Introduction
Actinomycetes are Gram-positive filamentous bacteria having high mol % of the base guanine plus cytosine content in their genomes (Stackebrandt et al., 1997). They have been well known as the valuable economically importance microorganisms for a long time because of their ability to produce a large number of bioactive secondary metabolites (Berdy, 2005). In general, actinomycetes are widely distributed in terrestrial habitats, mainly in soils, organic materials, and plant materials (Goodfellow et al., 1988; Qin et al., 2012). Since they have been isolated from these habitats for a century, numerous redundant isolates were obtained. Three-quarters of the planet earth covers with the oceans which contain a huge biological diversity. Since the discovery of the true obligate marine actinomycetes in the genus Salinispora in the last decade, the ocean has been accepted for the existence of the actinomycetes. Moreover, the distribution of actinomycetes in the marine environment are largely unexplored (Bister et al., 2004; Jensen et al., 2007; Kwon et al., 2006). The aim of this study is to isolate, identify and screen for antimicrobial activities of the marine actinomycetes isolated from sand, sediments and marine sponges collected in Thailand.
Materials and Methods
Samples collection and isolation methods
The marine samples, including sand, sediments and marine sponges, were collected from Chumphon (Chumphon beach and Koh Khai), Chonburi (the nature education center for mangrove conservation and ecotourism, and Bangsaen beach), Phuket (Phanwa beach), Trang (Koh Rok Nork, Koh Rok Nai, and Koh Mah) and Krabi Province (mangrove forest) using scuba diving gears. They were isolated using the standard dilution-plating method. One gram of samples was suspended in 9 ml of sterile natural seawater to make 10-fold dilution series to 10–4. Each diluted suspension (0.1 ml) was spread on M1, M2 (Zhang et al., 2006) and seawater-proline media (Inahashi et al., 2011; modified with seawater) supplemented with nalidixic acid 25 μg ml–1 and cycloheximide 50 μg ml–1. The plates were incubated at 28oC for 30 days. The colonies of marine actinomycetes were observed using a light microscope and were transferred to ISP2 agar plates (Shirling and Gottlieb, 1966). The purified cultures were maintained on ISP2 medium at 4oC. All isolates were preserved using freeze-drying and freezing at –80oC in 15% (v/v) glycerol solution.
Identification methods
Phenotypic, chemotaxonomic, and genotypic characterization
The marine actinomycete isolates were identified using the phenotypic and genotypic characteristics. Morphological characteristics of the isolates were observed on the culture grown on ISP2 medium at 28oC for 14 days. Cultural characteristics were determined using 14-day cultures grown at 28oC on yeast extract-malt extract medium (ISP2 medium). The isomers of diaminopimelic acid were analyzed using the standard TLC method (Staneck and Robert, 1974).
The genomic DNA for 16S rRNA gene amplification was extracted from the cells using the method as described by Tamaoka (1994). The amplification of 16S rRNA gene was carried out using two primers 20F (5’-GAGTTTGATCCTGGCTCAG-3’, positions 9-27) and 1500R (5’-GTTACCTTGTTACGACTT-3’, positions 1492-1509). The PCR (Polymerase chain reaction) mixture (final volume 100 μl) contained 4 μl each of primers (10 pmol/μl), 2 μl of dNTP (10 mM), 10 μl of 10x Taq buffer, 8 μl of MgCl (25 mM), 0.5 μl of Taq DNA polymerase, 61.5 μl of dH2O, and 10 μl of template DNA. The amplification was performed with an initial denaturation at 94oC for 3 minutes, followed by 30 cycles with denaturation at 94oC for 1 minute, annealing at 50oC for 1 minute and extension at 72oC for 2 minutes, followed by the last step at 72oC for 3 minutes (Suriyachadkun et al., 2009). The PCR product was purified using the PCR purification kit (Gene aid). The sequencing of nucleotides was performed using universal primers, 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 800R (5’-TACCAGGGTATCTAATCC-3’) (Lane, 1991) (Macrogen; Seoul, Korea).
Antimicrobial activities
The culture broth library was prepared by using four different production media including 301 medium, 54 medium (2 g soluble starch, 0.5 g glycerol, 1 g defatted wheat germ, 0.3 g meat extract, 0.3 g yeast extract, 0.3 g CaCO3, 500 ml tap water, 500 ml artificial seawater, pH 7.4–7.8), 51 medium, and yeast-dextrose (YD) broth (Sripreechasak et al., 2013; 2014). The inoculum of each strain was cultured in yeast extract-dextrose broth for 4–7 days. Then, 0.1 ml of the culture was transferred to 10 ml of the screening production media and incubated in a shaking condition at 180 rpm at 30oC for 7–14 days. After incubation, 10 ml of 95% ethanol were added into the culture broth and shaked at 180 rpm for 2 hours. The extract solution was centrifuged at 3,400 rpm for 15 minutes and preserved at –20 oC. The production media without the culture was used as the negative control.
The screening of antimicrobial activities were performed using agar disc diffusion method (Qin et al., 2009). Each of paper disc (8 mm) was soaked into the extract solution and air-dried. After drying, the discs were put onto the surface of the agar plate containing a tested microorganism and cooled at 4oC for 30 minutes before incubation. The bacterial plates were incubated at 37oC for 24 hours while yeast and filamentous fungus were incubated at 30oC for 48 hours. The inhibition zones (mm) were measured using a Vernier caliper. Three bacteria, Bacillus subtilis ATCC 6633, Staphylococcus aureusATCC 25923, Escherichia coliNIHJ KB213, one yeast, Candida albicans KF1, and one filamentous fungi Mucor racemosus IFO 4581o were used as the tested microorganisms.
Results and Discussion
Identification
On the basis of 16S rRNA gene sequence analysis and morphological characteristics, the actinomycete isolates were belonged to the family Micromonosporaceae, Nocardiaceae and Streptomycetaceae (Table 1). They were identified as Salinispora (13 isolates), Micromonospora (11 isolates), Nocardia(1 isolate), Verrucosispora (2 isolates), and Streptomyces (14 isolates).
Group I, the members of this group exhibited a monomeric spore on the substrate mycelia but lacked aerial mycelia. They contained meso-diaminopimelic acids. They were divided into three genera based on the key morphological and chemotaxonomic characteristics and16S rRNA gene sequence analysis including phylogenetic tree relationship.
Group IA comprised 13 isolates (KRN2-1, SPM3-5, SPM3-1, SPM3-3, SPM3-6, SPM3-7, SPM9-1, SPM9-2, SRK1-1, SRK1-2, SRK1-3, SRK2-1 and SRK2-3) (Table 1). All strains required sea water for the growth. The 16S rRNA gene analysis revealed that these isolates showed the highest similarity with Salinispora arenicola CNH-643o (100% similarity). Therefore, they were identified as S. arenicola (Maldonado et al., 2005).
Group IB consisted of 11 isolates (BS-003, BS-007, CH7-4m, CPB1-11, CPB1-21, KK1-10, PW-001, PW-002, PW-004, PW-006 and PWB-012) (Table 1). Almost all isolates showed orange to brown substrate mycelia which changed into a dark brown or black color when cultured more than 10 days. Based on BLASTn and phylogenetic analyses, these strains showed the highest 16S rRNA gene sequence similarities with type strains of the members of the genus Micromonospora (Kawamoto, 1989).
Group IC Verrucosisporacomprised two isolates including KK2-1 and SPM3-8 (Table 1). The representative isolate KK2-1 produced hairy spores which born singly on the substrate mycelia. BLASTn analysis of 16S rRNA gene sequences revealed that the isolates KK2-1 and SPM3-8 were the closest similar to Verrucosispora gifhornensis DSM 44337T (99.7%) and Verrucosispora sediminis MS426T (99.5%), respectively. On the basis of 16S rRNA gene sequence, they were identified as Verrucosispora (Rheims et al., 1998).
Group II consisted of one isolate, PWB-002. This isolate showed the fragmentation on the substrate mycelia and contained meso-diaminopimelic acid in cell-wall peptidoglycan. It showed the phylogenetic relationship within the genus Nocardia. Based on the fragmentation of substrate mycelia and 16S rRNA gene sequence analysis, this isolate was identified as Nocardia(Kageyama et al., 2004).
Group III, 14 isolates including SRN1-2, LKB1-4, KK5-10, KRN1-1, PWB-011, BM2-4, CPB1-13, LKB1-6, PWB-010, PWB-016, SRN1-1, LKB1-5, LKB1-7 and LKB1-11 (Table 1). They produced extensively branch aerial and substrate mycelia. Almost all isolates produced spiral spore chains, while long straight spore chains were occasionally observed. The chemotaxonomic analysis revealed that all of these isolates contained LL-diaminopimelic acids. In addition, the 16S rRNA gene sequence analysis (ranged from 98.2% to 99.9% similarity), these isolates shared the clade within the genus Streptomyces(Mao et al., 2007).
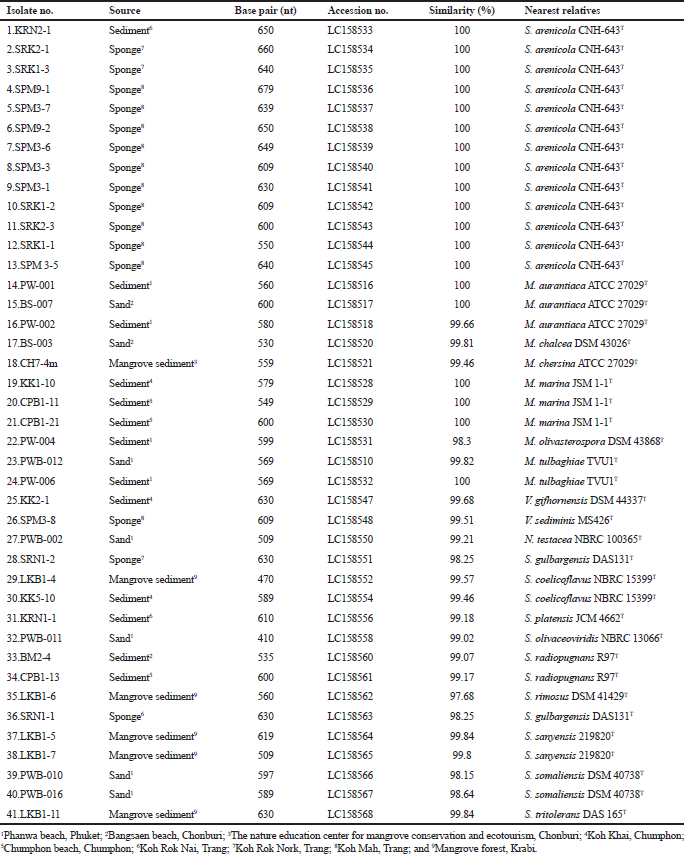 | Table 1. Isolate number, sources of isolation, length of nucleotide sequence, accession no., 16S rRNA gene sequence similarity (%) and nearest relatives. [Click here to view] |
Antimicrobial activities
Fourteen isolates of the family Micromonosporaceae showed antimicrobial activities against tested microorganisms. Twelve isolates from genus Salinispora including the isolates SRK2-1, SRK1-3, SPM9-1, SPM3-7, SPM9-2, SPM3-6, SPM3-1, SRK1-2, SRK2-3, SRK1-1 and SPM3-5 showed antimicrobial activities against S. aureus ATCC 25923, Kocuria rhizophila ATCC 9341 and B. subtilis ATCC 6633, while one isolate, KRN2-1 showed activities against S. aureus ATCC 25923 Tables 2 and 3. All of these isolates were identified as S. arenicola.
Two isolates of the genus Micromonospora including the isolate CH7-4m and PW-004 showed antimicrobial activities against tested microorganisms. The isolate CH7-4m identified as M. chersina ATCC 27029T, exhibited activities against S. aureus ATCC 25923, K. rhizophila ATCC 9341, B. subtilis ATCC 6633. In addition, the isolate PWB-004 identified as Micromonospora olivasterospora, exhibited activities against B. subtilis ATCC 6633.
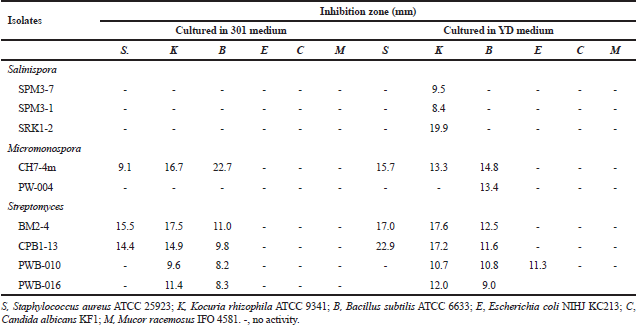 | Table 2. Antimicrobial activities of Salinispora, Micromonospora and Streptomycesisolates when cultured in 301 and YD media. [Click here to view] |
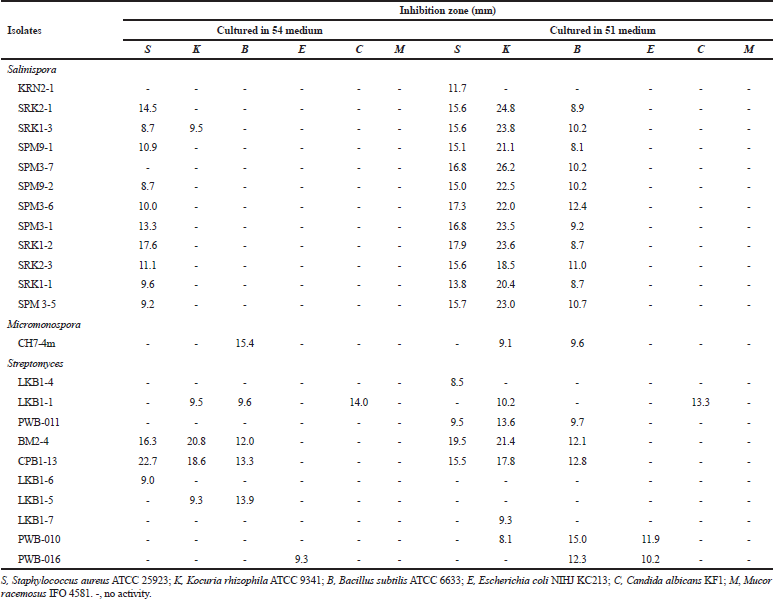 | Table 3. Antimicrobial activities of Salinispora, Micromonospora and Streptomycesisolates when cultured in 54 and 51 media. [Click here to view] |
Ten Streptomyces isolates showed the antimicrobial activities against tested microorganisms. However, six isolates (SRN1-2, KK5-10, KRN1-1, CH3-1, SRN1-1 and LKB1-11) did not show any antimicrobial activities when cultured in four different media. Among the active isolates, anti-Gram-positive bacteria activity could be observed in most isolates, while anti-Gram-negative bacteria, anti-yeast, and anti-mold activities were observed only 5, 8, and 4 isolates, respectively. The detailed antimicrobial activities of the Streptomyces isolates are shown in Tables 2 and 3. No antimicrobial activities were observed for the members of the genus Nocardia and Verrucosispora.
According to the screening of their antimicrobial activities, the good example could be observed in the members of the Salinispora group (Tables 2 and 3). No activities were observed when they were grown in 301 medium, but almost all isolates showed the activities against tested Gram-positive bacteria when they were grown in 51 medium. Besides this, the difference between strains within the same species but different isolates are other factors for the determination of the antimicrobial activity. For example, isolate SRK1-2 identified as S. arenicola showed activities in 54 and 51 media. On the basis of this study, it suggested that the screening should be determined using various different media. Moreover, the same species of the isolates does not mean the same activities will be observed. In addition, the further elucidation of bioactive compounds of S. arenicola, Micromonosporaand Streptomycesisolates mentioned should be done as previously reported (Abdelmohsen et al., 2014; Supong et al., 2012).
Conclusion
In this study, the marine actinomycetes, S. arenicola, Micromonospora aurantiaca, Micromonospora chalcea, Micromonospora chersina, Micromonospora marina, M. olivasterospora, Micromonospora tulbaghiae, V. gifhornensis, V. sediminis, Nocardia testacea, Streptomyces coelicoflavus, Streptomyces gulbargensis, Streptomyces olivaceoviridis, Streptomyces platensis, Streptomyces radiopugnans, Streptomyces rimosus, Streptomyces sanyensis, Streptomyces somaliensis, and Streptomyces tritoleransisolates were distributed in sand, mangrove sediments, marine sediments, and marine sponges collected from Chumphon, Chonburi, Phuket, Trang, and Krabi provinces in Thailand. Micromonospora, Salinispora, and Streptomyces isolates showed antimicrobial activity against S. aureus ATCC 25923, K. rhizophila ATCC 9341, B. subtilis ATCC 6633, E. coli NIHJ KC213, C. albicans KF1, and M. racemosus IFO 4581.
Acknowledgments
We thank Prof. Dr. Yoko Takahashi, Prof. Dr. Kazuro Shiomi, Dr. Atsuko Matsumoto, and the staffs of Kitasato Institute for Life Sciences, Kitasato University, Tokyo, Japan for supporting the screening and sequencing of the isolates.
Financial Support and Sponsorship
This study was supported by the Thailand Research Fund via a 2011 Royal Golden Jubilee Ph. D. Program as a scholarship to W. P. and we are grateful to the Plant Genetic Conservation Project under the Royal Initiative of Her Royal Highness Princess Maha Chakri Sirindhorn for the marine sediment samples collection.
Conflict of Interest
There are no conflicts of interest.
REFERENCES
Abdelmohsen UR, Bayer K, Hentschel U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat Prod Rep, 2014; 31:381–99. CrossRef
Berdy J. Bioactive microbial metabolites. J Antibiot, 2005; 58:1–26. CrossRef
Bister B, Bischoff D, Ströbele M, Riedlinger J, Reicke A, Wolter F, Bull AT, Zähner H, Fiedler HP, Süssmuth RD. Abyssomicin C-A polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew Chem Int Ed Engl, 2004; 43(19):2574–6. CrossRef
Goodfellow M, Williams ST, Mordarski M. Actinomycetes in biotechnology. Academic Press Inc, London, UK, pp 1–88, 1988.
Inahashi Y, Matsumoto A, Omura S, Takahashi Y. Streptosporangium oxazolinicum sp. nov., a novel endophytic actinomycete producing new antitrypanosomal antibiotics, spoxazomicins. J Antibiot (Tokyo), 2011; 64(4):297–302. CrossRef
Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol, 2007; 73(4):1146–52. CrossRef
Kageyama A, Yazawa K, Nishimura K, Mikami Y. Nocardia testaceus sp. nov. and Nocardia senatus sp. nov., isolated from patients in Japan. Microbiol Immunol, 2004; 48:271–6. CrossRef
Kawamoto I. Genus Micromonospora. In: Williams ST, Sharpe ME, Holt JG (eds.). Bergey’s manual of systematic bacteriology, vol. 4, Williams & Wilkins, Baltimore, MD, pp 2442–50, 1989.
Kwon HC, Kauffman CA, Jensen PR, Fenical W. Marinomycins A-D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora”. J Am Chem Soc, 2006;128(5):1622–32. CrossRef
Lane DJ. 16S/23S rRNA sequencing. In Strackbrandt E, Goodfellow M (eds.). Nucleic acid techniques in bacterial systematics, Wiley, Chichester, UK, pp 115–48, 1991.
Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC, Bull AT, Goodfellow M. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol, 2005; 55:1759–66. CrossRef
Mao J, Tang Q, Zhang Z, Wang W, Wei D, Huang Y, Liu Z, Shi Y, Goodfellow M. Streptomyces radiopugnans sp. nov., a radiation-resistant actinomycete isolated from radiation-polluted soil in China. Int J Syst Evol Microbiol, 2007; 57:2578–82. CrossRef
Qin S, Chen H-H, Zhao G-Z, Li J, Zhu W-Y, Xu L-H, Jiang J-H, Li W-J. Abundant and diverse endophytic actinobacteria associated with medicinal plant Maytenus austro- yunnanensis in Xishuangbanna tropical rain forest revealed by culture-dependent and culture-independent methods. Envion Microbiol Report, 2012; 4:522–31. CrossRef
Qin S1, Li J, Chen HH, Zhao GZ, Zhu WY, Jiang CL, Xu LH, Li WJ. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl Environ Microbiol, 2009; 75(19):6176–86. CrossRef
Rheims H, Schumann P, Rohde M, Stackebrandt E. Verrucosispora gifhornensis gen. nov., sp. nov., a newmember of the actinobacterial family Micromonosporaceae.Int J Syst Bacteriol, 1998; 48:1119–27. CrossRef
Shirling EB, Gottlieb D. Methods for characterization of Streptomycesspecies. Int J Syst Bacteriol, 1966; 16:313–40. CrossRef
Sripreechasak P, Tanasupawat S, Matsumoto A, Inahashi Y, Suwanborirux K, Takahashi Y. Identification and antimicrobial activity of actinobacteria from soils in southern Thailand. Trop Biomed, 2013; 30(1):46–55.
Sripreechasak P, Suwanborirux K, Tanasupawat T. Characterization and antimicrobial activity of Streptomycesstrains from soils in southern Thailand. J App Pharm Sci, 2014; 4(10):024–031.
Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol, 1974; 28:226–31.
Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol, 1997; 47:479491. CrossRef
Suriyachadkun C, Chunhametha S, Thawai C, Tamura T, Potacharoen W, Kirtikara K, Sanglier JJ. Planotetraspora thailandica sp. nov., isolated from soil in Thailand. Int J Syst Evol Microbiol, 2009; 59:1632–7. CrossRef
Supong K, Thawai C, Suwanborirux K, Choowong W, Supothina S, Pittayakhajonwut P. Antimalarial and antitubercular C-glycosylated benz[a]anthraquinones from the marine-derived Streptomyces sp. BCC45596. Phytochem Lett, 2012; 5:651–6. CrossRef
Tamaoka J. Determination of DNA base composition. In Goodfellow M, O’Donnel AG (eds.). Chemical methods in prokaryotic systematics, Wiley, Chichester, UK, pp 463–70, 1994.
Zhang H, Lee YK, Zhang W, Lee HK. Culturable actinobacteria from the marine sponge Hymeniacidon perleve: isolation and phylogenetic diversity by 16S rRNA gene-RFLP analysis. Antonie Van Leeuwenhoek, 2006; 90(2):159–69. CrossRef