INTRODUCTION
Base on International Agency for Research on cancer, with emphasis on geographical differences in 20 countries in the world, The GLOBOCAN 2018 estimates of cancer mortality and incidence (Bergman et al., 2000). The first prevalence of cancer in women is breast cancer, the second is colorectal and lung cancer (for incidence), followed by vice versa (for mortality), and the fourth, for mortality and incidence, is cervical cancer (Ferlay et al., 2010).
Breast tissues differentiation and development influence by ovarian hormones (Bernstein and Press, 1998). Breast is one of the estrogen-responsive target tissue induces of cancer development. Tamoxifen is one of the nonsteroidal compounds (Bentrem and Craig Jordan, 2002), which has been studied, whose effect is varied as agonists or antagonists based on the investigation of a gene or particular organ system (Rojas and Stuckey, 2016).
Estrogen has a play role in growth, development, and in the pathology of bones, breast, and uterus. Estrogen receptor is classified into two subtypes, ERα (Estrogen Receptor-α) and Erβ (Payne et al., 2008). The role of ERα is in cell proliferation (Fox et al., 2008), and ERα found in endometrial, mammary epithelial cells which are the origin cell for growth in most breast cancers, ovarian stromal cell, and hypothalamus (Levin, 2005). ERα plays an important and responsible role as the most common trigger of breast cancer (Narod, 2011). Various molecules have been an investigation to find out compounds that bind well with ERα, as a crucial receptor for breast cancer (Cragg et al., 1997). The most widely used as hormonal therapy of breast cancer is tamoxifen. The risk of recurrence and death of breast cancer are reduced by tamoxifen when given as adjuvant therapy. It also provides effective palliation for patients with metastatic breast cancer (Yang et al., 2013). Tamoxifen is a Selective Estrogen Receptor Modulator (SERM) that has antagonist activity to breast cancer but agonist activity to other receptors, especially in the uterus (Fisher et al., 2005). Tamoxifen is given to women who have stopped menstruation with ERα+ tumors. Tamoxifen plays a crucial role in breast cancer therapy. Tamoxifen can reduce breast cancer relapse significantly (Davies et al., 2013). Tamoxifen interacts with co-repressors, thus inhibiting expression of estrogen-dependent response genes (Chang, 2012). Besides the benefit of tamoxifen, there are adverse effects in the uterus (Fisher et al., 2005). Depending on the results of the study, endometrial cancer risk increased from 1.5 to 6.9 fold (Cohen, 2004). The risk of adverse effect in the uterus increases with accumulative usage and longer duration of tamoxifen therapy (Bergman et al., 2000). The risk of endometrial malignancy increases significantly with an increased body weight of postmenopausal females. Besides that, patients with ER+ show the intrinsic resistance to SERM not depend on ER increased (Fan et al., 2015). The mechanism of tamoxifen resistance occurs by loss of ERα expression, which leads to the removal of ligand for tamoxifen, change mechanism of co-activators or co-regulators, stimulate kinases and ER phosphorylation, change profile pharmacokinetic of metabolites active of tamoxifen, regulation of apoptosis, and antioxidant protein-mediated cell survival (Chang, 2012). Because of these cases, an alternative treatment was needed through the natural compound. Drug discovery from medicinal plants has played an important role in cancer treatment and, indeed, the newest therapy of herbal medicine practiced in fighting cancer (Prasad et al., 2006).
Rational drug discovery and development of new active agents or leads is utilizing in silico study. Ligand-based drug design and structure-based drug design (SBDD) were used as a modern method in drug discovery (Dror et al., 2004). One of the natural compound, which has potential as anti-breast cancer, is chalcone. Chalcone is one of the flavonoid groups secondary metabolite that is found in many plants (Prasad et al., 2006). In the previous study, 2’,4’-dihydroxy-6-methoxy-3,5-dimethylchalcone was isolated from leaves of Eugenia aquea and was further investigated on to breast cancer therapy (Subarnas et al., 2015). The results showed that the chalcone isolates reduce cell proliferation against Michigan Cancer Foundation-7 human breast cell using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide bioassay in a dose-dependent manner with the half maximal inhibitory concentration (IC50) of 74.5 μg/ml (250 μM) and induce apoptosis via the activation of poly (adenosine diphosphate-ribose) polymerase (Subarnas et al., 2015). However, this IC50 was categorized in moderate potential and this compound cannot compete with tamoxifen due to its lack of hydrophobic tail. So, we need an effort to improve 2’,4’-dihydroxy-6-methoxy-3,5-dimethylchalcone potency and the aim of this study is to find the best-modified chalcone that binds well with ERα by replacement of the carbonyl group of that chalcone that binds well with the ERα.
MATERIALS AND METHODS
Molecular docking simulation
3ERT taken from protein data bank (PDB) used as a standard was complex ERα with 4-hydroxytamoxifen (4-OHT). The ligand and macromolecule structures were separated using Discovery Studio 4.0. The SBDD using molecular docking simulation methods has been carried out in the previous study (Muchtaridi et al., 2014; 2017). Using AutoDockTools 1.5.6, all the ligands and receptor were prepared for docking simulation and protonated. The solvation and default Kollman charge parameters were designate to the macromolecule atoms. Addition of Gasteiger charges to molecule as a ligand atom A grid box comprised of 40 × 40 × 40 points distance by 0.375 Å and was focused on the ER binding site (x = 30.282, y = −1.913, and z = 24.207). The bond strength of the atom in the ligand was calculated using an Autogrid (Morris et al., 2009). The Lamarckian genetic algorithm (LGA) specifications were 100 runs, elitism of 1, the mutation rate of 0.02, the population size of 100, and a crossover rate of 0.08 band 10,000,000 energy evaluations (Ikram et al., 2015). A root means square deviation was used for clustering the results of docked conformation, tolerance of 1.0 Å. The docking outcomes were imaged using Discovery Studio Visualizer 4.0.
Pharmacophore modeling
Ligand-based drug design using pharmacophore fit score calculated the qualified element of the corresponding site of the compound to the features of pharmacophore model and was used to interpret the value of the corresponding site from the pharmacophore model. A 3-D pharmacophore model using LiganScout 4.1 was derived from the X-ray derived structure of ERα that binds with 4-OHT (Wolber and Langer, 2005).
preADMET and Toxtree
In silico, pharmacokinetic properties and toxicities were predicted using preADMET and Toxtree software, which are available online (Lee et al., 2003). preADMET (v2.0) online at https://preadmet.bmdrc.kr/ is a web-based application for predicting absorption and distribution data using in silico method. The Toxtree v2.3.16 that we used was available online at https://sourceforge.net/projects/toxtree/files/toxtree/Toxtree-v.2.6.13/Toxtree-v2.6.13-setup.exe/download, used to predict the toxicity of compounds.
RESULTS AND DISCUSSION
Modification of chalcone
Molecular docking of protein that binds with ligand is most widely used as Structure-Based Virtual Screening method. It is estimated affinity of the ligand and protein derived on its intermolecular interactions in the binding site. Chalcone derivatives are simple compounds, have ease of replacing hydrogen atom, easy and simple synthesis, with a numerous prospective effect as a new drug (Todorova, 2010). However, it is necessary to develop chalcone as a new drug based on its chemical properties. The in silico study base on computer-aided drug design, especially to chalcone is needed for more valuable research (Gomes et al., 2017).
A number of compounds of pyrazolic chalcone derivatives were shown to have anticancer activity, based on 50% inhibitory concentration (IC50) values (Hawash et al., 2017). Extension of the functional group to ring A and/or ring B (Fig. 1) can induce the activity of chalcone (Lahsasni et al., 2014).
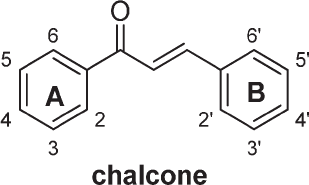 | Figure 1. Basic structure of chalcone. [Click here to view] |
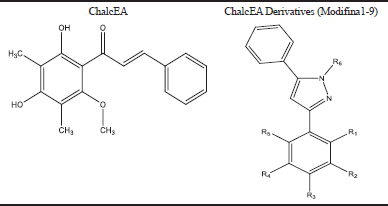 | Table 1. Modification of chalcone. [Click here to view] |
In our study, in order to find the best-modified chalcone, we were replaced the carbonyl group of ChalcEA, to increase hydrophobicity, which is the important pharmacophore using hydrazine to produce pyrazole derivatives, to increase selectivity and activity against the ERα using rational drug design. The design of chalcone modifications is presented in Table 1.
Pharmacophore modeling
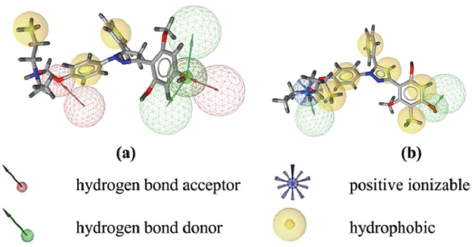
The important feature that provides to biological activity is represented by pharmacophore (Wolber, 2008). Complexes of ligand-protein in PDB, the compound as a ligand separated from the protein and elucidated chemical properties of the ligand. Pharmacophore modeling representing the interaction of ligand and receptor which are generated from every one of these molecules and its surrounding them (Wolber and Langer, 2005). Figure 2a shows the pharmacophore of 4-OHT. The interaction between ERα (PDB code: 3ERT) with 4-OHT forms hydrophobic interactions predominantly with aromatic rings, hydrogen bond interactions, and positive ionizable interaction. The 2-D (Fig. 2b) pharmacophore modeling illustrates the interaction between a hydrophobic pocket with amino acids residue.
Pharmacophore fit scores indicate that chemical properties of the ligands are suitable to the feature of the 4-OHT structure-based pharmacophore model. Table 2 showed that Modifina1 and Modifina3 were the best two pharmacophores fit score, which means the chemical properties of that ligands are most suitable with the features of the 4-OHT pharmacophore model. However, Modifina5 and Modifina7 do not have pharmacophore fit score, which means that chemical features of those compounds are not aligned to the feature of the 4-OHT pharmacophore modeling.
Molecular docking results
SBDD is a method that depends on possessing the knowledge of the 3-D structure of the receptor as a biological target (Kalyaanamoorthy and Chen, 2011). Structural determination of biological macromolecules by using X-Ray crystallographic is currently the most favored method (Smyth and Martin, 2000). Crystallographic ERα structure a that binds with 4-OHT from PDB code: 3ERT was preferred for molecular docking simulation of chalcone and its derivatives since they have suitable criteria for research resolution (1.9 Å). Molecular docking is commonly used for prediction of biology molecules complexes in molecular design. The fast results of free energy binding in Autodock come from combining a force field of empirical free energy with an LGA (Morris et al., 2009).
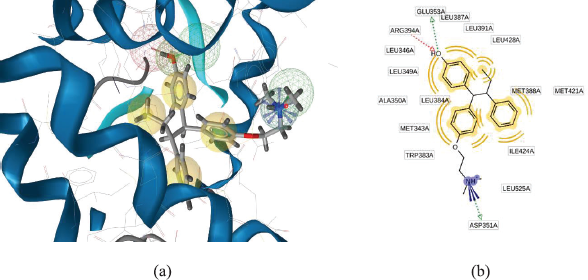 | Figure 2. (a) The 3-D pharmacophore modeling using LiganScout 4.1 based of 4-OHT that complexed with ERα f (PDBid: 3ERT). Yellow spheres, blue star, green and red arrows were illustrating of hydrophobic, positive ionizable, hydrogen bond donor, and acceptor interaction, respectively. (b) The 2-D pharmacophore modeling illustrates the interaction between hydrophobic pockets with the binding site residues. [Click here to view] |
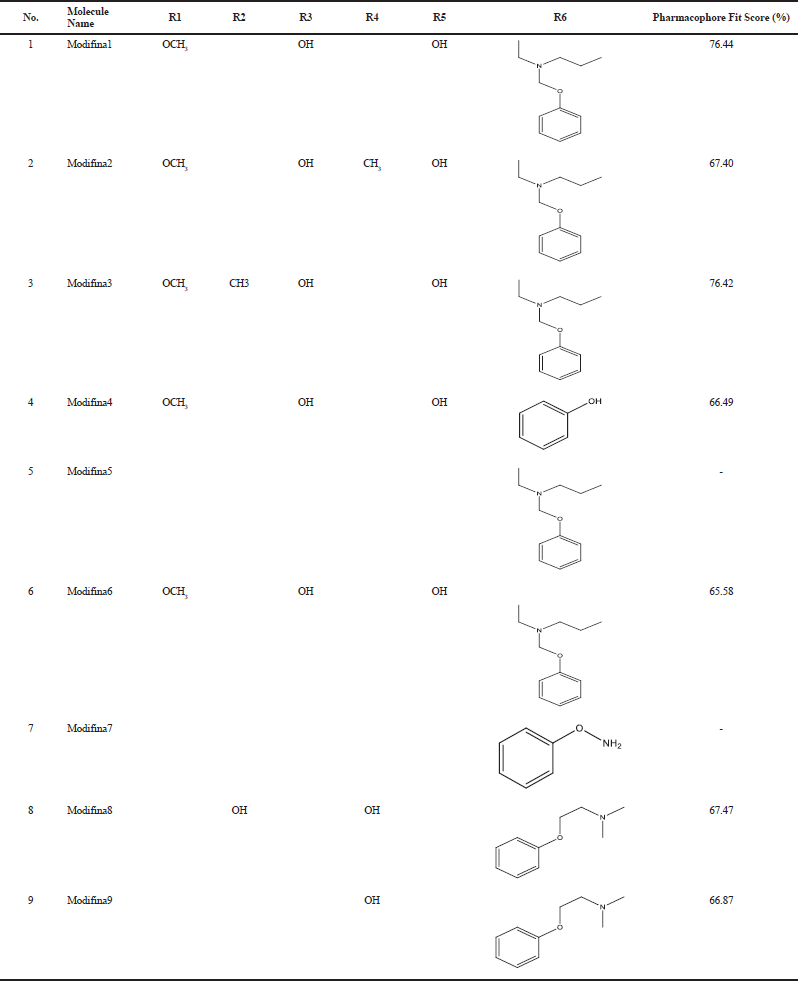 | Table 2. Pharmacophore fit score result. [Click here to view] |
Validation of molecular docking is done first, before performing molecular docking simulation of ligand that will be tested, conducted by separating 4-OHT from ERα in PDB and docking it into active site to verify that the method running well as the bioactive conformation antagonist of 4-OHT. The best-docked antagonist bioactive ligand conformation is shown in Figure 3. Chalcone is a secondary metabolite and its derivatives have several anticancer activities. In addition, other advantages of chalcone are ic hardly interact with DNA and less mutagenic (Xu et al., 2015). Both synthesis and natural products chalcones have been proven in the various studies for important pathway or molecular targets in cancers. Chalcone has the advantages of being inexpensive, easily available, and less toxic. Moreover, chalcones are not difficult to synthesize, which makes them an attractive drug scaffold (Jandial et al., 2014). Chalcone was discovered in recent time as a potential and specific inhibitor. However, chalcone has cytotoxic activity. Replacement at positions 3, 4, and 5 of chalcone induced cytotoxic activity (Rangel et al., 2013). Modifying the carbonyl group of the chalcone with pyrazole group was proved to induce better cytotoxicity against many cancer cell line (Hawash et al., 2017). The aim of this study was to find the best-modified chalcone that binds well with the ERα and to focus on the modification at the position of the carbonyl group by pyrazole derivate to increase the hydrophobicity.
Chalcone modification based on ERα interaction with 4-OHT (Fig. 2a), molecular bond acceptors, and less than five hydrogen bond donor base on Lipinski’s Rule of Five. Table 3 below shows molecular docking results using Autodock 4.2. The dimethylamino ethoxy group of 4-OHT elongated than the carbonyl group of ChalcEA. This unlikeness makes it possible for higher computer free energy of binding (ΔG) of 4-OHT lower than ChalcEA (Muchtaridi et al., 2017). All of the modified chalcones have a free energy of binding (ΔG) lower than ChalcEA, except Modifina8 and 9, formed hydrogen bonds with Arg394 similar to 4-OHT. Modifina3 was the best compound with lowest free energy binding, so it has the highest affinity to bind properly with the ERα and interacted with Arg394, Leu387, Leu346, Glu353, Leu525, Leu536, Trp383, Leu354, Ala350, Leu384, Leu349, and Leu391. However, Modifina3 has more Leusin, which is hydrophobic amino acids residue, and amino acids residue polar Met421, Met343, and Asp351 interacted with 4-OHT which made the free energy binding (ΔG) of Modifina3 lower than 4-OHT which cause Modifina3 has the highest affinity to ERα. The hydrogen bonds formation with Arg394 and Glu353 is essential to ERα.
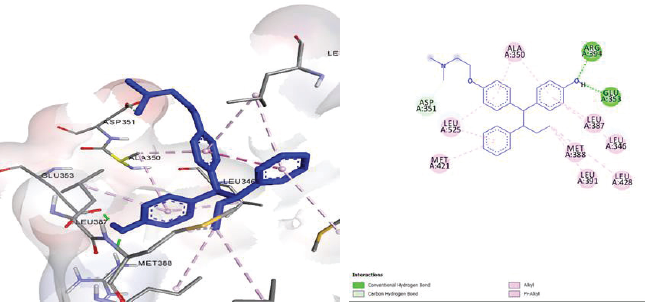 | Figure 3. Result of molecular docking method validation. The best conformation of docking pose of 4_OHT with ERα (3 ERT) using Autodock 4.2. [Click here to view] |
 | Table 3. Molecular docking result. [Click here to view] |
Interpretation of the in silico results
Modifina1 and Modifina3 show the best two pharmacophore fit scores (Fig. 4) of derivatives and Modifina3 has lowest ΔG free energy of binding (−11.07 kcal.mol−1) than Modifina1 (−10.51 kcal.mol−1) and 4-OHT (−10.15 kcal.mol−1), respectively.
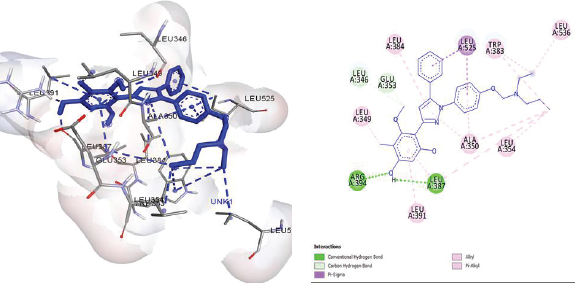
Figure 3 shows that the modification of carbonyl groups on Modifina1 blocking complete interaction with all three hydrophobic features of 4-OHT (Fig. 4a). On the other hand, the meta position of methyl groups on the ring of Modifina3 enables better alignment with the center of the hydrophobic features, thus resulting in a better pharmacophore fit score (Fig. 4b)
Modifina3 formed 10 hydrophobic interactions with Leu387, Leu346, Leu525, Leu536, Leu354, Ala350, Leu384, Leu349, Tryptopan383, and Leu391, and two hydrogen bonds with Leu387 and Arg394 (Fig. 5). The interaction of hydrogen bonds with Glu353 and Arg394 is necessary for binding to ER (Wang et al., 2010), which is in suitable with the results of previous docking studies requiring chalcone (Vasanthi, Reuben and Usha, 2016). Hydrophobic interactions were the most important site of binding of ChalcEA derivatives with ERα. Role of aromatics ring in binding interaction was shown in Fig. 4.
 | Figure 4. Fit of the (a) Modifina1 and (b) Modifina3 were derived from 4-OHT pharmacophore models with ERα (3ERT) by LigandScout 4.1 Advanced. Virtual screening was conducted leaving at least two features out. [Click here to view] |
 | Figure 5. Molecular docking result of Modifina3. There is hydrogen bond with Arg394, Glu353, and Leu387 amino acids residues. [Click here to view] |
The cyclic compound of Modifina3 formed CH-pi hydrophobic binding with Ala350, Met421, and Leu525 (Fig. 6). In the previous study, the substituent of ring A and B of chalcone plays a role in the activity (Wang et al., 2010).
In silico prediction of absorption, distribution, and toxicity
In the latest decades, in silico absorption, distribution, metabolism, excretion/toxicity (ADME/T) modeling as a computation method for rational drug design with various models has been used by pharmaceutical scientists. The high analysis results and the efficient cost of the method made the simultaneous investigations of the compounds, including pharmacokinetic profile, safety, and activity. The compounds that used as a drug must have good ADME properties. The complex mechanism of in vivo process of a drug made the ADME prediction method more simple using major component or as several single processes (Thomas et al., 2008).
Determination of permeation across a monolayer of the human adenocarcinoma cell line, Caco-2 is a popular surrogate for ligands permeation across the human intestinal epithelium. Human intestinal absorption (HIA%) of compounds was predicted very important for identifying a potential drug candidate. Among HIA of all the modified chalcones, more than 90% represented well-absorbed compounds (70–100%) in the intestines. The parameter of CaCO2 cell permeability capability shows all the compounds have a medium permeability (20–70%) (Thomas et al., 2008).
Protein plasma binding (PPB) is a significantly pharmacokinetic property of compounds in drug discovery and design. Effective and efficient in silico method for a pharmacokinetic profile of compounds. PPB is exactly related to drug distribution, metabolism, and clearance, which influence the efficacy and potency of drugs (Sun et al., 2018). A degree of PPB of a compound influences on the drug disposition, its action, and efficacy. PPB of all the compounds in Table 4 was more than 90%, which means all compounds will be well distributed in the body, chemical’s strongly bound. However, transport across cell membranes or diffusion, and bind with a pharmacological target (receptor) only requires the unbound drug. If there is reversible protein plasma-drug binding, there will be a chemical balance between protein plasma-drug binding with the unbound drug. The bound part of the drug will become a reservoir and then release as unbound drug to maintain equilibrium.
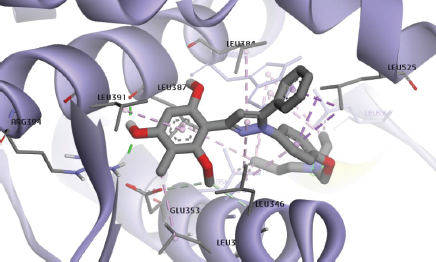 | Figure 6. Interaction of Modifina3 within the binding site of ERα. Ion-ion interaction, hydrogen bond, and pi-alkyl interactions are represented in purple and green colored dashed lines, respectively. [Click here to view] |
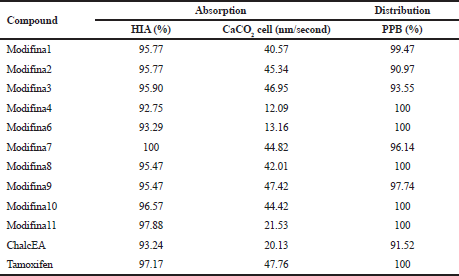 | Table 4. ADME prediction result. [Click here to view] |
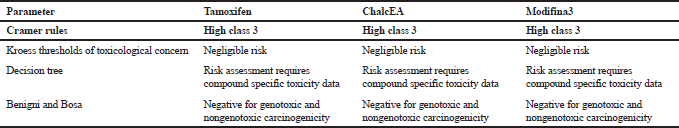 | Table 5. Toxtree result. [Click here to view] |
In silico toxicity prediction
Prediction toxicity using a computational method is needed in the early step of drug development. Toxicity is the concentration level of the compounds which disturbs an organism or its substructure (Wang et al., 2015).
In the drug design and development, genotoxicity is the critical point in the in vitro toxicity assay. Information Technology development and growing experimental data made in silico screening and toxicity prediction interesting (Wang et al., 2015).
In silico toxicity risk was performed to check the genotoxic and carcinogenic effect of the compound. In Table 5 shown that the Modifina3 has nongenotoxic and non-carcinogenicity properties.
In order to improve the patient’s quality of life through reduced side effect of the chemotherapeutic agents for breast cancer, Modifina3 is possibly potent for that activity. The molecular docking results showed that most of the residue which interacted with Modifina3 and 4-OHT were Arg394, Glu353, Leu387, Ala350, Leu346, and Leu525 almost similar. Agonistic effect of 4-OHT can be eliminated by Modifina3. In the previous study, molecular dynamic simulations on ERα showed that between Helix-11 and Helix-12 was very adjustable which made support the different conformations (for examples: apo-, agonist-, and antagonist-form) (Musfiroh et al., 2013). A hydrophobic cavity of the ligand binding domain (LBD) of ERα consists of residues from helices 3, 6, 7, 8, 11, and 12 (Morris et al., 2009). Based on the previous study, residues 536-544 in Helix-12 of the ER is the important thing responsible for the activity of agonist or antagonist of a ligand. For example, an antagonist such as 4-OHT is accommodated by helix-12 of the ERα LBD occludes the co-activator recognition channel resulting in antagonist activity.
As shown in Fig. 4, Modifina3 lacks hydrogen bond with His524. Interestingly, the loss of hydrogen bonding with His524 when 4-OHT is bound while agonist activity existing when the hydrogen bonding with His524 and the ERα agonist estradiol (Muchtaridi et al., 2014; 2017; Musfiroh et al., 2013). It is predicted that the Modifina3 has no agonist effect on ERα due to the loss of the hydrogen bonding with His524 when Modifina3 is bound.
CONCLUSION
Modifina3 has the potential to be a novel therapeutic compound for targeted breast cancer treatment due to its highest affinity, with high-class category toxicity but still can be used as a compound for drugs. Based on the further biological investigation, Modifina3 represents rational computationally designed compound prioritized.
ACKNOWLEDGMENTS
This study was supported by The Directorate General of Higher Education of the Ministry of Research and Technology of Indonesia through International Research Collaborations and Publications Grants 2017 no. 006/ADD/SP2H/LT/DRPM/VIII/2017.
REFERENCES
Bentrem DJ, Craig Jordan V. Tamoxifen, raloxifene and the prevention of breast cancer. Minerva Endocrinol, 2002; 27(2):127–39.
Bergman L, Beelen ML, Gallee MP, Hollema H, Benraadt J, Van Leeuwen FE. Risk and prognosis of endometrial cancer after tamoxifen for breast cancer. Comprehensive cancer centres’ ALERT Group. Assessment of liver and endometrial cancer risk following tamoxifen. Lancet, 2000; 356(9233):881–7. CrossRef
Bernstein L, Press MF. Does estrogen receptor expression in normal breast tissue predict breast cancer risk? J Natl Cancer Inst, 1998; 90(1):5–7. CrossRef
Chang M. Tamoxifen resistance in breast cancer. Biomol Therap, 2012; 20(3):256–67. CrossRef
Cohen I. Endometrial pathologies associated with postmenopausal tamoxifen treatment. Gynecol Oncol, 2004; 94(2):256–66. CrossRef
Cragg GM, Newman DJ, Snader KM. Natural products in drug discovery and development. J Nat Prod, 1997; 60(1):52–60. CrossRef
Davies C, Pan H, Godwin J, Gray R, Arriagada R, Raina V, Abraham M, Medeiros Alencar VH, Badran A, Bonfill X, Bradbury J, Clarke M, Collins R, Davis SR, Delmestri A, Forbes JF, Haddad P, Hou MF, Inbar M, Khaled H, Kielanowska J, Kwan WH, Mathew BS, Mittra I, Muller B, Nicolucci A, Peralta O, Pernas F, Petruzelka L, Pienkowski T, Radhika R, Rajan B, Rubach MT, Tort S, Urrutia G, Valentini M, Wang Y, Peto R. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet, 2013; 381(9869):805–16. CrossRef
Dror O, Shulman-Peleg A, Nussinov R, Wolfson HJ. Predicting molecular interactions in silico: I. A guide to pharmacophore identification and its applications to drug design. Curr Med Chem, 2004; 11(1):71–90. CrossRef
Fan W, Chang J, Fu P. Endocrine therapy resistance in breast cancer: current status, possible mechanisms and overcoming strategies. Fut Med Chem, 2015; 7(12):1511–9. CrossRef
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer, 2010; 127(12):2893–917. CrossRef
Fisher B, Costantino JP, Wickerham DL, Cecchini RS, Cronin WM, Robidoux A, Bevers TB, Kavanah MT, Atkins JN, Margolese RG, Runowicz CD, James JM, Ford LG, Wolmark N. Tamoxifen for the prevention of breast cancer: current status of the national surgical adjuvant breast and bowel project P-1 study. J Natl Cancer Inst, 2005; 97(22):1652–62. CrossRef
Fox EM, Davis RJ, Shupnik MA. ERβ in Breast Cancer—Onlooker, Passive Player, or Active Protector? Steroids, 2008; 73(11): 1039–51. CrossRef
Gomes MN, Muratov EN, Pereira M, Peixoto JC, Rosseto LP, Cravo PVL, Andrade CH, Neves BJ. Chalcone derivatives: Promising starting points for drug design. Molecules, 2017; 22(8):1–25. CrossRef
Hawash MMA, Kahraman DC, Eren F, Cetin Atalay R, Baytas SN. Synthesis and biological evaluation of novel pyrazolic chalcone derivatives as novel hepatocellular carcinoma therapeutics. Eur J Med Chem, 2017; 129:12–26. CrossRef
Ikram NKK, Durrant JD, Muchtaridi M, Zalaludin AS, Purwitasari N, Mohamed N, Rahim ASA, Lam CK, Normi YM, Rahman NA, Amaro RE, Wahab HA. A virtual screening approach for identifying plants with anti H5N1 neuraminidase activity. J Chem Inform Model, 2015; 55(2):308–16. CrossRef
Jandial DD, Blair CA, Zhang S, Krill LS, Zhang YB, Zi X. Molecular targeted approaches to cancer therapy and prevention using chalcones. Curr Cancer Drug Targets, 2014; 14(2):181–200. CrossRef
Kalyaanamoorthy S, Chen Y-PP. Structure-based drug design to augment hit discovery. Drug Discov Today, 2011; 16(17):831–9. CrossRef
Lahsasni SA, Al Korbi FH, Aljaber NA. Synthesis, characterization and evaluation of antioxidant activities of some novel chalcones analogues. Chem Cent J, 2014; 8:32. CrossRef
Lee S, Lee I, Kim H, Chang G, Chung J, No K. The PreADME approach: web-based program for rapid prediction of physico-chemical, drug absorption and drug-like properties. Paper read at EuroQSAR 2002 Designing Drugs and Crop Protectants: processes, problems and solutions, Massachusetts, MA, 2003.
Levin ER. Integration of the extranuclear and nuclear actions of estrogen. Mol Endocrinol, 2005; 19(8):1951–9. CrossRef
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and autodocktools4: Automated docking with selective receptor flexibility. J Comput Chem, 2009; 30(16):2785–91. CrossRef
Muchtaridi M, Syahidah H, Subarnas A, Yusuf M, Bryant S, Langer T. Molecular docking and 3-D-pharmacophore modeling to study the interactions of chalcone derivatives with estrogen receptor alpha. Pharmaceuticals, 2017; 10(4):81. CrossRef
Muchtaridi M, Yusuf M, Diantini A, Choi S, Al-Najjar B, Manurung J, Subarnas A, Achmad T, Wardhani S, Wahab H. Potential activity of fevicordin-A from phaleria macrocarpa (Scheff) boerl. Seeds as estrogen receptor antagonist based on cytotoxicity and molecular modelling studies. Int J Mol Sci, 2014; 15(5):7225. CrossRef
Musfiroh I, Muchtaridi M, Muhtadi A, Diantini A, Hasanah AN, Udin LZ, Susilawati Y, Mustarichie R, Kartasasmita RE, Ibrahim S. Cytotoxicity studies of xanthorrhizol and its mechanism using molecular docking simulation and pharmacophore modelling. J Appl Pharm Sci, 2013; 3(06):007–15.
Narod SA. Hormone replacement therapy and the risk of breast cancer. Nat Rev Clin Oncol, 2011; 8(11):669–76. CrossRef
Payne SJ, Bowen RL, Jones JL, Wells CA. Predictive markers in breast cancer–the present. Histopathology, 2008; 52(1):82–90. CrossRef
Prasad YR, Kumar PR, Deepti CA, Ramana MV. Synthesis and antimicrobial activity of some novel chalcones of 2-Hydroxy -1-Acetonapthone and 3-Acetyl coumarin. E-J Chem, 2006; 3(4):236–41. CrossRef
Rangel LP, Winter E, Gauthier C, Terreux R, Chiaradia-Delatorre LD, Mascarello A, Nunes RJ, Yunes RA, Creczynski-Pasa TB, Macalou S, Lorendeau D, Baubichon-Cortay H, Ferreira-Pereira A, Di Pietro A. New structure-activity relationships of chalcone inhibitors of breast cancer resistance protein: polyspecificity toward inhibition and critical substitutions against cytotoxicity. Drug Design Develop Ther, 2013; 7:1043–52.
Rojas K, Stuckey A. Breast cancer epidemiology and risk factors. Clin Obstet Gynecol, 2016; 59(4):651–72. CrossRef
Smyth MS, Martin JHJ. X-ray crystallography. Mol Pathol, 2000; 53(1):8–14. CrossRef
Subarnas A, Diantini A, Abdulah R, Zuhrotun A, Hadisaputri YE, Puspitasari IM, Yamazaki C, Kuwano H, Koyama H. Apoptosis induced in MCF-7 human breast cancer cells by 2’, 4’-dihydroxy-6-methoxy-3, 5-dimethylchalcone isolated from Eugenia aquea Burm f leaves. Oncol Lett, 2015; 9(5):2303–6. CrossRef
Sun L, Yang H, Li J, Wang T, Li W, Liu G, Tang Y. In silico prediction of compounds binding to human plasma proteins by QSAR models. Chem Med Chem, 2018; 13(6):572–81. CrossRef
Thomas S, Brightman F, Gill H, Lee S, Pufong B. Simulation modelling of human intestinal absorption using Caco-2 permeability and kinetic solubility data for early drug discovery. J Pharm Sci, 2008; 97(10):4557–74. CrossRef
Todorova IT, Batovska DI. Trends in utilization of the pharmacological potential of chalcones. Curr Clin Pharmacol, 2010; 1–29; doi: 10.2174/157488410790410579.
Wang Z, Li Y, Ai C, Wang Y. In silico prediction of estrogen receptor subtype binding affinity and selectivity using statistical methods and molecular docking with 2-arylnaphthalenes and 2-arylquinolines. Int J Mol Sci, 2010; 11(9):3434–58. CrossRef
Wang Y, Xing J, Xu Y, Zhou N, Peng J, Xiong Z, Liu X, Luo X, Luo C, Chen K, Zheng M, Jiang H. In silico ADME/T modelling for rational drug design. Q Rev Biophys, 2015; 48(4):488–515. CrossRef
Wolber G. Efficient 3-D pharmacophore alignment as a tool for structure-based modeling and scoring. Chem Cent J, 2008; 2(1):49. CrossRef
Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model, 2005; 45(1):160–9. CrossRef
Xu S, Chen M, Chen W, Hui J, Ji J, Hu S, Zhou J, Wang Y, Liang G. Chemopreventive effect of chalcone derivative, L2H17, in colon cancer development. BMC Cancer, 2015; 15:870.
Yang G, Nowsheen S, Aziz K, Georgakilas AG. Toxicity and adverse effects of Tamoxifen and other anti-estrogen drugs. Pharmacol Ther, 2013; 139(3):392–404. CrossRef