INTRODUCTION
Sponges are the most well-studied animals among the marine invertebrates, from which a vast array of bioactive compounds have been isolated (Hu et al., 2015). According to the review by Mehbub et al. (2014), Indonesia is the highest contributor of new compounds from marine sponges in tropical countries, and new compounds are continuously being found from the Indonesian marine sponges, as exemplified by tetradehydrohalicyclamine B from Acanthostrongylophora ingens (Kato et al., 2019), (+)-jasplakinolide Z6 from Jaspis splendens (Ebada et al., 2019), and nakijiquinone V from Dactylospongia elegans (Balansa et al., 2019). Intensive natural product screening from marine sponges in these several decades, however, makes it difficult to discover new structures. On the other hand, marine microorganisms are considered to have a potent capability of producing novel bioactive compounds that were acquired through the evolutionary pressure they suffer under unique physical, chemical, and biological conditions of the marine environment (Romano et al., 2017). Therefore, our interest in the screening source is shifting from marine sponges to the symbiotic microorganisms residing in sponges (Indraningrat et al., 2016).
In recent years, sponge-associated fungi are attracting attention as a productive source of new chemical entities (Liu et al., 2018; Pang et al., 2018; Wang et al., 2018). Furthermore, sponges are suggested to give influences to the diversity of compounds produced by associated fungi (Imhoff, 2016; Proksch et al., 2008). In Indonesia, the giant-barrel Xestospongia spp. is one of the most well-known genera for unique secondary metabolites such as aaptamine, isoaaptamine, demethyl(oxy)aaptamine, 2-(3H-diazirine-3-yl)benzaldehyde, manzamine C, xestosaprols, and N-methylniphatyne A (Agustina et al., 2018; Arai et al., 2016; Calcul et al., 2003; Dai et al., 2010; Millán-Aguiñaga et al., 2010; Murtihapsari et al., 2018). It is thus assumed that sponge-associated fungi from Xestospongia might have the potential to produce structurally diverse compounds. Unfortunately, only a few studies reported secondary metabolites from Indonesian Xestospongia-derived fungi to date. The first example is the antimicrobial xestodecalactones from Xestospongia-derived Penicillium cf. montanense reported by Edrada et al. (2002). Another case is a series of antibacterial chromones reported by Lin et al. (2003) from Aspergillus versicolor isolated from Xestospongia exigua. Since then, no additional studies on bioactive compounds have been reported from fungi associating with Xestospongia sponges.
Exploration of antibacterial compounds is becoming a core of natural product screening in this past few years, along with the increasing emergence of multidrug-resistant (MDR) bacteria (Asagabaldan et al., 2017; Ayuningrum et al., 2019; Goncalves et al., 2016; Kristiana et al., 2017; Magairakos et al., 2011; Malhotra et al., 2016; Sibero et al., 2018a; 2018b). Salmonella enterica ser. Typhi is one of such MDR bacteria causing serious problematic infections (Crump et al., 2015; Stephens et al., 2019). This pathogen caused serious typhoid fever outbreak in several cities in China, Pakistan, Thailand, and Zimbabwe (Limpitikul et al., 2014; Muti et al., 2014; Yan et al., 2016; Wong et al., 2018). In this paper, we report the isolation and structure determination of secondary metabolites possessing antibacterial activity against MDR S. enterica ser. Typhi from a Xestospongia-derived fungus.
EXPERIMENTAL PROCEDURE
General experiment procedures
NMR spectra were recorded on a Bruker AVANCE II 500 spectrometer (Bruker Biospin K. K., Yokohama, Japan) and mass spectra were measured on a Bruker microTOF (Bruker Daltonics K. K., Yokohama, Japan). IR spectra were recorded on a Shimadzu FT-IR-300 spectrophotometer (Shimadzu Corp., Kyoto, Japan) and UV spectra on a Shimadzu UV-1800 (Shimadzu Corp., Kyoto, Japan). All compounds were analyzed by using reversed phase column Microsorb C18 (particle size 3 μm, pore size 50×4.6 mm). Compound 1 was eluted at tR 5.31 minute compound 2 at 8.87 minutes, compound 3 at 13.41 minutes, and compound 4 at 11.23 minutes. Purification was carried out by using COSMOSIL Cholester Packed Column (Nacalai Tesque, 10.0 mm i.d. x 250 mm).
Sponge
A giant barrel sponge was purposively collected from Karimunjawa National Park, Central Java Indonesia with permission number 1096/T.34/TU/SIMAKSI/7/2017 by scuba diving. The sponge was identified as Xestospongia sp. KJMT.SP.04 by the specific giant-barrel shape. Approximately 3 × 3 cm tissues were taken using a blade and put into a zip-lock plastic bag then kept in a chilling temperature for fungal isolation.
Microorganism
The fungal strain KJMT FP 4.1 was isolated and identified as Daldinia with 99.9% similarity in the internal transcribed spacer (ITS) rRNA gene sequence (548 nucleotides; accession number MG972929.1 in GeneBank) to Daldinia eschscholtzii (GeneBank accession number KU304335).
Fermentation
This step was carried out according to the reported procedure (Shimoyama et al., 2018). Strain KJMT FP 4.1 growing on malt extract agar medium was inoculated into 500 mL K-1 flasks, each containing 100 mL of V-22 medium [1% soluble starch, 0.5% glucose, 0.3% NZ-case, 0.2% yeast extract (Kyokuto Pharmaceutical Industrial, Co. Ltd, Tokyo Japan), 0.2% Tryptone (Difco Laboratories, Sparks, USA), 0.1% K2HPO4, 0.05% MgSO4·7H2O, and 0.3% CaCO3 in distilled water] at pH 7.0 for seed culture. The inoculated flasks were placed on a rotary shaker (200 rpm) at 30°C for 3 days. Then, 5 ml of the seed culture was transferred into 500 mL K-1 flasks each containing 100 mL of production medium (A-11M) consisting of 2% glucose, 2.5% soluble starch, 0.5% yeast extract, 0.5% polypeptone, 0.5% NZ-amine, 0.5% CaCO3, and 1% Diaion HP-20 (Mitsubishi Chemical Co., Yokohama, Japan) at pH 7.0. The inoculated flasks were placed on a rotary shaker (200 rpm) at 30°C for 7 days.
Extraction and isolation
After fermentation, 100 ml 1-butanol was added to each K-1 flask and shaken on a rotary shaker (200 rpm) for 1 hour. The mixture was centrifuged at 6,000 rpm for 15 minutes, and the organic layer was separated from the aqueous layer containing the mycelium (Shimoyama et al., 2018). The organic solvent was removed by evaporation with a rotary vacuum evaporator, providing 4.44 g of crude extract. It was then subjected to silica gel column chromatography with a step gradient of CHCl3-MeOH (1:0, 20:1, 10:1, 4:1, 2:1, 1:1, and 0:1 v/v). The selected fractions were further purified using an ODS column chromatography with a step gradient of MeCN-0.1% HCO2H (20:80, 30:70, 40:60, 50:50, 60:40, 70:30, and 80:20 v/v). All compounds were purified by reversed-phase preparative high performance liquid chromatography (HPLC) using a Cosmosil Cholester Packed Column (Nacalai Tesque Inc., 20 × 250 mm). Compound 1 (88.8 mg) was obtained from an ODS 60:40 fraction of silica gel 20:1 fraction after HPLC purification using an isocratic condition (40% MeCN) at tR 19.4 minutes. Compounds 2 (10.5 mg, tR 18.6 minutes) and 3 (5.6 mg, tR 23.2 minutes) were obtained from an ODS 40:60 fraction of silica gel 10:1 fraction, after HPLC purification with a gradient condition (0–25 minutes: 25%–40% MeCN). Compound 4 (17.1 mg, tR 19.8 minutes) was isolated from an ODS 30:70 fraction of silica gel 4:1 fraction after HPLC purification with a gradient condition (0–25 minutes: 15%–30% MeCN in 0.1% HCO2H).
Antimicrobial assay
Compounds 1, 2, 3, and 4 were tested for antibacterial activity with microdilution method (Igarashi et al., 2011). Salmonella enterica ser. Typhi were obtained from Dr. Kariadi General Hospital in Semarang and confirmed as a multi-drug resistant organism. The pathogen was refreshed from stock 24 hours before diluted into physiological saline solution with a density value of 0.5 McFarland. All compounds were diluted into mixture of Mueller Hinton Broth (MHB) and dimethyl sulfoxide (DMSO) (1:2) and delivered into a microplate with 250, 125, 62.5, 31.3, 15.6, 7.8, 3.9, and 2.0 μg to a volume of 50 μl in each well. Amoxicillin (PT. Graha Farma) was used as a positive control and DMSO as a negative control. Each well was then loaded with 145 μL MHB and 5 μL pathogen containing liquid, and incubated at 32°C for 24 hours. At the end of incubation, 10 μL of WST-1 indicator was added into each well and incubated for 1 hour at 32°C. Living bacteria were detected by the changing of color to yellow. The lowest concentration that inhibits pathogen growing was determined as minimum inhibitory concentration (MIC) value. Then 10 μl of the solution in each well was taken and spread onto Mueller-Hinton Agar and incubated at 32°C for 24 hours.
RESULTS AND DISCUSSION
Fungal strain KJMT FP 4.3 was isolated from a sponge Xestospongia sp. collected in Karimunjawa National Park, Central Java. This fungus was identified as a member of Daldinia on the basis of 99.9% similarity of ITS gene sequence to D. eschscholtzii. In the HPLC/UV analysis of the crude extract, four major peaks were detected and thus purified for structural analysis. One unknown major peak was characterized as a new compound designated karimanone (1), which is a diastereomeric mixture of α-D-ribofuranoside of (2R)- and (2S)-5,7-dihydroxy-2-methyl-4-chromanone. The other three compounds (2, 3, and 4) were identified as known compounds (Fig. 1).
Compound 1 was isolated as a yellow powder that showed a deprotonated molecule [M-H]- ion peak at m/z 325.0911 in the high resolution electrospray ionization mass spectrometry (HRESIMS) analysis, corresponding to a molecular formula of C15H18O8 (calcd for C15H17O8, 325.0923) with seven degrees of unsaturation. This compound showed IR absorption bands at 3,415 and 1,642 cm−1, suggesting the presence of hydroxy and carbonyl groups. Investigation of the 1H and 13C NMR and heteronuclear single-quantum correlation (HSQC) spectra revealed 15 carbon resonances (Table 1), consisting of seven sp2 carbons (δC 198.8, 166.9, 165.0, 164.6, 104.7, 98.1, and 97.2; including one carbonyl and three oxygenated), five oxygenated methine carbons (δC 101.7, 88.2, 75.7, 73.6, and 71.2), two methylene carbons (δC 63.2 and 44.3, one oxygenated), and one methyl carbon (δC 21.1).
The planar structure of 1 was assigned by the analysis of 1D and 2D NMR spectral data recorded in CD3OD (Fig. 2). From the correlation spectroscopy (COSY) analysis, two 1H-1H spin systems, 2-Me/H2/H3 and H11/H12/H13/H14/H15 were identified. The oxygenated methine H2 showed heteronuclear multiple bond coherence (HMBC) correlations to C4 and C9 and methylene protons H3 were correlated to C4 and C10. These correlation data established the ether bridge between C2 and C9 and the presence of a ketone group being located between C3 and C10. Meanwhile, aromatic methine protons H6 and H8 were mutually HMBC-correlated and both protons were correlated to C10. Additionally, HMBC correlations were shown from H6 to C5 and C7 and from H8 to C7 and C9. Together with these HMBC correlations, high-field shifted chemical shifts for the carbons C6, C8, and C10 suggested that these carbons were positioned ortho to the oxygenated carbons C5, C7, and C9, thereby establishing the tetrasubstituted benzene ring comprising six carbons from C5 to C10. Therefore, along with the aforementioned correlations from H2 to C9 and H3 to C10, the 2-methyl-4-chromanone substructure was confirmed. Another COSY-defined fragment from H11 to H15 was deduced to form a pentofuranosyl moiety by the mutual HMBC correlations between H11 and H14. Finally, this sugar part was connected at C7 through the ether bond by an HMBC correlation from H11 to C7, completing the planar structure of 1 (Fig. 2).
The relative configuration of the pentose moiety was established by NOESY analysis. NOE correlations were observed for H11/H12, H12/H13, and H13/H15, confirming that H11, H12, and H13 were on the same side, and H14 was on the opposite side (Fig. 3). The coupling constant 3JHH of the anomeric proton H11 showed 4.8 Hz, a typical value for the α-ribofuranoside. β-Ribofuranosides usually give a singlet proton signal for the anomeric proton with J values near 0 Hz (Du et al., 2008; Sharma et al., 2012). Therefore, the α-ribofuranosyl configuration was established.
The absolute configuration of 1 was estimated by comparing its specific rotation with the reported values of α-ribofuranosides (Fig. 6) (Walker and Hogenkamp, 1974; Du et al., 2008; Sharma et al., 2012). Since α-D-ribofuranosides always provide positive [α]D values, the positive specific rotation value shown by 1 ([α]22D +181 (c 1.0, MeOH)) suggested the α-D-ribofuranoside with 11R, 12R, 13S, and 14R-configurations. L-Ribofuranosides have not been reported from natural products to date.
 | Figure 1. Structures of compounds 1,2,3, and 4 [Click here to view] |
The remaining chiral center C2 was deduced to be racemic based on the following observations. Several unexpected splitting patterns were found in 1H and 13C NMR spectra. The anomeric proton H11 appeared as two doublet signals and the carbon signals for 2-Me, C2, C3, C4, C5, and C7 displayed weak splitting (Table 1, Figs. 4–5). These carbons were located in the chromanone moiety, indicating that compound 1 was obtained as a diastereomeric mixture regarding the C2 chiral center. This conclusion was further supported by the isolation of compound 3 as a racemic mixture from this strain, an aglycon part of which might be a biosynthetic precursor of 1. Therefore, 1 was determined to be a mixture of (2R)- and (2S)-isomers of 7-α-D-ribofuranosyl-5-hydroxy-2-methyl-4-chromanone.
In this study, 1 was obtained as an inseparable mixture of two diastereomers containing (2R)- and (2S)-isomers of 1. (2R)-Isomer is known as a metabolite of an endophytic Daldinia but (2S)-isomer has not been reported to date (Hu et al., 2017). (S)-3 was isolated from a botanical source Diospyros maritima (Chang et al., 2009), while (R)-3 is not found from nature to date. This is the first finding of 3 from microorganisms. (S)-2 was isolated from Daldinia (Nadeau and Sorensen, 2011), and (R)-2 from Nodulisporium, Cryptosporiopsis, and Daldinia (Dai et al., 2006; Hu et al., 2017; Pathania et al., 2015; Zilla et al., 2013). (R)-2 has potent anti-leukemic activity, while no obvious biological activity was found from (S)-2. Compound 4 was previously isolated from Nodulisporium and Daldinia with herbicidal activity (Hu et al., 2017; Igarashi et al., 1993; Wang and Liu, 2004). Antibacterial activity of the single compounds is shown in Table 2.
It is noted that compound 2 had MIC (minimum inhibitory concentration) value of 62.5 μg/mL and compounds 1, 3 and 4 had MIC value of 125 μg/mL, while the MIC for amoxicillin as the positive control was 250 μg/mL (Table 2). Enantiomerically pure compound (2R)-1 was reported to has antibacterial activity against non-MDR Staphylococcus aureus with MIC value of 64 μg/mL (Hu et al., 2017). CLSI (2016) stated that Salmonella spp. could be stated as resistant to amoxicillin if the MIC value is > 36 μg/ml. Table 2 shows that the S. enterica ser. Typhi that used in this study was resistant to amoxicillin. Moreover, the result of antibacterial activity indicated that these compounds had stronger antibacterial activity against MDR S. enterica ser. Typhi than the positive control.
 | Figure 2. COSY and key HMBC correlations of compound 1. [Click here to view] |
 | Figure 3. NOESY correlations of the ribose part of compound 1. [Click here to view] |
Karimanone (1): yellow powder; [α]22D + 181 (c 1.0, MeOH); UV (MeOH) λmax (log ε) 284 (5.48), 325 (4.71) nm; IR νmax 3,415, 1,642 cm−1; see Table 1 for 1H NMR and 13C NMR data; HR-ESITOFMS [M - H]- m/z 325.0911 (calcd for C15H17O8, 325.0923).
(S)-5-Hydroxy-2-methyl-4-chromanone (2): pale pink powder; [α]22D -1.9 (c 1.0, MeOH) {lit. (R)-2: [α]25D +6 (c 0.19, CH2Cl2),11 (S)-2: [α]22D -1.1915}; UV (MeOH) λmax (log ε) 204 (5.41), 220 (sh, 5.26), 271 (5.15), 348 (4.67) nm; IR νmax 1,622, 1214, 1,056 cm-1; 1H NMR (500 MHz, CDCl3) δ 11.70 (1H, s, OH), 7.32 (1H, t, J = 8.2 Hz, H7), 6.47 (1H, d, J = 8.3 Hz, H6), 6.40 (1H, d, J = 8.3 Hz, H8), 4.54 (1H, m, H2), 2.67 (2H, m, H3),1.49 (3H, d, J = 6.4 Hz, H11); 13C NMR (125 MHz, CDCl3) δ 198.6 (C4), 162.2 (C5), 161.8 (C9), 138.3 (C7), 109.2 (C6), 108.1 (C10), 107.4 (C8), 73.9 (C2), 43.9 (C3), 20.9 (C11); HR-ESITOFMS [M - H]- m/z 177.0521 (calcd 177.0552 for C10H9O3).
5,7-Dihydroxy-2-methyl-4-chromanone (3): pale yellow powder; [α]22D -6.6 (c 1.0, MeOH) {lit. (S)-3: [α]D -58.6 (c 1, MeOH) (Rao et al., 2017)}; UV (MeOH) λmax (log ε) 211 (5.40), 228 (sh, 5.20), 288 (5.35), 330 (sh, 4.56) nm; IR νmax 3,163, 1,601, 1,301, 1,164 cm−1; 1H NMR (500 MHz, CD3OD) δ 5.84 (1H, s, H6), 5.84 (1H, s, H8), 4.50 (1H, m, H2), 2.66, 2.57 (each 1H, dd, J = 12.3, 17.1 Hz, dd, J = 3.1, 17.1 Hz, H3), 1.44 (3H, d, J = 6.3 Hz, H11); 13C NMR (125 MHz, CD3OD) δ 198.1 (C4), 168.4 (C7), 165.6 (C5), 165.1 (C9), 103.4 (C10), 97.0 (C8), 96.1 (C6), 75.5 (C2), 44.2 (C3), 21.1 (C11); HR-ESITOFMS [M - H]- m/z 193.0545 (calcd for C10H9O4, 193.0501).
 | Table 1. NMR spectroscopic data (500 MHz, CD3OD) for Karimanone (1). [Click here to view] |
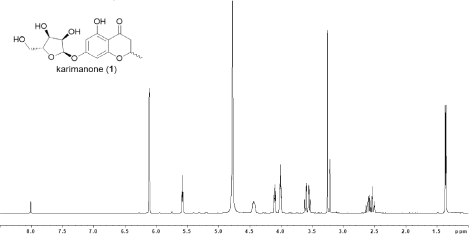 | Figure 4. 1H NMR spectrum of 1 (500 MHz, CD3OD) [Click here to view] |
 | Figure 5. 13C NMR spectrum of 1 (125 MHz, CD3OD) [Click here to view] |
 | Figure 6. Specific rotation values of α-ribofuranosides [Click here to view] |
 | Table 2. Antibacterial activity of the pure compounds against S. enterica ser. Typhi. [Click here to view] |
AB5046A (4): yellow oil; UV (MeOH) λmax (log ε) 229 (4.89), 273 (4.86), 340 (3.72) nm; IR νmax 3364, 2963, 1549, 1061 cm-1; 1H NMR (500 MHz, CDCl3) δ 4.40 (1H, br, H5), 2.77, 2.65 (each 1H, o, dd, J = 5.8, 16.3 Hz, H4), 3.01 (2H, t, J = 7.3 Hz, H8), 2.96, 2.80 (each 1H, dd, J = 2.0, 18.0 Hz, o, H6), 1.64 (2H, sextet, J = 7.3 Hz, H9), 0.99 (3H, t, J = 7.4 Hz, H10); 13C NMR (125 MHz, CDCl3) δ 205.7 (C7), 196.2 (C3), 193.2 (C1), 113.2 (C2), 63.7 (C5), 47.4 (C4), 42.4 (C8), 41.7 (C6), 18.2 (C9), 14.1 (C10); HR-ESITOFMS [M + Na]+ m/z 221.0745 (calcd 221.0790 for C10H14O4Na).
CONCLUSION
Four compounds (1–4) were successfully isolated from D. eschscholtzii KJMT FP 4.1, an Indonesian sponge-associated fungus. Karimanone (1) showed an interesting structure due to the glycosylated aromatic structure. Moreover, 1 was obtained as an inseparable mixture of two diastereomers containing (2R)- and (2S)-isomers for the chromanone part. (2R)-Isomer is known as a metabolite of an endophytic Daldinia, while (2S)-isomer has not been reported to date. All compounds were active against the multidrug-resistant strain of S. enterica ser. Typhi with an MIC of 62.5 μg/ml for 2 and 125 μg/ml for 1, 3, and 4.
CONFLICT OF INTEREST
The authors declare that they do not have any conflict of interest regarding the publication of this work.
ACKNOWLEDGMENT
We wish to acknowledge the Ministry of Research, Technology and Higher Education, Indonesia who funded this work under the Program Pendidikan Magister Menuju Doktor untuk Sarjana Unggul (PMDSU) with contract number 315-06/UN7.5.1/PP/2017; Peningkatan Kualitas Publikasi Internasional (PKPI) Program. We also acknowledge Balai Taman Nasional Karimunjawa for the research permit with number 1096/T.34/TU/SIMAKSI/7/2017 and USAID-PEER Science grant cycle 5 Subaward number 2000007644 for the support of publication payment.
REFERENCES
[CLSI] Clinical Laboratory Standard Institue. Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. CLSI supplement M100S, 2016.
Agustina S, Karina S, Kurnianda V, Rahmi R, Khairunnisa K. Manzamine C, an alkaloid indole as an inhibitor of the cancer cells adapted to nutrient starvation, from an Indonesian marine sponge of Xestospongia muta. IOP Conf Ser: Earth Environ Sci, 2018; 216:012006.
Arai M, Kamiya K, Shin D, Matsumoto H, Hisa T, Setiawan A, Kotoku N, Kobayashi M. N-Methylniphatyne A, a new 3-Alkylpyridine alkaloid as an inhibitor of the cancer cells adapted to nutrient starvation, from an Indonesian marine sponge of Xestospongia sp.. Chem Pharma Bull, 2016; 64:766–71.
Asagabaldan MA, Ayuningrum D, Kristiana R, Sabdono A, Radjasa OK, Trianto A. Identification and antibacterial activity of bacteria isolated from marine sponge Haliclona (Reniera) sp. against multi-drug resistant human pathogen. IOP Conf Ser: Earth Environ Sci, 2017; 55:012019.
Ayuningrum D, Liu Y, Riyanti, Sibero MT, Kristiana R, Asagabaldan MA, Wuisan ZG, Trianto A, Radjasa OK, Sabdono A, Schäberle TF. Tunicate-associated bacteria show a great potential for the discovery of antimicrobial compounds. PLos One, 2019; 14:e0213797.
Balansa W, Mettal U, Wuisan ZG, Plubrukarn A, Ijong FG, Liu Y, Schäberle TF. A new sesquiterpenoid aminoquinone from an Indonesian marine sponge. Mar Drugs, 2019; 17:158.
Calcul L, Longeon A, Mourabit AA, Guyot M, Bourguet-Kondracki ML. Novel alkaloids of the aaptamine class from an Indonesian marine sponge of the genus Xestospongia. Tetrahedron, 2003; 59:6539–44.
Chang CI, Chen CR, Chiu HL, Kuo CL, Kuo YH. Chemical constituents from the stems of Diospyros maritima. Molecules, 2009; 14:5281–8.
Crump JA, Sjölund-Karlsson M, Gordon MA, Parry CM. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clinical Microbiol Rev, 2015; 28:901–37.
Dai J, Krohn K, Flörke U, Draeger S, Schulz B, Kiss-Szikszai A, Antus S, Kurtán T, Ree T. Metabolites from the endophytic fungus Nodulisporium sp. from Juniperus cedre. Eur J Org Chem, 2006: 15:3498–506.
Dai J, Sorribas A, Yoshida WY, Kelly M, Williams PG.. Xestosaprols from the Indonesian marine sponge Xestospongia sp. J Nat Prod, 2010; 73:1188–91.
Du L, Zhu T, Liu H, Fang Y, Zhu W, Gu Q. Cytotoxic polyketides from a marine-derived fungus Aspergillus glaucus. J Nat Prod, 2008; 71:1837–42.
Ebada SS, Müller WEG, Lin W, Proksch P. New acyclic cytotoxic jasplakinolide derivative from the marine sponge Jaspis splendens. Mar Drugs, 2019; 17:100.
Edrada RA, Heubes M, Brauers G, Wray V, Berg A, Gräfe U, Wohlfarth M, Mühlbacher J, Schaumann K, Sudarsono, Bringmann G, Proksch P. Online analysis of xestodecalactones A - C , novel bioactive metabolites from the fungus Penicillium cf. montanense and their subsequent isolation from the sponge Xestospongia exigua. J Nat Prod, 2002; 65:1598–604.
Goncalves LF, Martins-Junior PDO, de Melo ABF, da Silva RCRM, Martins VP, Pitondo-Silva A, de Campos TA. Multidrug resistance dissemination by extended-spectrum β-lactamase-producing Escherichia coli causing community-acquired urinary tract infection in the Central-Western Region, Brazil. J Glob Antimicrob Resist, 2016; 6:1–4.
Hu M, Yang XQ, Zhou QY, Li SQ, Wang BY, Ruan BH, Yang YB, Zhang ZX, Zhou H, Ding ZT. Benzopyran derivatives from endophytic Daldinia eschscholzii JC-15 in Dendrobium chrysotoxum and their bioactivities. Nat Prod Res, 2017; 22:1–5.
Hu Y, Chen J, Hu G, Yu J, Zhu X, Lin Y, Chen S, Yuan J. Statistical research on the bioactivity of new marine natural products discovered during the 28 years from 1985 to 2012. Mar Drugs, 2015; 13:202–21.
Igarashi M, Tetsuka Y, Mimura Y, Takahashi A, Tamamura T, Sato K, Naganawa H, Takeuchi T. AB5046A AND B, novel chlorosis-inducing substances from Nodulisporium sp. J Antibiot, 1993; 46:1843–8.
Igarashi Y, Ogura H, Furihata K, Oku N, Indananda C, Thamchaipenet A. Maklamicin, an antibacterial polyketide from an endophytic Micromonospora sp. J Nat Prod, 2011; 74:670–4.
Imhoff JF. Natural products from marine fungi—still an underrepresented resource. Mar Drugs, 2016; 14(1):19.
Indraningrat AAG, Smidt H, Sipkema D. Bioprospecting sponge-associated microbes for antimicrobial compounds. Mar Drugs, 2016; 14:87.
Kato H, El-Desoky AH, Takeishi Y, Nehira T, Angkouw ED, Mangindaan REP, de Voogd NJ, Tsukamoto S. Tetradehydrohalicyclamine B, a new proteasome inhibitor from the marine sponge Acanthostrongylophora ingens. Bioorg Med Chem Lett, 2018; 29:8–10.
Kristiana R, Ayuningrum D, Asagabaldan MA, Nuryadi H, Sabdono A, Radjasa OK, Trianto A. Isolation and partial characterization of bacteria activity associated with gorgonian Euplexaura sp. against methicillin-resistant Staphylococcus aureus (MRSA). IOP Conf Ser: Earth Environ Sci, 2017; 55:012056.
Limpitikul W, Henpraserttae N, Saksawad R, Laoprasopwattana K. Typhoid outbreak in Songkhla, Thailand 2009-2011: Clinical outcomes, susceptibility patterns, and reliability of serology tests. PLoS One, 2014; 9:7–12.
Lin W, Brauers G, Ebel R, Wray V, Berg A, Sudarsono, Proksch P. Novel chromone derivatives from the fungus Aspergillus versicolor isolated from the marine sponge Xestospongia exigua. J Nat Prod, 2003; 66:57–61.
Liu Y, Ding L, Fang F, He S. Penicillilactone A, a novel antibacterial 7-membered lactone derivative from the sponge-associated fungus Penicillium sp. LS54. Nat Prod Res, 2018; 19:1–5.
Magairakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Bacteria : an international expert proposal for interim standard definitions for acquired resistance. Microbiology, 2011; 18:268–81.
Malhotra R, Sikka R, Chaudhary U, Malhotra R, Med JR, Feb S. Antimicrobial sensitivity pattern among clinical isolates of Escherichia coli in tertiary care centre of Northern Indi. Int J Res Med Sci, 2016; 4:639–42.
Mehbub MF, Lei J, Franco C, Zhang W. Marine sponge derived natural products between 2001 and 2010: trends and opportunities for discovery of bioactives. Mar Drugs 2014; 12:4539–77.
Millán-Aguiñaga N, Soria-Mercado IE, Williams P. Xestosaprol D and E from the Indonesian marine sponge Xestospongia sp. Tetrahedron Lett, 2010; 51:751–3.
Murtihapsari M, Suruwaky AM, Kadarusman K, Kurnia D, Herlina T, Supratman U. A new antiplasmodial compound from the Papuan marine sponge Xestospongia sp. J Penelitian Dan Pengembangan Ilmu Kimia, 2018; 4:1–6.
Muti M, Gombe N, Tshimanga M, Takundwa L, Bangure D, Mungofa S, Chonzi P. Typhoid outbreak investigation in Dzivaresekwa, suburb of Harare City, Zimbabwe, 2011. Pan Afr Med J, 2014; 18:1–7.
Nadeau AK, Sorensen JL. Polyketides produced by Daldinia loculata cultured from Northern Manitoba. Tetrahedron Lett, 2011; 52:1697–99.
Pang X, Lin X, Tian Y, Liang R, Wang J, Yang B, Zhou X, Kaliyaperumal K, Luo X, Yu Z, Liu Y. Three new polyketides from the marine sponge-derived fungus Trichoderma sp. SCSIO41004. Nat Prod Res, 2018; 32:105–11.
Pathania AS, Guru SK, Ashraf NU, Riyaz-Ul-Hassan S, Ali A, Tasduq SA, Malik F, Bhushan S. A novel stereo bioactive metabolite isolated from an endophytic fungus induces caspase dependent apoptosis and STAT-3 inhibition in human leukemia cells. Eur J Pharmacol, 2015; 765:75–85.
Proksch P, Ebel R, Edrada R, Riebe F, Liu H, Diesel A., Bayer M, Li X, Lin WH, Grebenyuk V, Muller WEG, Draeger S, Zuccaro A, Schulz B. Sponge-associated fungi and their bioactive compounds: The Suberites case. Bot Mar, 2008; 51:209–18.
Rao AR, Gaitonde AS, Prakash KRC, Rao SP. A concise synthesis of chiral 2-methyl chroman-4-ones: stereo selective build-up of the chromanol moiety of anti-HIV agent calanolide A. Tetrahedron Lett, 1994, 35:6347–50.
Romano G, Costantini M, Sansone C, Lauritano C, Ruocco N, Ianora A. Marine microorganisms as a promising and sustainable source of bioactive molecules. Mar Environ Res, 2017; 128:58–69.
Sharma RK, Singh S, Tiwari R, Mandal D, Olsen CE, Parmar VS, Parang K, Prasad AK. O-Aryl α,β-d-ribofuranosides: synthesis & highly efficient biocatalytic separation of anomers and evaluation of their Src kinase inhibitory activity. Bioorg Med Chem, 2012; 20:6821–30.
Shimoyama T, Miyoshi M, Nehira T, Motojima A, Oikawa T, Laurence O, Igarashi, Y. Two new secondary metabolites from a fungus of the genus Robillarda. J Antibiot, 2018; 71:432–7.
Sibero MT, Radjasa OK, Sabdono A, Trianto A, Triningsih DW, Hutagaol ID. Antibacterial activity of indonesian sponge associated fungi against clinical pathogenic multidrug resistant bacteria. J App Phar Sci, 2018a; 8:88–94.
Sibero MT, Herdikiawan D, Radjasa OK, Sabdono A, Trianto A, Triningsih DW. Antibacterial activity of sponge associated fungi against vibriosis agents in shrimp and its toxicity to Litopenaeus vannamei. AACL Bioflux 2018b; 11:10–8.
Stephens KC, Medalla F, Hughes M, Appiah GD, Aubert RD, Caidi H, Angelo KM, Walker AT, Hatley N, Masani S, Nash J, Belko J, Ryan ET, Mintz E, Friedman CR. Emergence of extensively drug-resistant Salmonella typhi infections among travelers to or from Pakistan — United States, 2016–2018. MMVWR Morb Mortality Wkly Rep, 2019; 68:11–3.
Walker T, Hogenkamp H. A new synthesis of L-ribofuranose derivatives. Carbohyd Res, 1974; 32:413–7 .
Wang CY, Hao JD, Ning XY, Wu JS, Zhao DL, Kong CJ, Shao CL, Wang CY. Penicilazaphilones D and E: two new azaphilones from a sponge-derived strain of the fungus Penicillium sclerotiorum. RSC Adv, 2018; 8:4348–53.
Wang F, Liu JK. A pair of novel heptentriol stereoisomers from the ascomycete Daldinia concentrica. Helv Chim Acta, 2004; 87:2131–4.
Wong W, Rawahi HA, Patel S, Yau Y, Eshaghi A, Zittermann S, Tattum L, Morris SK. The first Canadian pediatric case of extensively drug-resistant Salmonella typhi originating from an outbreak in Pakistan and its implication for empiric antibiotic choices. IDCases, 2018; 15:1–4.
Yan M, Li X, Liao Q, Li F, Zhang J, Kan B. The emergence and outbreak of multidrug-resistant typhoid fever in China. Emerg Microbes Infect, 2016; 5:e62.
Zilla MK, Qadri M, Pathania AS, Strobel GA, Nalli Y, Kumar S, Guru SK, Bhushan S, Singh SK, Vishwakarma RA, Riyaz-Ul-Hassan S. Bioactive metabolites from an endophytic Cryptosporiopsis sp. inhabiting Clidemia hirta. Phytochemistry 2013; 95:291–7.