INTRODUCTION
Respiratory tract infections (RTIs) are one of the most common infectious diseases globally imposing a significant increase in morbidity and mortality in the developing countries. Most of the infections affect the upper respiratory tract and merely 5% constitute the lower RTIs (LRTIs). However, LRTIs are more serious and debilitating than upper RTIs (URTIs) (Shanmugam et al., 2015; Sharma et al., 2015; Taura et al., 2013). URTIs are common in individuals of all the age group and include Pharyngitis, Nasopharyngitis, Tonsillitis, Otitis Media, and Sinusitis. These infections are primarily caused by viral assault, secondary infection with bacteria, such as Haemophilus influenzae, Streptococcus pyogenes, Moraxella catarrhalis, Staphylococcus aureus, and Streptococcus pneumoniae, can complicate the illness (Kousalya et al., 2010; Wang et al., 2016). The LRTIs mainly affect bronchi and alveoli causing bronchitis and pneumonia. Gram positive bacteria such as Staphylococcus aureus, Streptococcus pneumonia and gram negative organisms such as H. influenzae, Pseudomonas species, Klebsiella species, Acinetobacter are the major culprits of LRTIs. Factors such as age, gender, season, smoking, immunosuppressive states link to the causatives of RTIs (Khan et al., 2014; Kumar et al., 2015).
There is an exponential increase in antibiotic resistance attributed to indiscriminate use of antibiotics, lack of monitoring of antibiotic susceptibility patterns, and cross infections. In India, most hospitals follow empirical therapy, which fails to work upon the emergence of resistance and adds additional cost to the patient and the healthcare sector. Therefore, it is of utmost importance to assess the antibiotic susceptibility pattern of local bacterial species which aids in developing an antibiotic policy. The aim of this study is to determine the antibiotic susceptibility pattern of bacterial isolates in patients with RTIs in our region.
MATERIALS AND METHODS
The study was conducted at Pediatrics, Intensive Care Unit, Female, Male medical ward and Outpatient department of two secondary care hospitals in the area. The study is an Open Label, Prospective, and Cross-sectional study. The Pediatrics and adult population of age 6–65 years old with a clear diagnosis of RTI were included in our study. Patients who have been taking antibiotics for infections other than respiratory infections and patients who are not willing to provide the informed consent were excluded from the study.
The Institutional Review Board approval was obtained (approval no: JSSCP/DPP/IRB/03/2017-18, approval date: 03/02/2018) before initiation of the study. Sputum and throat swab samples were collected from clinically diagnosed RTI in 50 patients by simple random sampling after obtaining the informed consent and the patient data were entered using the patient profile form. Out of the 50 samples collected by purposive sampling, 35 samples were throat swab and 15 were sputum samples. Sputum and Throat swab samples were collected using their respective standard procedures. Proper labeling was done and the samples collected were brought to the Microbiology lab within 20 minutes and inoculated in the nutrient broth. Following which they were incubated at 35°C ± 2°C for 24 hours for observation of bacterial growth (Fig. 1) (Balaji, 2015; Sharma, 2008). Out of the 50 samples, 41 were found to be positive of bacterial growth and nine samples showed no growth. The selective Media used were Cetrimide agar medium, Mannitol salt agar medium, Mac Conkey agar medium , and Blood agar medium (Fig. 2) (Balaji, 2015; Gazi et al., 2004; Kousalya et al., 2015). These Media were selected because of their ability to facilitate the growth of P. aeruginosa, S. aureus, Klebsiella pneumoniae, Escherichia coli, and S. pneumoniae, which are the causative organism for most RTIs. The Media were then incubated at 35°C ± 2°C for 24 hours to isolate the desired bacterial species. The identified organisms were tested for Antibiotic susceptibility using Kirby–Bauer Disc Diffusion method as per the standards of Clinical Laboratory Standards Guidelines (CLSI) (Balaji, 2015; Hudzicki, 2009).
 | Figure 1. Inoculation and incubation of samples in nutrient broth. [Click here to view] |
The readymade antibiotic discs were selected based on the commonly used antibiotics to treat RTIs in hospitals in Ooty. To increase precision, similar concentrations were used. The panel of antibiotic discs chosen for the isolated organisms includes Amikacin (AK 30 μg), Gentamicin (HLG 120 μg), Cefotaxime (CTX 30 μg), Ceftriaxone (CTR 30 μg), Azithromycin (AZM 15 μg), and Ciprofloxacin (CIP 1 μg) (Balaji, 2015; Hudzicki, 2009).
The Zone of inhibition was measured using a metal calliper and the results were interpreted according to the standard reference CLSI (Fig. 3). The results were then reported as sensitive, intermediate, and resistant according to the criteria set by CLSI. Descriptive statistics were used to present the distribution of samples, the growth of bacteria, gender, and age distribution (Balaji, 2015).
RESULTS
In our study, we collected about 50 samples, out of which 35 (70%) throat swab samples are collected from the patients with URTI and 15 (30%) sputum samples are collected from the patients with LRTI.
From the collected samples, 41 (82%) showed growth and 9 (18%) samples did not show any growth. Figure 4 shows equal gender distribution among male and female.
The highest predominant age group with RTI is patients between the age group 19 and 35 years as depicted in Figure 5.
Around three samples were collected in the month of January, 38 samples in the month of February and nine samples during March. Out of the male patients, 17 (34%) were smokers and all female patients, including 33 pediatric patients (66%), were non-smokers.
About 45 organisms were isolated in our study, among which 36 (80%) are Gram-positive and 9 (20%) Gram-negative organisms.
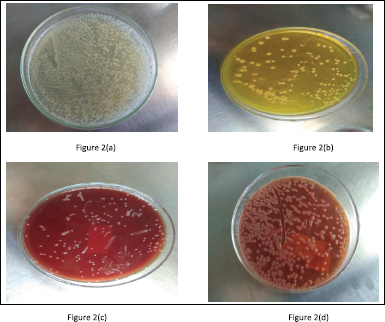 | Figure 2. Isolation of bacterial species. (a) Isolation of P. aeruginosa on Cetrimide agar. (b) Isolation of S. aureus on Mannitol Salt Agar medium. (c) Isolation of K. pneumoniae on blood agar. (d) Isolation of K. pneumoniae on MacConkey agar [Click here to view] |
The overall distribution of isolated bacterial species in our study is shown in Figure 6.
Antibiotic susceptibility testing
The most common causative organism found in our study is S. aureus and the sensitivity pattern of various antibiotics are as follows: AK (94.12%) is the most sensitive, followed by HLG (88%), AZM (82%), CTR (58.82%), CTX (47%), and the highly resistant antibiotic is CIP (53%). Isolated E. coli is sensitive to HLG (60%) followed by CIP (40%), AK (20%) and highly resistant to CTX (100%) followed by CTR (80%). Pseudomonas aeuroginosa is highly sensitive to AK (100%), HLG (100%), followed by CTR (50%), and resistance pattern observed in them were as follows; CIP (50%) and CTX (50%). Klebsiella pneumoniae had intermediate susceptibility to CTX (100%) and CTR (50%) is sensitive to this organism (Table 1). Even if Staphylococcus epidermidis has 38% occurrence, it is of no significance in our study as it is not a causative organism of RTI, but it is sensitive to almost all the antibiotics used in our study.
DISCUSSION
RTIs are one of the most common and the biggest threats to global health, which can affect anyone of any age in any country. It includes both upper and lower RTIs among which URTI is most common. Even though LRTIs contribute to a lesser percent of infections they are potentially fatal (Sharma et al., 2015; Taura et al., 2013). The rapid emergence of antibiotic resistant bacteria is making the antibiotic therapy ineffective and prolonging the hospital stay thereby causing economic burden and fatality in the patient. Our study, focus mainly on collecting data on the risk factors associated with RTIs, the prevalence of the causative organism responsible for RTIs and observing their susceptibility pattern to various antibiotics, such as AK, HLG, CIP, AZM, CTX, and CTR. During the 6 months of the study period, 50 samples were collected from respiratory tract infected patients, out of which, 70% were swab (URTIs) and 30% were sputum samples (LRTIs), this is contrary to a study conducted by Manikandan and Amsath (2013) study where the pattern was vice versa. This shows a marked difference in the distribution of URTI and LRTI. From the collected samples, 41 had growth, which is in accordance with sample distribution in Taura et al. (2013) study. The distribution of samples among male patients was 42%, female patients were 46%, and 12% in pediatrics is in accordance with El-Mahmood et al. (2010) and is opposing Taura et al. (2013). The prevalence of RTI in various age groups were as follows: 52% in 19–35 years, 22% in 50–65 years, 14% in 36–49 years, and 12% in 5–18 years, this similar pattern is seen in a study conducted by Taura et al. (2013). The highest prevalence of RTI in young and middle age group can be due to the high exposure of these age groups to the risk factors, such as smoking, alcoholism, and work exposure. All the samples were collected in the autumn season.
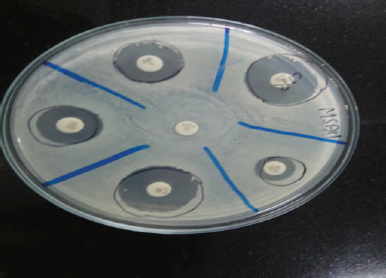 | Figure 3. Zone of inhibition by Kirby Bauer disc diffusion method. [Click here to view] |
From the 41 positive samples, 45 bacteria were isolated, out of which 80% was Gram-positive and 20% were Gram-negative which is slightly similar to the result observed by Manikandan and Amsath (2013) study. The overall prevalence of isolated bacterial species was 38% S. aureus, 11% E. coli, 9% P. aeruginosa, and 4% K. pneumoniae. In LRTI, the highest pathogenic bacterium is S. aureus, which is dissimilar to the results obtained in studies conducted by Khan et al. (2015), Saxena et al. (2015), and Srivasthava et al. (2013). The same bacterial species were isolated by Bajpai et al. (2013) and Manikandan and Amsath (2013) in their study conducted on LRTI samples. Other than these isolated organisms, 38% S. epidermidis was obtained in our study, which is not a causative organism of RTI, and, therefore, is not considered in our study. This can be attributed to the patient suffering from other infections which were not ruled out. In URTI, the most common organism is S. aureus followed by P. aeuroginosa and S. epidermidis, a similar result was detected by Kousalya et al. (2010).
Antimicrobial sensitivity pattern of S. aureus observed in our study is as follows: AK is the highest sensitive antibiotic which is similar to the sensitivity pattern observed in Manikandan and Amsath (2013) study, followed by AZM, CTR, and CTX, whereas in study conducted by Kousalya et al. (2010), HLG was highly sensitive to this organism and it was highly resistant to CIP. In our study, E. coli is highly resistant to CTX and CTR. It is sensitive to HLG and CIP. Pseudomonas aeuroginosa is highly sensitive to AK and HLG followed by CTR; it is found to be resistant to CIP and CTX. Our study is in line with the reports of Wang et al. (2015), K. pneumoniae had intermediate sensitivity to CTX. CTR was sensitive to this organism, which is in accordance with Bajpai et al. (2013) study.
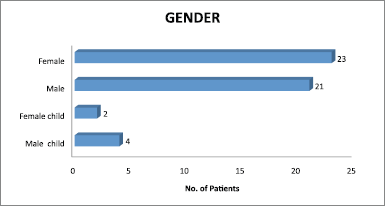 | Figure 4. Gender distribution. [Click here to view] |
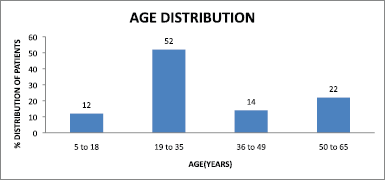 | Figure 5. Age distribution of patients. [Click here to view] |
.png) | Figure 6. Isolated bacterial species. [Click here to view] |
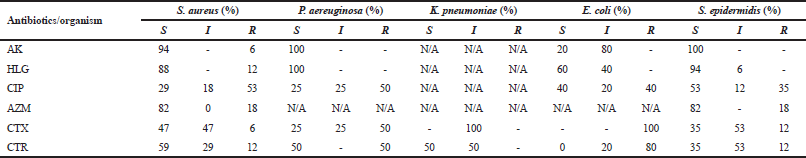 | Table 1. Antibiotic susceptibility (%). [Click here to view] |
From our study, AK, HLG, and CTR were highly sensitive to almost all the organisms although other third-generation cephalosporin and AZM had intermediate sensitivity pattern (Yadav et al., 2014). CIP is resistant against most of the organisms. This susceptibility pattern observed in our study highlights the irrational use of these antibiotics as empirical therapy for RTIs in most of the clinical settings in Ooty, Nilgiris District.
CONCLUSION
From our study, we got S. aureus as the most prevalent bacteria responsible for infections. Our study shows that AK, HLG, and CTR were highly sensitive to most of the organisms which are in concordance with the most widely used antibiotics in hospitals in Nilgiris. However, CIP was found to be the highly resistant antibiotic in our study which is surprisingly not the case in practical settings. A high level of morbidity was seen in the majority of patients in our study.
RECOMMENDATIONS AND FUTURE OUTLOOK
Existing management strategies seems insufficient for the control of RTIs as many of the hospitals lack antibiotic susceptibility testing facilities and the treatment is purely empirical. This leads to a huge level of discomfort for both the public and the government. Regional differences in prevalence and resistance patterns of bacteria make it mandatory to establish the treatment guidelines based on antibiotic susceptibility testing.
ACKNOWLEDGMENTS
The authors express their gratitude to all the staffs of the Department of Biotechnology, JSS College of Pharmacy, Ooty for their support and assistance in performing the microbiological experiments.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest in the study.
FUNDING
None.
REFERENCES
Bajpai T, Shrivastava G, Bhatambare GS, Deshmukh AB, Chitnis V. Microbiological profile of lower respiratory tract infections in neurological intensive care unit of tertiary care center from Central India. J Basic Clin Pharma, 2013; 4(3):51–5. CrossRef
Balaji V, Parija S, Kapil A, Gautam V. Standard operating procedures bacteriology [Internet]. Icmr.nic.in. 2015. Available via https://icmr.nic.in/sites/default/files/guidelines/Standard_Operating_Procedures.pdf (Accessed 10 November 2017).
El-Mahmood AM, Isa H, Mohammed A, Tirmidhi AB. Antimicrobial susceptibility of some respiratory tract pathogens to commonly used antibiotics at the Specialist Hospital, Yola, Adamawa State, Nigeria. J Clin Med Res, 2010; 2(8):135–42.
Gazi H, Kurutepe S, SürücüoÄŸlu S, Teker A, OzbakkaloglÅ B. Antimicrobial susceptibility of bacterial pathogens in the oropharynx of healthy school children in Turkey. Indian J Med Res, 2004; 120:489–94.
Hudzicki J. Kirby-Bauer disk diffusion susceptibility test protocol [Internet]. Asmscience.org. 2009. Available via http://www.asmscience.org/content/education/protocol/protocol.3189 (Accessed 10 November 2017).
Khan S, Singh P, Ankit S. Bacteria etiological agents causing lower respiratory tract infections and their resistance patterns. Iran Biomed J, 2015; 19(4):240–6.
Khan S, Singh P, Ansari M, Gurung K. Bacteria etiological agents causing lower respiratory tract infections in the Western Part of Nepal. Ibnosina J Med BS, 2014; 6(1):3–8. CrossRef
Kousalya K, Thirumurugu S, Arumainayagam DC, Manavalan R. Antimicrobial resistance of bacterial agents of the upper respiratory tract in South Indian population. J Adv Pharm Tech Res, 2010; 1(2):207–15.
Kumar SV, Kumar GPV, Kandati J, Pathapati RM, Buchineni M. A surveillance study of microbial pathogens and their antibiotic sensitivity of respiratory tract infections in a tertiary care hospital. Int J Curr Microbiol Appl Sci, 2015; 4(11):35–44.
Manikandan C, Amsath A. Antibiotic susceptibility of bacterial strains isolated from patients with respiratory tract infections. Int J Pure Appl Zool, 2013; 1(1):61–9.
Saxena S, Banerjee G, Garg R, Singh M, Verma SK, Kushwaha RAS. Bacterial colonization in patients with lower respiratory tract specimens: demographic profile and microbiological pattern. Int J Med Sci Public Health, 2015; 4(11):1498–503. CrossRef
Shanmugam S, Thekkinkattil M, Kanayalal M, Mohan CM, Mohnot C, Monisha K. Surveillance data on micro-organisms in respiratory tract infections at a tertiary care teaching hospital in South India. Int J Res Med Sci, 2015; 3(9):2366–75. CrossRef
Sharma K. Manual of microbiology: tools and techniques. 2nd edition, Ann Books Gopaljee Enterprises, New Delhi, India, 2008.
Sharma S, Srivastava P, Kumar A. Respiratory tract infection (RTI) and its treatment. Sch J Appl Med Sci, 2015; 3(8E):3118–22.
Srivasthava P, Kumar P, Nirwan PS, Sharma M. Bacteriological profile and antibiogram pattern of lower respiratory tract infections in a tertiary care hospital in Northern India. IJPRBS, 2013; 2(3):225–33.
Taura DW, Hassan A, Yayo AM, Takalmawa H. Bacterial isolates of the respiratory tract infection and their current sensitivity pattern among patients attending Aminu Kano Teaching Hospital Kano-Nigeria. Int Res J Microbiol, 2013; 4(9):226–231.
Wang LM, Qiao XL, Ai L, Zhai JJ, Wang XX. Isolation of antimicrobial resistant bacteria in upper respiratory tract infections of patients. 3 Biotech, 2016; 6(166):1–7. CrossRef
Yadav N, Mishra R, Singh V, Mandloi S, Dudey S. Isolation and identification of upper respiratory tract pathogens. Nanobiotechnic Univ, 2014; 5(1):1–7.