INTRODUCTION
Gout is a common chronic arthritis with every 9.5% to 13.5% per 1,000 persons getting affected (Saigal et al., 2015). The pathophysiology of the disease is mainly due to the improper uric acid metabolism leading to the precipitation and accumulation of uric acid crystals in joints, bones, tissues, and many other organs. Hence, gout is also known as hyperuricemia (Ragab et al., 2017). Gout is more common in male compared to female with a ratio of 4:1. However, the incidence increases in women after menopause (Doherty, 2009). In addition to inflammation in joints, bones persons with gout are at the risk of developing chronic kidney disease, hypertension, cardiovascular diseases, metabolic disorders, and psychosis (Chiu et al., 2012; Feig et al., 2008; Gerald and Falasca, 2006; Grayson et al., 2011; Mohandas and Johnson, 2008; Soltani et al., 2013).
Hypoxanthine and xanthine formed during purine metabolism are metabolized in the liver to uric acid by the enzyme xanthine oxidase (Kostalova et al., 2015). Uricase, an enzyme that degrades the uric acid to soluble allantoin but humans lack this enzyme. Hence, uric acid is not degraded leading to the accumulation of insoluble uric acid crystals in joints, bones, and many other organs like kidney (El Ridi and Tallima, 2017; Roddy and Doherty, 2010). Xanthine oxidase is the primary target to inhibit uric acid formation and in the treatment of Gout. Other main biomarkers in gout are interleukin (IL) 17A and IL18, proinflammatory cytokines that are upregulated in the serum of gout patients (Cavalcanti et al., 2016; Liu et al., 2018). These cytokines are also targets in many other diseases; IL17A is the major target in many other diseases, such as psoriasis, rheumatoid arthritis, systemic lupus erythematosus, and also in many different cancers (Kirkham et al., 2014). IL18 is mainly involved in many autoimmune diseases (Sedimbi et al., 2013).
In gout patients, IL17A binds to its receptor leading to chemokine release (Zhou et al., 2016). Monoclonal antibody, secukinumab is the only one approved IL-17A inhibitor at present (Frieder et al., 2018). Drugs to treat IL18, another major proinflammatory cytokine upregulated in gout are currently under investigation. Targetting these cytokines with chemical drug can be effective, however, may lead to many side effects. Hence, identification of plant-based compounds with IL17A and IL18 inhibitory potential can be an important breakthrough in the treatment of Gout. This is the first In silico study to find the binding potential of plant-based medicinal compounds to proinflammatory cytokines IL17A and IL18, important biomarkers in gout. The results from this study can be a foundation for developing new drugs for Gout treatment.
MATERIALS AND METHODS
Macromolecules structure retrieval
The structure of IL17A-PDB ID:4HR9 and IL18-PDB ID: 3WO2 were retrieved from Protein Data Bank (PDB) (Huang et al., 2015). PDB is a database that contains the data of experimental structures of proteins and nucleic acids. Structure of IL7A was a homodimer (chain A and B) and IL18 3WO2 was homotetramer (chain A, B, C, and D). Only chain A was used for docking studies. Other chains and water molecules were removed using PyMol (Fig. 1). PyMol is a useful open source software tools to perform molecular graphics (Yuan et al., 2017)
Ligand structure retrieval
Eleven plant-based molecules: carvacrol, thymol, thiamine, riboflavin, limonene, diadzein, genistein, canthin-6-one, syriacusin A, syringaresinol, and gossypin with potential pharmaceutical and medicinal benefits were chosen for the ligand protein docking study. The docking study was performed against a control drug Allopurinol, prescribed to gout patients to reduce gout attacks. The structures of the ligand molecules and the control drug Allopurinol were retrieved from Pubchem database. Pubchem contains information about chemical compounds their structure, molecular formula, molecular weight, etc. (Kim et al., 2015). The structures were retrieved in SDF format and were changed to PDB format using PyMol. The structure and medicinal properties of the ligands are listed in Table 1.
Drug scan
All the ligands were tested for their drug potential based on Lipinski’s rule of five. Molinspiration (http://www.molinspiration.com/cgi-bin/properties) was used for calculating Lipinski’s properties. Lipinski’s rule mainly determine the molecular attributes, such as molecular weight, logP, number of hydrogen bond acceptors, and number of hydrogen bond donors. Any ligand showing violations in Lipinski’s rule were eliminated from further studies (Lipinski et al., 2012).
Active site prediction
Amino acids involved in active pocket formation were determined using Computed Atlas for Surface Topography of proteins (CASTp). CASTp is a very easy and useful web-based tool for determining the topology and active site pockets in the proteins (Dundas et al., 2006). Active site determination is important to set the grid box at before docking.
Docking and visualization
AutoDock 4.2 was used for docking of ligands to the proteins. AutoDock is the fully automated docking software tool most widely used to study the protein ligand binding and interactions (Morris et al., 2009). Protein molecule was initially fixed by adding polar hydrogen, kollman charges (Azam and Abbasi, 2013). Ligand molecule was added with gasteiger charges (Karunakar et al., 2014). Autodock 4.2 allows setting a specific target site with the help of grid box. The X, Y, and Z dimensions were set to 40*40*40, the X center, Y center, and Z center were adjusted based on the active site of the proteins (Adejoro et al., 2016). The output for ligand conformations were analyzed using stochastic Lamarckian genetic algorithm (Morris et al., 1998). The least negative ΔG indicates a strong binding and favorable conformation for the ligand and the protein interaction (Afriza et al., 2018). The 3D visualization of docked structures was performed using graphical user interface, Discovery studio (Kemmish et al., 2017). Protein-ligand interactions, structure and ligand designing are few facilities available in Discovery studio (Meenambiga et al., 2018)
ADME/T prediction
Absorption, distribution, metabolism, excretion, and toxicity (ADMET) of the ligands are their pharmacokinetic properties and are needed to be evaluated to determine their activity inside the body. The ADMET properties of the ligands were analyzed using admetSAR, an online ADMET prediction tool (http://lmmd.ecust.edu.cn:8000/) (Cheng et al., 2012).
RESULTS AND DISCUSSION
Globally, Gout incidence and prevalence is continuously increasing with people aged above 30 years which is getting greatly affected. In addition to joint pains, Gout is also associated with organ failure due to the accumulation of uric acid crystals in the organs. Serum levels of proinflammatory cytokines IL17a and IL18 are upregulated in Gout patients, leading to inflammation and pain. Many plant-based herbal solutions are used for gout treatments for many decades. In this study, 11 plant compounds with potential medicinal and pharmaceutical potential were chosen for their ability to inhibit IL17A and IL 18. The structure of these ligands was obtained from pubchem (Table 1). The structure of IL17a and IL18 were retrieved from PDB. Any extra chains and water molecules were removed from the proteins using PyMol and the resulting protein file was saved as PDB file (Fig. 1).
.png) | Figure 1. Chain-A Structure of A) IL17A and B) IL18 viewed using PyMol. [Click here to view] |
For in silico analysis of plant compounds, the drug potential of all the ligands was tested using Lipinski’s rule of five. Molinspiration server was used to analyze the molecular parameters of the ligands in the Lipinski’s rule. The results include the molecular weight, no of hydrogen donors, acceptors, and lipophilicity of the ligand molecules. Out of the 11 compounds studied, 10 compounds cleared Lipinski’s rule of five, except gossypin, which showed two violations; hence, the compound was eliminated from further docking analysis. Log p value indicates the hydrophilic and hydrophobic nature of the ligands. Only thiamine, riboflavin had negative log p score indicating that these compounds are highly hydrophilic in nature (Table 2).
.png) | Table 1. Structure and medicinal uses of plant phytochemicals. [Click here to view] |
.png) | Table 2. Lipinski properties of plant compounds analyzed using molinspiration. [Click here to view] |
.png) | Figure 2. Results of amino acids involved in forming active site for IL17A. Letters highlighted in blue indicates active site residues. [Click here to view] |
.png) | Figure 3. Results of amino acids involved in forming active site for IL18. Letters highlighted in blue indicates active site residues. [Click here to view] |
.png) | Table 3. List of amino acids in the active site pocket of IL17A obtained from CASTp. [Click here to view] |
Active site pockets in IL17A and IL18 were determined using CASTp. CASTp is a web-based tool to determine the amino acid residues in the active pocket of the proteins. CASTp results are depicted in Figure 2 for IL17A and Figure 3 for IL18. From CASTp results, only the amino acids in the active site and their positions are listed as Table 3 for IL17A and Table 4 for IL18. Autodock4.2 was used for performing docking analysis for all the 10 compounds and their binding potential with both IL17a and IL18 was analyzed. Autodock4.2 is used to analyze the affinity, binding conformations, best orientation of ligand and target (Morris et al., 2009). The binding energy, number of hydrogen bond interactions, and amino acids involved in the interactions are tabulated in Table 5 for IL17A and Table 6 for IL18.
.png) | Table 4. List of amino acids in the active site pocket of IL18 obtained from CASTp. [Click here to view] |
The docking studies of the compounds with IL17A revealed that syringaresinol a polyphenolic ligand showed the lowest binding energy of −6.05 kcal/mol. With IL17A, syringaresinol formed four hydrogen bonds with Ala69, Glu68, Cys71, and Ser118 (Fig. 4A). The anti-inflammatory potential of syringaresinol was also proved by their inhibition of iNOS, COX-2, TNF-α, IL-1β, and IL-6 by in vitro studies (Bajpai et al., 2018). Diadzein, an isoflavone and a potential inhibitor of TNF-α (Soumyakrishnan et al., 2011), showed a good binding energy of -5.23 kcal/mol and formed hydrogen bonds with GLU68, SER89, CYS71 with IL17A (Fig. 4B). Canthin-6-one also proved to inhibit TNF-α and monocyte chemoattractant protein from in vitro studies (Cho et al., 2018). In this study, canthin-6-one, showed a binding affinity of −5.06 kcal/mol with the protein IL17A (Fig. 4C). With IL17A compounds Syriacusin A, Riboflavin, Genistein, and Thiamine showed a binding energy in the range of −4.96 kcal/mol and −4.1 kcal/mol. The ligands and their docking interactions with IL17A amino acids are shown in Figure 4D–G, respectively. Thymol and its isomer carvacrol are proved for their inhibitory actions against IL1β and TNF-α (Gholijani et al., 2014). In this study, docking of Thymol and Carvacrol with IL17A showed binding energies in the same range with values −3.70 kcal/mol and −3.75 kcal/mol, respectively. Figure 4H and I show the interactions of thymol and carvacrol with IL17A. All the compounds used in the study showed a binding energy greater than the standard drug Allopurinol, that showed a binding energy of −3.32 kcal/mol with IL17A (Fig. 4J)
With IL18 again syringaresinol showed the least binding energy of −6.32 kcal/mol and formed one hydrogen bond with Ser 118, in the active site of the protein (Fig. 5A). Docking of Diadzein with IL18 showed a binding energy of −5.6 kcal/mol forming hydrogen bonds at ARG 104, SER118 (Fig. 5B). Canthin-6-one, showed a binding affinity of −5.15 kcal/mol with IL18 (Fig. 5C). Syriacusin A, Riboflavin, Genistein, and Thiamine showed a binding energy in the range between −5.14 and −3.75 kcal/mol with IL18. Figure 5D–G depicts the association of these compounds with IL18, respectively. With IL18, thymol and carvacrol gave a binding energy of −3.56 kcal/mol and −3.51 kcal/mol. Figure 5H and I show thymol and carvacrol interactions with IL18. Allopurinol showed a binding energy of −3.18 kcal/mol with IL18 (Fig. 5J).
All the compounds except limonene formed bonds with the amino acids in the active site pockets of IL17A and IL18. Though limonene did not form any hydrogen bond with both IL17A and IL18, it formed other interactions with the 4 amino acids in IL17A and 11 amino acids in IL18 protein. The ADMET properties of the ligands are needed to be known for determining their drug likeliness. The ADMET properties of the ligands were analyzed using admetSAR. ADMET properties for the compounds in the study were analyzed using admetSAR. All the compounds showed good human intestinal solubility (HIA), blood brain barrier (BBB) penetration. No drug was carcinogenic. Salmonella typhimurium reverse mutation assay (AMES) toxicity is a preliminary drug screening test to analyze whether the drug causes any mutation in bacteria Salmonella typhimurium. All the compounds except Canthin-6-one and Syriacusin A were AMES negative. The lethal dose (LD50) in rats was also analyzed. The results of HIA, BB penetration, LD50 values for the compounds are listed in Table 7.
.png) | Table 5. Molecular docking analysis of plant compounds with IL17A. [Click here to view] |
.png) | Table 6. Molecular docking analysis of plant compounds with IL18. [Click here to view] |
.png) | Figure 4. 3D interaction residues of IL17A with (A) Syringaresinol, (B) Diadzein, (C) Canthine 6 one, (D) Syriacusin A, (E) Riboflavin, (F) Genistein, (G) Thiamine, (H) Thymol, (I) Carvacrol, and (J) Allopurinol. [Click here to view] |
.png) | Figure 5. 3D interaction residues of IL18 with (A) Syringaresinol, (B) Diadzein, (C) Canthine 6 one, (D) Syriacusin A, (E) Riboflavin, (F) Genistein, (G) Thiamine, (H) Thymol, (I) Carvacrol, and (J) Allopurinol. [Click here to view] |
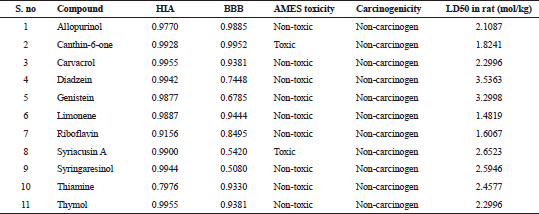 | Table 7. ADMET properties of the ligands. [Click here to view] |
CONCLUSION
Currently, the drugs available can only prevent gout attacks but does not treat gout. Hence, the identification of new drugs to target and treat gout is of utmost importance. In this study, many drug potential compounds from plants were studied using docking analysis against Gout inflammatory cytokines IL17a and IL18. All the compounds had a good inhibitory potential with both the cytokines. Among them, Syringaresinol, Diadzein, and Canthine 6 one showed good binding affinity with both IL17a and IL18. Hence, these compounds can be analyzed by further in vitro studies and can be a lead in the designing of potential drug in the treatment of Gout disease.
ACKNOWLEDGMENTS
The author would like to thank the Vels Institute of Science Technology and Advanced Studies management for their support toward the successful completion of the research work.
FINANCIAL SUPPORT
None.
CONFLICTS OF INTEREST
The author declares no conflicts of interest.
REFERENCES
Adejoro IA, Waheed SO, Adeboye OO. Molecular docking studies of Lonchocarpus cyanescens triterpenoids as inhibitors for malaria. J Phys Chem Biophys, 2016; 6(2):1–4. CrossRef
Afriza D, Suriyah WH, Ichwan SJ. In silico analysis of molecular interactions between the anti-apoptotic protein survivin and dentatin, nordentatin, and quercetin. J Phys Conf Ser, 2018; 1073(3):1–7. CrossRef
Azam SS, Abbasi SW. Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routines. TBioMed, 2013; 10(1):1–16. CrossRef
Bajpai VK, Alam MB, Quan KT, Ju MK, Majumder R, Shukla S, Huh YS, Na M, Lee SH, Han YK. Attenuation of inflammatory responses by (+)-syringaresinol via MAP-Kinase-mediated suppression of NF-κB signaling in vitro and in vivo. Sci Rep, 2018; 8(1):9216:1–10. CrossRef
Cavalcanti NG, Marques CD, Lins e Lins TU, Pereira MC, Rêgo MJ, Duarte AL, Pitta ID, Pitta MG. Cytokine profile in gout: inflammation driven by IL-6 and IL-18? Immunol Invest, 2016; 45(5):383–95. CrossRef
Chandrashekhar VM, Ganapaty S, Ramkishan A, Narsu ML. Neuroprotective activity of gossypin from Hibiscus vitifolius against global cerebral ischemia model in rats. Indian J Pharmacol, 2013; 45(6):575. CrossRef
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model, 2012; 3099–105. CrossRef
Chiu CC, Chen CH, Huang MC, Chen PY, Tsai CJ, Lu ML. The relationship between serum uric acid concentration and metabolic syndrome in patients with schizophrenia or schizoaffective disorder, J Clin Psychopharmacol, 2012; 32(5):585–92. CrossRef
Chung BH, Kim S, Kim JD, Lee JJ, Baek YY, Jeoung D, Lee H, Choe J, Ha KS, Won MH, Kwon YG. Syringaresinol causes vasorelaxation by elevating nitric oxide production through the phosphorylation and dimerization of endothelial nitric oxide synthase. Exp Mol Med, 2012; 44(3):191. CrossRef
Dai J, Li N, Wang J, Schneider U. Fruitful decades for canthin-6-ones from 1952 to 2015: biosynthesis, chemistry, and biological activities. Molecules, 2016; 21(4):493. CrossRef
de Andrade JA, Gayer CR, de Almeida Nogueira NP, Paes MC, Bastos VL, Neto JD, Alves SC, Coelho RM, da Cunha MG, Gomes RN, Águila MB. The effect of thiamine deficiency on inflammation, oxidative stress and cellular migration in an experimental model of sepsis. J Inflamm, 2014; 11(1):11. CrossRef
Doherty M. New insights into the epidemiology of gout. Rheumatology, 2009; 48:ii2–8. CrossRef
Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res, 2006; 34(suppl_2):W116–8. CrossRef
El Ridi R, Tallima H. Physiological functions and pathogenic potential of uric acid: a review. J Adv Res, 2017; 8(5):487–93. CrossRef
Feig DI, Kang DH, Johnson RJ. Uric acid and cardiovascular risk. N Engl J Med, 2008; 359(17):1811–21. CrossRef
Frieder J, Kivelevitch D, Menter A. Secukinumab: a review of the anti-IL-17A biologic for the treatment of psoriasis. Ther Adv Chronic Dis, 2018; 9(1):5–21. CrossRef
Gerald F and Falasca MD. Metabolic diseases: gout. Clin. Dermatol, 2006; 24(6):498–508. CrossRef
Gholijani N, Gharagozloo M, Farjadian S, Amirghofran Z. Modulatory effects of thymol and carvacrol on inflammatory transcription factors in lipopolysaccharide-treated macrophages. J Immunotoxicol, 2016; 13(2):157–6. CrossRef
Grayson PC, Kim SY, LaValley M, Choi HK. Hyperuricemia and incident hypertension: a systematic review and meta-analysis. Arthritis Care Res (Hoboken), 2011; 63(1):102–10. CrossRef
Huang YH, Rose PW, Hsu CN. Citing a data repository: a case study of the protein data bank. PloS one, 2015; 10(8):e0136631. CrossRef
Karunakar P, Girija CR, Krishnamurthy V, Krishna V, Shivakumar KV. In silico antitubercular activity analysis of benzofuran and naphthofuran derivatives. Tuberc Res Treat, 2014; 2014:1–10. CrossRef
Kemmish H, Fasnacht M, Yan L. Fully automated antibody structure prediction using BIOVIA tools: validation study. PLoS One, 2017; 12(5):1–26. CrossRef
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J. PubChem substance and compound databases. Nucleic Acids Res, 2015; 44(D1):D1202–13. CrossRef
Kirkham BW, Kavanaugh A, Reich K. Interleukin-17A: a unique pathway in immune-mediated diseases: psoriasis, psoriatic arthritis and rheumatoid arthritis. Immunology, 2014; 141(2):133–42. CrossRef
Kostalova E, Pavelka K, Vlaskova H, Musalkova D, Stiburkova B. Hyperuricemia and gout due to deficiency of hypoxanthine-guanine phosphoribosyltransferase in female carriers: new insight to differential diagnosis. Clin Chim Acta, 2015; 440:214–7. CrossRef
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliver Rev, 2012; 64:4–17. CrossRef
Liu Y, Zhao Q, Yin Y, McNutt MA, Zhang T, Cao Y. Serum levels of IL-17 are elevated in patients with acute gouty arthritis. Biochem Biophys Res Commun, 2018; 497(3):897–902. CrossRef
Marashly ET, Bohlega SA. Riboflavin has neuroprotective potential: focus on Parkinson’s disease and migraine. Front Neurol, 2017; 8:333. CrossRef
Meenambiga SS, Venkataraghavan R, Biswal RA. In silico analysis of plant phytochemicals against secreted aspartic proteinase enzyme of Candida albicans. J Appl Pharm Sci, 2018; 8(11):140–50. CrossRef
Mohandas R, Johnson RJ. Uric acid levels increase risk for new-onset kidney disease. J Am Soc Nephrol, 2008; 19(12):2251–3. CrossRef
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem, 1998; 19(14):1639–62. CrossRef
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem, 2009; 30(16):2785–91. CrossRef
Naghdi Badi H, Abdollahi M, Mehrafarin A, Ghorbanpour M, Tolyat M, Qaderi A, Ghiaci Yekta M. An overview on two valuable natural and bioactive compounds, thymol and carvacrol, in medicinal plants. J Med Plant Res, 2017; 3(63):1–32.
Ragab G, Elshahaly M, Bardin T. Gout: an old disease in new perspective—a review. J Adv Res, 2017; 8(5):495–511. CrossRef
Roddy E, Doherty M. Epidemiology of gout. Arthritis Res Ther, 2010; 12(223):1–11. CrossRef
Saigal R, Agrawal A. Pathogenesis and clinical management of gouty arthritis. J Assoc Physicians India, 2015; 63:56–63.
Sedimbi SK, Hägglöf T, Karlsson MC. IL-18 in inflammatory and autoimmune disease. Cell Mol Life Sci, 2013; 70(24):4795–808. CrossRef
Shi LS, Wu CH, Yang TC, Yao CW, Lin HC, Chang WL. Cytotoxic effect of triterpenoids from the root bark of Hibiscus syriacus. Fitoterapia, 2014; 97:184–91. CrossRef
Soltani Z, Rasheed K, Kapusta DR, Reisin E. Potential role of uric acid in metabolic syndrome, hypertension, kidney injury, and cardiovascular diseases: is it time for reappraisal? Curr Hypertens Rep, 2013; 15(3):175–81. CrossRef
Soumyakrishnan S, Sudhandiran G. Daidzein attenuates inflammation and exhibits antifibrotic effect against Bleomycin-induced pulmonary fibrosis in Wistar rats. Biomed Prev Nutr, 2011; 1(4):236–44. CrossRef
Sun MY, Ye Y, Xiao L, Rahman K, Xia W, Zhang H. Daidzein: a review of pharmacological effects. Afr J Tradit Complement Altern Med, 2016; 3(3):117–32. CrossRef
Verdrengh M, Jonsson IM, Holmdahl R, Tarkowski A. Genistein as an anti-inflammatory agent. Inflamm Res, 2003; 52(8):341–6. CrossRef
Yuan S, Chan HS, Hu Z. Using PyMOL as a platform for computational drug design. Wiley interdisciplinary reviews. Comput Mol Sci, 2017; 7(2):e1298. CrossRef
Zhou Z, Li X, Li H, Guo M, Liu S, Li C. Genetic analysis of IL-17 gene polymorphisms in gout in a male Chinese Han population. PLoS One, 2016; 11(2):e0148082. CrossRef