INTRODUCTION
Klebsiella pneumoniae that is resistant to carbapenem antibiotic has emerged as a global threat over the years, largely due to the dissemination of carbapenem-hydrolyzing beta-lactamases (Cheepurupalli et al., 2017). On medical equipment, K. pneumoniae can grow as a biofilm during an infectious state. This is not only fundamental for the establishment of infection but it also makes the treatment very difficult. K. pneumoniae remains one among the top eight significant pathogens causing nosocomial infection (Zowawi et al., 2015). Pneumonia caused by K. pneumoniae is characterized by an exacerbated inflammatory response, associated with excessive neutrophil and macrophage infiltration, high production of pro-inflammatory cytokines, and severe lung injury (Marwa et al., 2017).
Importantly, K. pneumoniae has emerged as a new multidrug-resistant (MDR) variety of human pathogens that can have drastic consequences on healthcare worldwide. All the modes of drug resistance have been exhibited by K. pneumoniae, producing extended-spectrum β-lactamase and carbapenemase enzymes (Zowawi et al., 2015). Imipenem is recommended as a first-line therapy for MDR Gram-negative bacteria (Marwa et al., 2017).
Hence, there is a dire need for novel molecules to fight against Carbapenem resistant - Klebsiella pneumoniae (CR-KP). Actinobacteria are Gram-positive filamentous bacteria, where the biotechnological interest of these microbes resides in their ability to produce different bioactive compounds. In fact, actinobacteria are the producer of about 45% of the total microbial bioactive products (Ayoubi et al., 2018). Notably, actinobacteria from rare ecosystems, like marine environment, are a reliable resource for novel anti-infective molecules. The present study is attempted to determine the in vitro anti-infective efficacy of marine actinobacterial strains against carbapenem-resistant K. pneumoniae ATCC BAA-1705.
MATERIALS AND METHODS
Description of actinobacterial strains
Totally, 98 marine-derived actinobacterial strains were obtained from the actinobacterial consortium of Centre for Drug Discovery & Development, Sathyabama Institute of Science and Technology, Tamil Nadu, India, for the current study. In that, 9 strains were isolated from Pichavaram mangrove sediments (Lat 11° 25′ 21.1944″ N; Long 79° 46′ 29.4024″ E), 5 strains were isolated from Vellar estuary (Lat. 11.8914° N; Long 77.9724° E), and 84 cultures were isolated from Andaman mangrove sediments (Lat 11.7401° N; Long 92.6586° E).
Screening of antibacterial activity by agar plug method
All the marine actinobacterial cultures were grown on International Streptomyces Project 2 (ISP2) agar medium for 7 days. Agar cylinders with 5 mm diameter were cut from the ISP2 agar medium inoculated with SACC E6 strain. Then, they were placed using sterile forceps over Muller–Hinton agar medium that was previously inoculated by the test pathogen. Plates were kept during the first 4 h at 4°C for a good diffusion of secondary metabolites that were produced, then incubated at 37°C for antibacterial activity against carbapenem-resistant K. pneumoniae ATCC BAA-1705 (Ahmed et al., 2018).
Effect of incubation period and fermentation method on bioactive metabolite production
The bioactive metabolite was produced by adopting submerged and solid state fermentation from strain SACC-E6. For the submerged production process, about 10% of strain SACC-E6 (48 hours old) inoculum was added to the ISP2 broth and kept in a shaking incubator for 15 days at 28°C. The supernatant Cell-free supernatant (CFS) from the fermentation broth was collected every 24 hours by centrifugation at 5,000 rpm for 10 minutes and its antibacterial activity was tested by a well diffusion method. In the agar surface fermentation, strain SACC-E6 was grown on Yeast Extract Malt Extract (YEME) agar plates for 15 days at 28°C. For every 24 hours, the agar plug from YEME agar medium was taken and its antibacterial activity was tested.
Production of bioactive metabolites and testing for activity
Agar surface fermentation was carried out for the production of bioactive metabolites from SACC-E6 on a large scale. After scrapping the mycelial growth, the YEME agar medium was cut into pieces and extracted by solid liquid extraction using n-Hexane, methanol, chloroform, and ethyl acetate. The crude extracts were collected, concentrated, and tested for its activity by the disk diffusion method at 100 µg/disk concentration (Radhakrishnan et al., 2014; Selvameenal et al., 2009).
Partial purification of the bioactive molecule and bioautography
The bioactive molecule was purified by using thin layer chromatography (TLC) using silica gel-coated chromatography sheets (50 × 20 cm in size). Organic solvents, viz. methanol, chloroform, ethyl acetate, n-butanol, and n-hexane, were used in different proportions to find the best solvent system for best separation of bioactive metabolites present in the crude extract.
Crude extract was placed as a spot using the glass capillary tube on the silica gel-coated sheet. The TLC sheet was then placed in the TLC tank and allowed to develop up to its three-quarter size and the spots were visualized with iodine vapors. The Rf value of the spot separated on the TLC plate was determined (Selvameenal et al., 2009; Irena and Edyta, 2010).
The active compounds separated in TLC were detected by the bioautography method (Selvameenal et al., 2009). The developed chromatogram was placed in a nutrient agar seeded with test pathogen and 0.1% TTC and incubated for 24 hours at 37°C.
Characterization of potential strain
Cultural properties, like rate of growth, consistency, color of aerial mycelium, and pigmentation of strain SACC E6, were studied ISP2 agar medium. Microscopic features like of the substrate and aerial mycelium and spore chain arrangement were studied under the bright field microscope at 40× magnifications.
Furthermore, cultural characteristics such as the growth of strain E6 on different cultural media (ISP1–ISP7), utilization of different carbon and nitrogen sources tolerance to different pH, temperature, and NaCl concentration were also studied (Mohana and Radhakrishnan, 2014; Shirling and Gottileb, 1966).
Sequencing of 16S rRNA gene and phylogenetic analysis
The total genomic DNA was extracted from the strain SACC-E6 using solute ready genomic DNA kit. The 16S rRNA of SACC-E6 was amplified by using the primer pairs: 27F 5′AGAGTTTGATCMTGGCTCAG3′ (forward) and 1481R 5′TACGGYTACCTTGTTACGACTT3′ (reverse) (Gothwal et al., 2007). The Polymerase Chain Reaction (PCR) product of SACC63 was sequenced and aligned with similar sequences available in GenBank using MEGA 6 program (Saitou and Nei, 1987). The phylogenetic tree was constructed using aligned sequences by following the neighbor-joining algorithm (Saitou and Nei, 1987) in MEGA 6 software. The bootstrap estimation (Felsenstein, 1985) was used to determine the confidence of different branches in the phylogenetic tree. The partial nucleotide sequence of SACC E6 16S rRNA gene was deposited in GenBank database (Meeta et al., 2018).
RESULTS
Preliminary screening
In the preliminary screening, 31.63% actinobacterial strains were inhibited by carbapenem-resistant K. pneumoniae. Five actinobacterial strains, viz. SACC-E6, SACC-96, SACC-97, SACC-160, and SACC-162, showed a maximum zone of inhibition of more than 15 mm (Fig. 1). The strain SACC-E6 which showed maximum activity (22.7 ± 1.1 mm) against CR-KP was selected as potential for further studies (Fig. 2A).
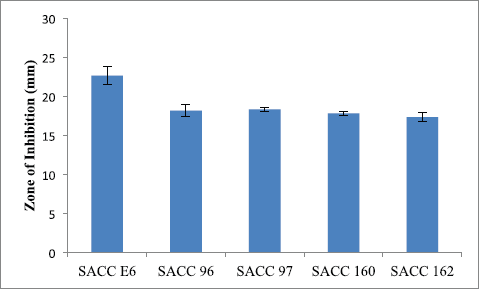 | Figure 1. Antagonistic activity of the potential cultures against carbapenem-resistant Klebsiella pneumonia. [Click here to view] |
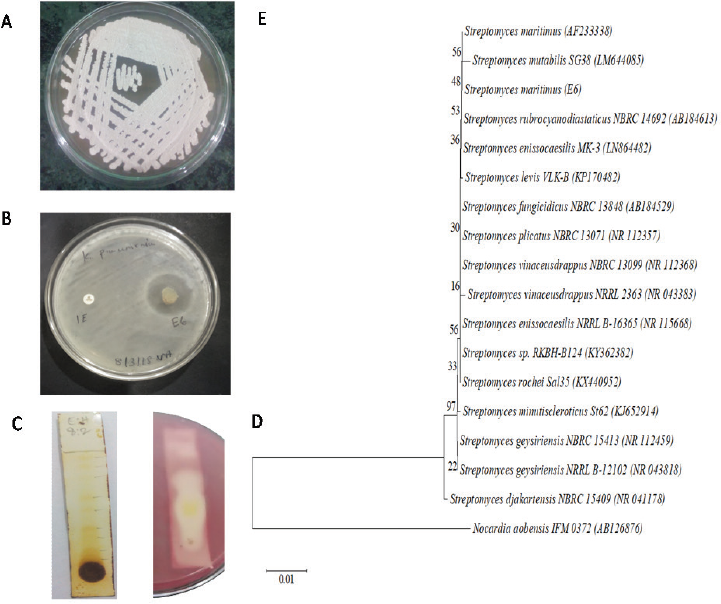 | Figure 2. (A) Colony morphology of the strain E6 on ISP2 agar. (B) Antimicrobial activity of E6 against CR-KP. (C) TLC separation of the crude methanol extract. (D) Detection of active compound by TLC bioautogram. (E) Phylogenetic tree of strain SACC-E6. [Click here to view] |
Extraction of the bioactive compound
Anti-infective molecules were best produced in agar surface fermentation (solid state fermentation) from the day 3 onward till the day 12, with a gradual increase in the diameter of zone of inhibition against CR-KP (Fig. 3).
Among the solvents used, the bioactive metabolites were extracted with methanol and produced a maximum 24 mm zone of inhibition against the test pathogen in 100 µg/disk concentration (Fig. 3).
Bioassay guided purification
The partially purified bioactive compound was found in two adjacent zones with Rf values of 0.52 and 0.41, respectively, against K. pneumoniae, in which the chromatogram was developed with the ratio of hexane : ethyl acetate in a 6:4 ratio, which were diffused together. Hence, further purification is needed to separate individual biomolecules (Fig. 2C).
Taxonomical investigation of SACC-E6
Strain SACC-E6 produced gray-colored powdery colonies with non-fragmented mycelium in ISP2 agar. This strain utilized a wide range of substrates and also grew well on different cultural media (Table 1). PCR amplification, Basic Local Alignment Search Tool (BLAST), phylogenetic analysis, and neighbor-joining method indicated that the SACC-E6 sequence showed 99.9% similarity with Streptomyces maritimus and received National Center for Biotechnology Information (NCBI) Genbank accession number MH171652 (Fig. 2D).
DISCUSSION
The discovery of new drugs is an indispensable process to combat those emerging resistant pathogens (Berdy, 2012). Actinobacteria from marine environment are documented as a promising but under explored resource for novel anti-infective molecules (Claverías et al., 2015). Marine-derived actinobacteria are recognized as the gifted source for novel bioactive metabolites with different biological activities (Velho-Pereira and Kamat, 2011). Apart from this, natural compounds were also synthesized and studied against CRKP, like synthetic peptides, TPA3, and TP4, which exhibited activity against CR-KP, indicating its potential to combat drug resistance (Pavan Kumar et al., 2018). According to Berdy (2012), a great number of antibiotics exhibit exclusively inhibited Gram-positive bacteria. But Gram-negative bacteria are inhibited by only 1.5% of antibiotics. However, in the present study, around 31.63% of marine actinobacterial strains showed activity against CR-KP. Similarly, Cheepurupalli et al., (2017) reported that 51 different actinobacteria were isolated from rhizosphere soil and Streptomyces sp. ASK2 was found to have a promising activity against CR-KP. Strain SACC-E6 showed good growth on ISP2 broth during submerged fermentation. But the supernatant showed no inhibition against the test organisms. However, in the agar surface fermentation, strain SACC-E6 showed good growth and the methanol extract obtained from the fermented medium showed activity against CR-KP. Similarly, Valan Arasu et al., (2014) reported that the methanolic extract of the actinobacteria strain exhibited more antimicrobial activity than other solvent extracts. Most of the secondary metabolites from actinobacteria were carried out using submerged fermentation processes, where they exhibited diverse morphological forms (Braun and Vechtlifshitz, 1991; Treskatis et al., 1997). Environmental factors like medium components and shear stress are reported to influence the morphology of actinobacteria (O’Cleirigh et al., 2005). Further optimization of the medium component and concentration may be needed to produce the compound from strain SACC-E6 by submerged fermentation. In the present study, the only the extracellular metabolites extracted in methanol showed antimicrobial activity. The results indicate the highly polar nature of the active compound produced by the actinobacterial strain SACC-E6. The methanol fraction of strain SACC-E6 showed promising activity against K. pneumoniae. This observation paves the way for the exploration of the strain SACC-E6 for applications in clinical settings.
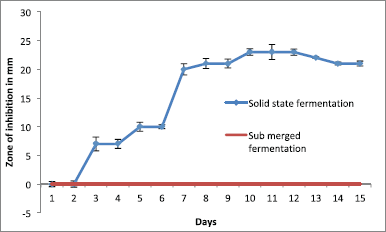 | Figure 3. Effect of fermentation method and incubation period on the bioactive metabolite. [Click here to view] |
The sequencing results of 16S ribosomal RNA (16S rRNA) gene has also revealed that the strain SACC-E6 belongs to a species of the genus Streptomyces. The BLAST analysis showed 99% similarity to S. maritimus. The findings of the present study shows that the S. maritimus SACC-E6 is a promising source for bioactive metabolite to fight against clinically important carbapenem-resistant K. pneumoniae.
 | Table 1. Morphological, cultural, and phenotypic characteristics of the potential strain. [Click here to view] |
CONCLUSION
The methanolic extract of S. maritimus SACC-E6 was found to be effective against CR-KP and also two bioactive molecules from SACC-E6 were purified through TLC and checked activity by bioautogram. However, further studies will be necessary to determine the structure of the active compounds.
ACKNOWLEDGMENTS
The authors thank the authorities of Sathyabama Institute of Science and Technology, Tamil Nadu, for the research facilities provided as well as the Department of Biotechnology, Government of India, New Delhi, for their support in the form of research grant (BT/PR5426/AAQ/3/599/2012 dated 16-03-15).
CONFLICT OF INTEREST
All the authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
ETHICAL APPROVALS
This study does not involve the use of animals or human subjects..
REFERENCES
Ahmed N, Ayoub K, Asma A, Yedir O, Lachen H. Endophytic actinobacteria of medicinal plant Aloe vera; isolation, antimicrobial, antioxidant, cytotoxicity assays and taxonomic study. Asian Pac J Trop Biomed, 2018; 8(10):513–8. CrossRef
Ayoubi H, Mouslim A, Moujabbir S, Amine S, AzougarIman, MJ, Menggad M. Isolation and phenotypic characterization of actinomycetes from Rabat neighborhood soil and their potential to produce bioactive compounds. Afr J Microbiol Res, 2018; 12(8):186–91. CrossRef
Berdy J. Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot, 2012; 65(8):385–95. CrossRef
Braun S, Vechtlifshitz SE. Mycelial morphology and metabolite production. Trends Biotechnol, 1991; 9(2):63–8. CrossRef
Cheepurupalli L, Raman T, Rathore SS, Ramakrishnan J. Bioactive molecule from Streptomyces sp. mitigates MDR Klebsiella pneumoniae in zebrafish infection model. Front Microbiol, 2017; 8:614. CrossRef
Claverías FP, Undabarrena A, González M, Seeger M, Cámara B. Culturable diversity and antimicrobial activity of actinobacteria from marine sediments in valparaíso bay, Chile. Front Microbiol, 2015; 6:737. CrossRef
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 1985; 39(4):783–91. CrossRef
Gothwal RK, Nigam VK, Krishna Mohan M. Extraction of bulk DNA from Thar Desert soils for optimization of PCR DGGE based microbial community analysis. Electron J Biotechnol, 2007; 10(3):400–8. CrossRef
Irena C, Edyta G. Bioautography detection in thin-layer chromatography. J Chromatogr A, 2010; 1218:2684–91. CrossRef
Marwa M, Abdel-Aziz, Mohamed Y, Basma HA. Control of imipenem resistant-Klebsiella pneumoniae pulmonary infection by oral treatment using a combination of mycosynthesised Ag-nanoparticles and imipenem. J Radiat Res Appl Sci, 2017; 10:353–60. CrossRef
Meeta M, Kunjukrishnan KS, Ekta M, Thangathurai T, Rangasamy A, Gaurav S, Natesan S, Solomon RDJ, Polpass AJ., Biosynthetic potential of bioactive streptomycetes isolated from arid region of the Thar Desert, Rajasthan (India). Front Microbiol, 2018; 9:687. CrossRef
Mohana S, Radhakrishnan M. Streptomyces sp MA7 isolated from mangrove rhizosphere sediment effective against Gram negative bacterial pathogens. Int J Pharm Tech Res, 2014; 6(4):1259–64.
O’Cleirigh C, Casey JT, Walsh PK, O’Shea DG. Morphological engineering of Streptomyces hygroscopicus var. geldanus: regulation of pellet morphology through manipulation of broth viscosity. Appl Environ Microbiol, 2005; 68:305–310. CrossRef
Pavan Kumar JG, Gomathi A, Gothandam KM, Vasconcelos V. Bioactivity assessment of Indian Origin—Mangrove actinobacteria against candida albicans. Mar Drugs, 2018; 16(2):60. CrossRef
Radhakrishnan M, Pazhanimurugan R, Gopikrishnan V, Balagurunathan R, Vanajakumar. Streptomyces sp D25 isolated from Thar Desert soil, Rajasthan producing pigmented antituberculosis compound only in solid culture. J Pure Appl Microbiol, 2014; 8(1):333–7.
Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol, 1987; 4:406–25.
Selvameenal L, Radakrishnan M, Balagurunathan R. Antibiotic pigment from desert soil actinomycetes; biological activity, purification and chemical screening. Indian J Pharm Sci, 2009; 71:499–504. CrossRef
Shirling EB, Gottileb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol, 1966; 16:313–40. CrossRef
Treskatis S-K, Orgeldinger V, Wolf H, Gilles ED. Morphological characterization of filamentous microorganisms in submerged cultures by on-line digital image analysis and pattern recognition. Biotechnol Bioeng, 1997; 53(2):191–201. CrossRef
Valan Arasu M, Rejiniemon TS, Al-Dhabi NA, Duraipandiyan V, Agastian P, Huxley VAJ, Song CE, Chol KC. In-vitro antimicrobial potential of organic solvent extracts of novel actinomycetes isolated from forest soil. Afr J Biotechnol, 2014; 13(18):1891–7. CrossRef
Velho-Pereira S, Kamat NM. Antimicrobial screening of actinobacteria using a modified cross-streak method. Indian J Pharma Sci, 2011; 73(2):223. CrossRef
Zowawi HM, Harris PNA, Roberts MJ, Tambyah PA, Schembri MA, PezzaniMD. The emerging threat of MDR Gram negative bacteria in urology. Nat Rev Urol, 2015; 12:570–84. CrossRef