cancer potential therapeutic target genes of sinensetin (PS). Further analysis of PS was conducted to explore the genetic alterations in more detail. The present study highlights the importance of sinensetin in overcoming chemoresistance in CRC.
MATERIAL AND METHODS
Data collection and processing
Targets of sinensetin were obtained from SwissTargetPrediction, (http://www.swisstargetprediction.ch) (Gfeller et al., 2014). SwissTargetPrediction is a public database containing updated data and information to effectively and efficiently predict the molecular target (Daina et al., 2019). Briefly, the sinensetin structure was drawn in the form of a simplified molecular-input line-entry system and submitted into the SwissTargetPrediction database. Homo sapiens were selected as target organisms. Predicted targets were selected with a cutoff value probability of > 0.1, resulting in 100 targets (Supplementary Table 1). Regulatory genes of human CRC cells were downloaded from PubMed with the keyword “human colorectal cancer cells” and resulted in 1,376 genes (Supplementary Table 2). Venn diagram of CRC regulatory genes and sinensetin targets was generated using Venny 2.1 (https://bioinfogpcnbcsices/tools/venny/indexhtml), resulting in 36 potential therapeutic targets of sinensetin against chemoresistance in colon cancer (PS) (Fig. 1B and Supplementary Table 3).
Analysis of GO and KEGG pathway enrichment
Analysis of GO was carried out to PS by WebGestalt, by using the default settings with a cutoff value of p < 0.05. KEGG pathway enrichment was conducted by the Database for Annotation, Visualization, and Integrated Discovery v6.7 (Huang da et al., 2009), by using the default settings with a cutoff value of p < 0.05.
PPI network and hub gene selection
PPI network analysis was executed with STRING-DB v11.0 (Szklarczyk et al., 2015). Briefly, PS was submitted into STRING-DB to build a PPI network, with a confidence score > 0.4 as the cutoff value and visualized with Cytoscape software (Shannon et al., 2003). Hub genes were selected from the CytoHubba plugin based on the highest degree score by using the default settings (Chin et al., 2014).
Analysis of genetic alterations of the PS
Genetic alteration analysis of the PS was carried out using cBioPortal (http://www.cbioportal.org) (Cerami et al., 2012; Gao et al., 2013). Briefly, selected PS was considered as a query and subjected to genetic alteration among 10 studies of CRC in cBioPortal. Selected CRC study was analyzed for OncoPrint, which depicts genome changes, that is, inframe, missense, and truncating mutation, and was chosen for further connectivity analysis using a one-sided Fisher’s exact test with a cutoff value of p < 0.05.
RESULTS AND DISCUSSION
GO and KEGG pathway enrichment analysis
This study aimed to determine the targets of sinensetin action, elucidate its molecular mechanism to overcome chemoresistance in colon cancer (PS), and reveal potential therapeutic targets of sinensetin. To identify gene classes and predict the function of PS, we carried out GO and KEGG pathway enrichment analysis. GO analysis was aimed at checking the role of PS in biological processes, cellular components, and molecular functions. The results of GO analysis revealed the regulation of the biological process response to stimulus and the metabolic process by PS (Fig. 1C). In addition, PS was located in the membrane and nucleus and served as a molecular function in protein, ion, and nucleotide binding. The results of the analysis of KEGG pathway enrichment revealed 21 pathways regulated by PS, including CRC, the vascular endothelial growth factor (VEGF) signaling pathway, the erbB signaling pathway, and the ATP-binding cassette (ABC) transporter (Table 1). Several PS were involved in the CRC pathway, including hosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (PIK3CG), B-Raf protooncogene, serine/threonine kinase (BRAF), Glycogen synthase kinase-3 beta (GSK3B), MET, and platelet-derived growth factor receptor beta (PDGFRB). The results of GO and KEGG pathway enrichment analysis from PS were further investigated for genetic alterations analysis using cBioPortal.
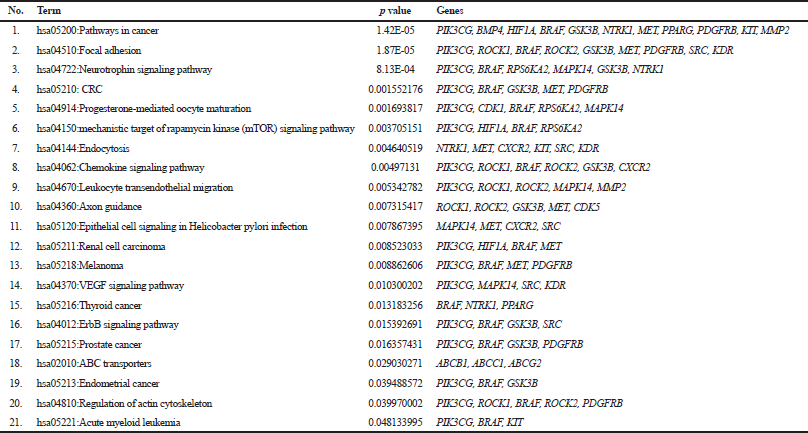 | Table 1. KEGG pathway enrichment analysis of the DEGs. [Click here to view] |
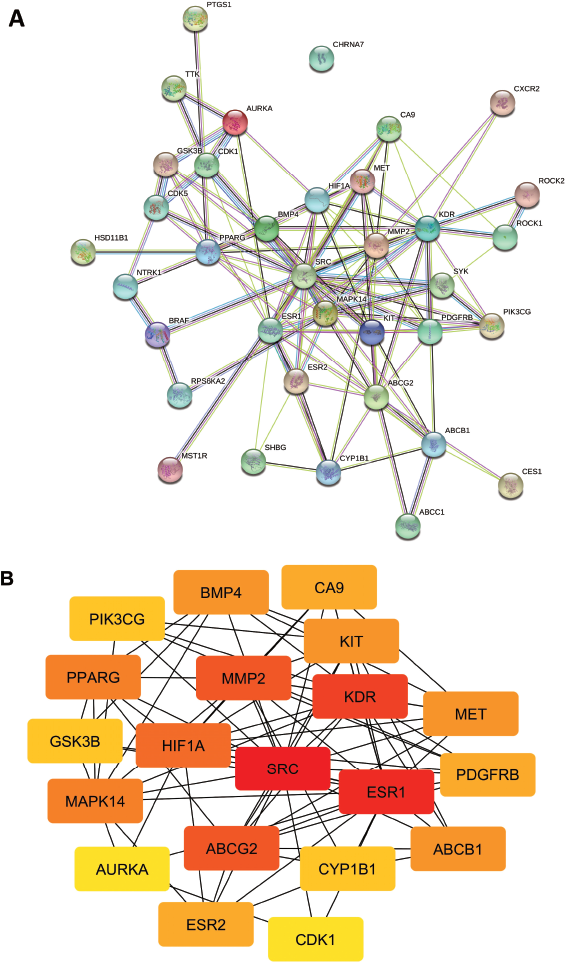 | Figure 2. (A) PPI network of potential target genes of sinensetin in overcoming CRC resistance, analyzed by STRING. (B) Top 20 hub genes based on highest degree score, analyzed by CytoHubba. [Click here to view] |
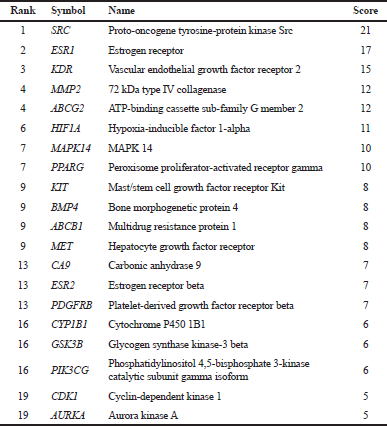 | Table 2. Top 20 in the network of sinensetin and CRC resistance, ranked by degree method of CytoHubba. [Click here to view] |
Analysis of the PPI network and hub gene selection
Analysis of the PPI network (confidence level of 0.4) was conducted on PS, which consist of 36 nodes, 113 edges, a PPI enrichment value of < 1.10e–16, and an average local clustering coefficient of 0.542 (Fig. 2A). The top 20 genes with the highest degree scores were identified, including SRC, estrogen receptor (ESR1), kinase insert domain receptor (KDR), matrix metallopeptidase 2 (MMP2), ATP-binding cassette sub-family G member 2 (ABCG2), and PIK3CG (Fig. 2B and Table 2). These results indicated that those genes have a pivotal role in the PPI network, making them strong candidates for target genes. In addition, the results are useful for selecting the candidate of target genes for further genetic alterations analysis using cBioPortal.
Analysis of genetic alterations of potential target genes
Analysis of genetic alterations was carried out to investigate the relationship between the resistance of CRC and sinensetin efficacy. Seven potential therapeutic targets of sinensetin against chemoresistance in colon cancer (PS), including ABCG2, ATP-binding cassette sub-family B member 1 (ABCB1), BRAF, MET, PDGFRB, GSK3B, and PIK3CG, were subjected to genetic alteration analyses using cBioPortal across CRC studies. PIK3CG, BRAF, GSK3B, MET, and PDGFRB were selected from KEGG pathway enrichment analysis. ABCG2, ABCB1, PDGFRB, GSK3B, and PIK3CG were selected on the basis of the highest degree score using CytoHubba. Out of 10 CRC studies, one study, namely the Dana-Farber Cancer Institute (DFCI) 2016 (Giannakis et al., 2016), was selected for further investigation (Fig 3A).
OncoPrint analysis of the DFCI 2016 study showed that genetic alterations in each target gene were found in 0.8%–21% of samples from patients with CRC, including ABCG2 (1.6%), ABCB1 (4%), BRAF (21%), MET (2.6%), PDGFRB (3%), GSK3B (0.8%), and PIK3CG (7%), in which most gene alterations were classified as missense mutations (putative driver) (Fig. 3B). OncoPrint provides an overview of genetic alterations in selected genes where the type of genetic alterations is highlighted per sample. A missense mutation is a change in a single base pair that causes amino acid substitution that produces different proteins and can render protein malfunctioning, including chemoresistance development (Zhang et al., 2017). The driver mutations can support the growth of cancer cells for their neoplastic transformation and chemoresistance development (Lønning and Knappskog, 2013). Activation of ABCG2 was found in CRC resistance to irinotecan (Tuy et al., 2016) and cisplatin (Chen et al., 2017a). Mutant BRAF, namely BRAFV600E, which occurs in approximately 8%–10% of patients with CRC, leads to dysregulation of the mitogen-activated protein kinase (MAPK) pathway (Zhang et al., 2018a). Mutation in BRAF is associated with resistance of metastatic CRC to anti-epidermal growth factor receptor antibodies (Sanz-Garcia et al., 2017). Mutation in PDGFR was in acute lymphoblastic leukemia resistance to tyrosine kinase inhibitor (Zhang et al., 2018b). Further research on the genetic alterations of PS in CRC patients is required.
Further analysis of mutual exclusivity showed that five gene pairs, including BRAF-MET, BRAF-PIK3CG, GSK3B-PIK3CG, ABCG2-PIK3CG, and ABCG2-ABCB1 exhibited significant cooccurrence (p < 0.05) in a CRC study according to the DFCI 2016 project (Table 3), which indicated the pivotal role of BRAF, PIK3CG, and ABCG2 in treatment with sinensetin. The role of each gene in CRC chemoresistance will be discussed in the following section. Moreover, genetic alterations of the query genes affected several pathways, including COADREAD-2012-RTK-RAS-PI(3)K, that regulate proliferation, cell survival, and translation (Fig. 3C). In addition, the results highlighted the importance of genetic alterations in BRAF in those signaling pathways. The results are supported by a previous study which showed that PI3K/AKT signaling is important in the development of CRC (Semba et al., 2002).
The role of PS in CRC chemoresistance
Seven potential therapeutic targets of sinensetin action against chemoresistance in colon cancer (PS), including ABCG2, ABCB1, BRAF, MET, PDGFRB, GSK3B, and PIK3CG, were identified. In the following sections, we will review the role of each gene in CRC chemoresistance. ABCG2, also known as adenosine triphosphate (ATP) binding cassette subfamily G member 2 or breast cancer resistance protein, and ABCB1, also known as ATP binding cassette subfamily B member 1 or P-glycoprotein, are members of the ABC transporter efflux transporter group whose function is to remove or pump drugs from within the cell to outside the cell, thereby reducing the concentration of intracellular drugs (Chen et al., 2016).
B-Raf protooncogene (BRAF) encodes a protein that belongs to the RAF proto-oncogene serine/threonine-protein kinase family of serine/threonine protein kinases (Cope et al., 2020). BRAF is one of the protein components of the MAPK pathway, also known as the extracellular-signal-regulated kinase (ERK) signaling pathway (McCain, 2013). The MAPK signaling pathway communicates with other pathways, for example, PI3K/AKT and mTOR (Burotto et al., 2014). MAPK signaling is important for the maintenance of cancer stem cell properties in CRC (Corcoran et al., 2018).
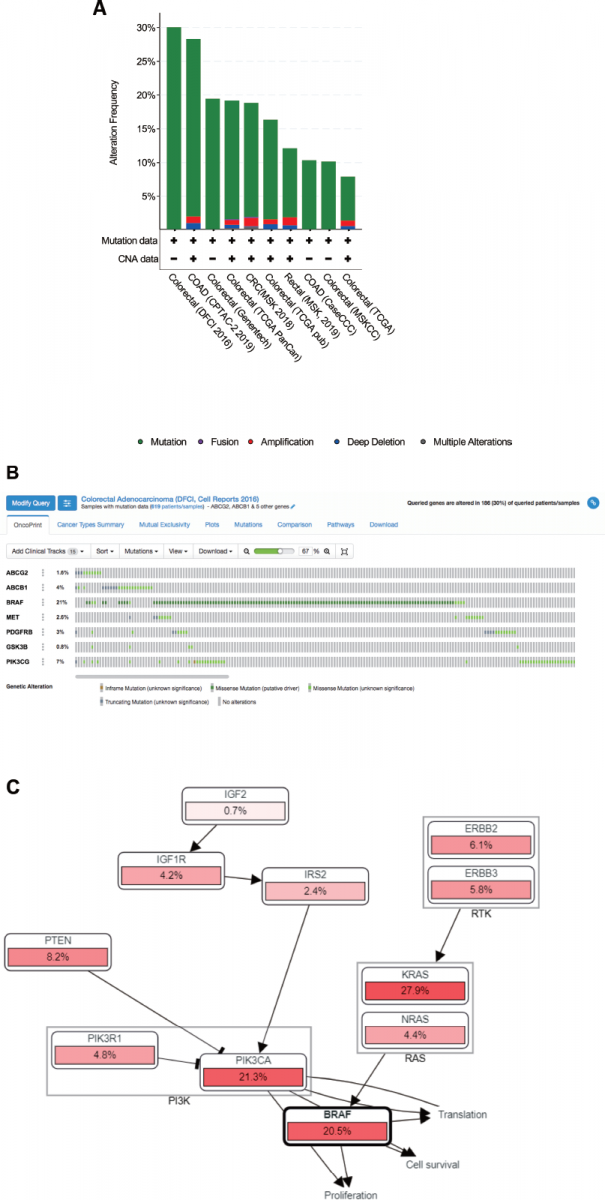 | Figure 3. (A) Overview of genetic changes in ABCG2, ABCB1, BRAF, MET, PDGFRB, GSK3B, and PIK3CG across 10 CRC studies, as analyzed by cBioportal. (B) Summary of alterations in ABCG2, ABCB1, BRAF, MET, PDGFRB, GSK3B, and PIK3CG across CRC patients using a study from Giannakis et al., 2016. (C) Pathway related to genetic alterations of ABCG2, ABCB1, BRAF, MET, PDGFRB, GSK3B, and PIK3CG across CRC patients using a study from Giannakis et al., 2016. [Click here to view] |
MET, also known as mesenchymal-epithelial transition factor gene or MET protooncogene, encodes a member of the receptor tyrosine kinase family of proteins (Gonzalez-Angulo et al., 2013). Upon binding to its ligand, namely hepatocyte growth factor (HGF), MET induces dimerization leading to activation of intracellular signaling, which is involved in cell proliferation, invasion, and migration (Organ and Tsao, 2011). Moreover, the same author stated that MET signaling also communicates with other intracellular signaling mechanisms, including the PI3K/ AKT and MAPK pathways (Organ and Tsao, 2011).
PDGFRB, which is expressed by tumor-associated fibroblasts, is involved in the cellular processes of angiogenesis, tumor growth, and metastasis in colon cancer cells (Takigawa et al., 2016). In addition, PDGFRB is involved in CRC progression via the mechanisms of platelet activation and transforming growth factor beta signaling (Steller et al., 2013). A recent study demonstrated that the platelet-derived growth factor receptor is required for epithelial to mesenchymal transition (EMT), and inhibition of PDGFRB with tyrosine kinase inhibitor effectively inhibits colon cancer progression (Olsen et al., 2019).
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma (PIK3CG), or p110 gamma, is a class I catalytic subunit of PI3K (Arthur and Uzairu, 2019). PIK3CG, a catalytic subunit of PI3K, is important for regulating PI3K/AKT signaling in the development of colon cancer (Semba et al., 2002).
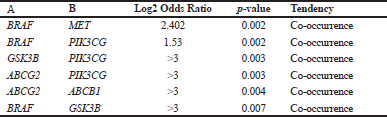 | Table 3. Additional mutual exclusivity analysis of selected genes. [Click here to view] |
Proposed mechanism of sinensetin action against chemoresistance in colon cancer
In the following section, we discussed the proposed mechanism of sinensetin against colon cancer chemoresistance. ABCB1 and ABCG2 are responsible for chemoresistance to selonsertib (Ji et al., 2019). Inhibition of ABC transporters is a strategy to overcome chemoresistance in colon cancer cells (Wang et al., 2015). A previous study showed that sinensetin overcame doxorubicin resistance in vincristine-resistant leukemic cells by reversing P-glycoprotein (Choi et al., 2002). Further research on the role of sinensetin in overcoming chemoresistance against colorectal cancer by targeting ABCB2 and ABCB1 is required.
A previous study showed that the upregulation of the MAPK signaling pathway was found in liver metastasis of CRC cells (Tang et al., 2019). Moreover, CRC resistance to BRAF inhibitor is mediated by the activation of Wnt/β-catenin signaling (Chen et al., 2018). However, the effect of sinensetin on overcoming CRC by targeting BRAF and MAPK signaling remains unclear.
MET not only plays an important role in the progression but is also a potential target for the treatment of CRC (Song et al., 2017). Overexpression of c-MET is an indicator of poor prognosis in patients with CRC (Lee et al., 2018). However, the effect of sinensetin on overcoming CRC by targeting MET remains unclear.
A study showed that activation of the Notch signaling pathway was found in PDGF-induced EMT and colon cancer progression (Chen et al., 2017b). The role of sinensetin in PDGFRB and its signaling awaits further investigation. A previous study showed that aberrant activation of Wnt/ß-catenin signaling was found in CRC cells (He et al., 2016). However, the effect of sinensetin on overcoming CRC by targeting GSK3B remains unclear.
A previous study showed that suppression of PIK3CG gene expression is important for blocking PI3K/AKT signaling, which leads to the inhibition of CRC progression (Semba et al., 2002). Another study showed that sinensetin can overcome chemoresistance in human gallbladder cancer cells by inhibiting the PTEN/PI3K/AKT signaling pathway (Liu et al., 2017). The effect of sinensetin on overcoming CRC chemoresistance by targeting PI3K/AKT awaits further investigation.
The present study revealed potential targets and molecular mechanisms of sinensetin in overcoming chemoresistance in CRC. Since the results of this study were derived from bioinformatics analysis, further in vitro and in vivo studies are required to validate the results. Further research using samples from patients with colorectal cancer is also needed to develop sinensetin as a combinatorial agent of chemotherapy in colorectal cancer treatment.
CONCLUSION
In conclusion, sinensetin potentially targets ABCG2, ABCB1, BRAF, MET, PDGFRB, GSK3B, and PIK3CG. Moreover, the PI3K/AKT and MAPK signaling pathways are potential target pathways of sinensetin action. The results of the present study await validation in subsequent experiments.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
FUNDING
This work was supported by the Penelitian Dasar Unggulan Perguruan Tinggi (PDUPT) 2020 from Ministry of Research and Technology, National Agency for Research and Innovation, Republic of Indonesia, Contract No. 1669/UN1/ DITLIT/DIT-LIT/PT/2020.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
REFERENCES
Androutsopoulos VP, Ruparelia K, Arroo RR, Tsatsakis AM, Spandidos DA. CYP1-mediated antiproliferative activity of dietary flavonoids in MDA-MB-468 breast cancer cells. Toxicology, 2009; 264(3):162–70. CrossRef
Arthur DE, Uzairu A. Molecular docking studies on the interaction of NCI anticancer analogues with human Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit. J King Saud Univ Sci, 2019; 31(4):1151–66. CrossRef
Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: a new perspective. Cancer, 2014; 120(22):3446–56. CrossRef
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov, 2012; 2(5):401–4. CrossRef
Chen B, Zhang D, Kuai J, Cheng M, Fang X, Li G. Upregulation of miR-199a/b contributes to cisplatin resistance via Wnt/β-catenin-ABCG2 signaling pathway in ALDHA1(+) colorectal cancer stem cells. Tumour Biol, 2017b; 39(6):1010428317715155. CrossRef
Chen G, Gao C, Gao X, Zhang DH, Kuan SF, Burns TF, Hu J. Wnt/β-catenin pathway activation mediates adaptive resistance to BRAF inhibition in colorectal cancer. Mol Cancer Ther, 2018; 17(4):806–13. CrossRef
Chen J, Yuan W, Wu L, Tang Q, Xia Q, Ji J, Liu Z, Ma Z, Zhou Z, Cheng Y, Shu X. PDGF-D promotes cell growth, aggressiveness, angiogenesis and EMT transformation of colorectal cancer by activation of Notch1/Twist1 pathway. Oncotarget, 2017a; 8(6):9961–73. CrossRef
Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, Hu T, Jiang L, Li J. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: a review of the past decade. Cancer Lett, 2016; 370(1):153–64. CrossRef
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol, 2014; 8 Suppl 4:S11. CrossRef
Choi CH, Sun KH, An CS, Yoo JC, Hahm KS, Lee IH, Sohng JK, Kim YC. Reversal of P-glycoprotein-mediated multidrug resistance by 5,6,7,3',4'-pentamethoxyflavone (Sinensetin). Biochem Biophys Res Commun, 2002; 295(4):832–40. CrossRef
Cope NJ, Novak B, Liu Z, Cavallo M, Gunderwala AY, Connolly M, Wang Z. Analyses of the oncogenic BRAF(D594G) variant reveal a kinase-independent function of BRAF in activating MAPK signaling. J Biol Chem, 2020; 295(8):2407–20. CrossRef
Corcoran RB, André T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, Hollebecque A, McRee AJ, Siena S, Middleton G, Muro K, Gordon MS, Tabernero J, Yaeger R, O'Dwyer PJ, Humblet Y, De Vos F, Jung AS, Brase JC, Jaeger S, Bettinger S, Mookerjee B, Rangwala F, Van Cutsem E. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF(V600E)-mutant colorectal cancer. Cancer Discov, 2018; 8(4):428–43. CrossRef
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucl Acids Res, 2019; 47(W1):W357–64. CrossRef
Danışman KF, Birman H, Candöken E, BilgiÅŸ GS, MelikoÄŸlu G, Kuruca S. Cytotoxic effects of some flavonoids and imatinib on the K562 chronic myeloid leukemia cell line: data analysis using the combination index method. Balkan Med J, 2019; 36(2):96–105. CrossRef
Deng Y. Rectal cancer in Asian vs. Western countries: why the variation in incidence? Curr Treat Options Oncol, 2017; 18(10):64. CrossRef
Dong Y, Ji G, Cao A, Shi J, Shi H, Xie J, Wu D. Effects of sinensetin on proliferation and apoptosis of human gastric cancer AGS cells. Zhongguo Zhong Yao Za Zhi, 2011; 36(6):790–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal, 2013; 6(269):l1. CrossRef
Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res, 2014; 42:W32–8. CrossRef
Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, Sukawa Y, Stewart C, Rosenberg M, Mima K, Inamura K, Nosho K, Nowak JA, Lawrence MS, Giovannucci EL, Chan AT, Ng K, Meyerhardt JA, Van Allen EM, Getz G, Gabriel SB, Lander ES, Wu CJ, Fuchs CS, Ogino S, Garraway LA. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep, 2016; 15(4):857–65. CrossRef
Gonzalez-Angulo AM, Chen H, Karuturi MS, Chavez-MacGregor M, Tsavachidis S, Meric-Bernstam F, Do KA, Hortobagyi GN, Thompson PA, Mills GB, Bondy ML, Blumenschein GR Jr. Frequency of mesenchymal-epithelial transition factor gene (MET) and the catalytic subunit of phosphoinositide-3-kinase (PIK3CA) copy number elevation and correlation with outcome in patients with early stage breast cancer. Cancer, 2013; 119(1):7–15. CrossRef
He F, Chen H, Yang P, Wu Q, Zhang T, Wang C, Wie J, Chen Z, Hu H, Li W, Cao J. Gankyrin sustains PI3K/GSK-3β/β-catenin signal activation and promotes colorectal cancer aggressiveness and progression. Oncotarget, 2016; 7(49):81156–71. CrossRef
Hu T, Li Z, Gao CY, Cho CH. Mechanisms of drug resistance in colon cancer and its therapeutic strategies. World J Gastroenterol, 2016; 22(30):6876–89. CrossRef
Huang B, Zhai M, Qin A, Wu J, Jiang X, Qiao Z. Sinensetin flavone exhibits potent anticancer activity against drug-resistant human gallbladder adenocarcinoma cells by targeting PTEN/PI3K/AKT signalling pathway, induces cellular apoptosis and inhibits cell migration and invasion. J BUON, 2020; 25:1251–56.
Huang da W, Sherman BT, Lempicki R. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res, 2009; 37(1):1–13. CrossRef
Ji N, Yang Y, Cai CY, Lei ZN, Wang JQ, Gupta P, Shukla S, Ambudkar SV, Kong D, Chen ZS. Selonsertib (GS-4997), an ASK1 inhibitor, antagonizes multidrug resistance in ABCB1- and ABCG2-overexpressing cancer cells. Cancer Lett, 2019; 440–441, 82–93. CrossRef
Kim SM, Ha SE, Lee HJ, Rampogu S, Vetrivel P, Kim HH, Venkatarame Gowda Saralamma V, Lee KW, Kim GS. Sinensetin induces autophagic cell death through p53-related AMPK/mTOR signaling in hepatocellular carcinoma HepG2 cells. Nutrients, 2020; 12(8):1–17. CrossRef
Kozovska Z, Gabrisova V, Kucerova L. Colon cancer: cancer stem cells markers, drug resistance and treatment. Biomed Pharmacother, 2014; 68(8):911–16. CrossRef
Lee SJ, Lee J, Park SH, Park JO, Lim HY, Kang WK, Park YS, Kim ST. c-MET Overexpression in colorectal cancer: a poor prognostic factor for survival. Clin Colorectal Cancer, 2018; 17(3):165–9. CrossRef
Liu K, Li J, Wu X, Chen M, Luo F, Li J. GSK-3β inhibitor 6-bromo-indirubin-3'-oxime promotes both adhesive activity and drug resistance in colorectal cancer cells. Int J Oncol, 2017; 51(6):1821–30. CrossRef
Lønning PE, Knappskog S. Mapping genetic alterations causing chemoresistance in cancer: identifying the roads by tracking the drivers. Oncogene, 2013; 32(46):5315–30. CrossRef
Luo J. Glycogen synthase kinase 3beta (GSK3beta) in tumorigenesis and cancer chemotherapy. Cancer Lett, 2009; 273(2):194– 200. CrossRef
Manoukian AS, Woodgett JR. Role of glycogen synthase kinase-3 in cancer: regulation by Wnts and other signaling pathways. Adv Cancer Res, 2002; 84:203–29. CrossRef
McCain J. The MAPK (ERK) pathway: investigational combinations for the treatment of BRAF-mutated metastatic melanoma. P T, 2013; 38(2):96–108.
O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol, 2016; 13(12): 691–706. CrossRef
Olsen RS, Dimberg J, Geffers R, Wågsäter D. Possible role and therapeutic target of PDGF-D signalling in colorectal cancer. Cancer Invest, 2019; 37(2):99–112. CrossRef
Organ SL, Tsao M-S. An overview of the c-MET signaling pathway. Ther Adv Med Oncol, 2011;3(1 Suppl):S7–19. CrossRef
Pereira CV, Duarte M, Silva P, Bento da Silva A, Duarte CMM, Cifuentes A, García-Cañas V, Bronze MR, Albuquerque C, Serra AT. Polymethoxylated flavones target cancer stemness and improve the antiproliferative effect of 5-fluorouracil in a 3D cell model of colorectal cancer. Nutrients, 2019;11(2):326. CrossRef
Polastro L, El Hachem G, Hendlisz A. Pseudoadjuvant chemotherapy in resectable metastatic colorectal cancer. Curr Opin Oncol, 2018; 30(4):269–75. CrossRef
Sanz-Garcia E, Argiles G, Elez E, Tabernero J. BRAF mutant colorectal cancer: prognosis, treatment, and new perspectives. Ann Oncol, 2017; 28(11):2648–57. CrossRef
Semba S, Itoh N, Ito M, Youssef EM, Harada M, Moriya T, Kimura W, Yamakawa M. Down-regulation of PIK3CG, a catalytic subunit of phosphatidylinositol 3-OH kinase, by CpG hypermethylation in human colorectal carcinoma. Clin Cancer Res, 2002; 8(12): 3824–31.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 2003; 13(11):2498–504. CrossRef
Song N, Qu X, Liu S, Zhang S, Liu J, Qu J, Zheng H, Liu Y, Che X. Dual inhibition of MET and SRC kinase activity as a combined targeting strategy for colon cancer. Exp Ther Med, 2017;14(2):1357–66. CrossRef
Steller EJ, Raats DA, Koster J, Rutten B, Govaert KM, Emmink BL, Snoeren N, van Hooff SR, Holstege FC, Maas C, Borel Rinkes IH, Kranenburg O. PDGFRB promotes liver metastasis formation of mesenchymal-like colorectal tumor cells. Neoplasia, 2013;15(2): 204–17. CrossRef
Szklarczyk D, Franceschini A, Wyder S, Forslund, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res, 2015;43(Database issue):D447–52. CrossRef
Takigawa H, Kitadai Y, Shinagawa K, Yuge R, Higashi Y, Tanaka S, Yasui W, Chayama K. Multikinase inhibitor regorafenib inhibits the growth and metastasis of colon cancer with abundant stroma. Cancer Sci, 2016;107(5):601–8. CrossRef
Tang B, Liang W, Liao Y, Li Z, Wang Y, Yan C. PEA15 promotes liver metastasis of colorectal cancer by upregulating the ERK/MAPK signaling pathway. Oncol Rep, 2019; 41(1):43–56. CrossRef
Tuy HD, Shiomi H, Mukaisho KI, Naka S, Shimizu T, Sonoda H, Mekata E, Endo Y, Kurumi Y, Sugihara H, Tani M, Tani T. ABCG2 expression in colorectal adenocarcinomas may predict resistance to irinotecan. Oncol Lett, 2016; 12(4):2752–60. CrossRef
Wang Z, Xu Y, Meng X, Watari F, Liu H, Chen X. Suppression of c-Myc is involved in multi-walled carbon nanotubes' down-regulation of ATP-binding cassette transporters in human colon adenocarcinoma cells. Toxicol Appl Pharmacol, 2015; 282(1):42–51. CrossRef
Zhang M, Zhuang G, Sun X, Shen Y, Wang W, Li Q, Di W. TP53 mutation-mediated genomic instability induces the evolution of chemoresistance and recurrence in epithelial ovarian cancer. Diagn Pathol, 2017; 12(1):16. CrossRef
Zhang P, Kawakami H, Liu W, Zeng X, Strebhardt K, Tao K, Huang S, Sinicrope FA. Targeting CDK1 and MEK/ERK overcomes apoptotic resistance in BRAF-mutant human colorectal cancer. Mol Cancer Res, 2018a; 16(3):378–89. CrossRef
Zhang Y, Gao Y, Zhang H, Zhang J, He F, Hnízda A, Qian M, Liu X, Gocho Y, Pui CH, Cheng T, Wang Q, Yang JJ, Zhu X, Liu X. PDGFRB mutation and tyrosine kinase inhibitor resistance in Ph-like acute lymphoblastic leukemia. Blood, 2018b; 131(20):2256–61. CrossRef