INTRODUCTION
The pathological consequences of chronic inflammation may be contributed by the generation of reactive oxygen species (ROS, e.g., OH• and O2•) and reactive nitrogen species (RNS), e.g., NO• and ONO2•. These ROS and RNS can induce DNA damage and modify essential cellular proteins that are involved in DNA repair, apoptosis, and cell cycle, either directly or indirectly (Hussain and Harris, 2007). NO• is produced by nitric oxide synthase (NOS), an enzyme that catalyzes the conversion of L-arginine to L-citrulline and uses molecular oxygen and NADPH as cofactors (Alderton et al., 2001; Marletta, 1994). Once reduced, the heme cofactor binds and activates molecular oxygen, which in turn reacts with a terminal guanidinium nitrogen of the substrate L-arginine that is bound at 4 Å distance from the heme iron (Tierney et al., 1998).
The classification of NOS inhibitors is based on its binding site to the enzyme, i.e., (1) the largest class that interacts with the arginine-binding site; (2) A set of compounds that mimic tetrahydrobiopterin cofactor; (3) inhibitors that directly interact with the heme; and (4) inhibitors that interact with calmodulin or flavine cofactors (Alderton et al., 2001). Quinazoline, spirocyclic quinazoline, and aminopyridine, three classes of NOS inhibitors, are potent (IC50 ranged from 10 nM to 1.2 μM). These inhibitors, which have a cis-amidine derived core in similar, are selective (2.7- to 3000-fold) for iNOS over eNOS and nNOS (Garcin et al., 2008). The crystal structures of human iNOS in complex with AR-C95791 (AT2) and S-ethylisothiourea (SEITU) reveal that the inhibitors are hydrogen-bonded to a conserved glutamate (Glu377) in a narrow cleft within the larger active-site cavity containing heme and tetrahydrobiopterin (Fischmann et al., 1999) Garcin et al., 2008). While SEITU binds in the active site of iNOS, L-arginine binds at the pterin site (Crane et al., 1998; Kadish et al., 2000). Glu371 in murine iNOS (corresponding to Glu377 in human iNOS) (Crane et al., 1998) has been found to be critical for substrate binding (Chen et al., 1997; Gachhui et al., 1997).
Berberine, palmatine, and jatrorrhizine, isolated from Arcangelisia flava (Menispermaceae), showed cytotoxic activity against MCF-7 cells (Keawpradub et al., 2005). Antidepressant effect of this plant has also been reported (Tiara et al., 2014). Butanol extract of Morus alba had been reported significantly reduced LPS-induced PGE2 production, TNF- α and COX-2 expression in RAW264.7 macrophages (Choi et al., 2005), whereas the methanol extract of the same plant might contain compounds which inhibit COX-2 and iNOS activity in cultured mouse macrophage cells (Hong et al., 2002). Moreover, cudraflavone B found in the roots of M. Alba could cause a significant inhibition on TNF-α by blocking the translocation of NF-kB from the cytoplasm to the nucleus, which resulted in the inhibition of COX-2 gene expression (Hošek et al., 2011). However, studies on the pharmacophores of the phytoconstituents in A. flava and M. alba and molecular docking of the phytoconstituents in those particular plants against NOS are very limited.
This work presents a pharmacophore modeling based on the interaction of AT2 and SEITU with human iNOS, followed by molecular docking of 29 phytoconstituents from A. flava and M. alba with human iNOS.
MATERIALS AND METHODS
Hardware used was a PC with OS Windows 10 64-bit, AMD A6-7310 APU processor with AMD Radeon R4 Graphic 2.00 GHz, and RAM 6 GB.
Softwares were ChemDraw professional 15.0 (Academic License), LigandScout 4.1.4 (Universitas Padjadjaran License), AutoDock 4.2.6 and AutoDockTools 1.5.6 (The Scripps Research Institute), BIOVIA Discovery Studio (Academic License).
Preparation and validation of the pharmacophore features
Protein targets, downloaded from https://www.rcsb.org/, were the crystal structures of human iNOS in complex with AT2 (PDB code: 3E7G ; resolution 2.2 Å; R-value free 0.213) deposited by Garcin et al. (2008) and SEITU (PDB code: 4NOS; resolution 2.25 Å; R-value free 0.289) deposited by Fischmann et al. (1999). LigandScout was employed to automatically activate the PDB interpretation algorithm and showed the complex in the macromolecule view. Their ligand-protein interaction was studied (Fig. 1) to obtain the pharmacophore features (Fig. 2). These features were validated against iNOS database at http://dude.docking.org/targets/nos1 by employing the decoy-set method. This method could be used to evaluate the discriminative ability of the best pharmacophore model and to distinguish the active compounds from the inactive compounds. A database screening was performed and a set of statistical parameters were calculated that include the total hits (Ht), % yield of actives, % ratio of actives, enrichment factor (EF), false negatives, false positives, goodness of hit score (GH), and the area under receiver operating characteristic (ROC) curve was calculated. The GH score ranges from 0 (indicates the null model) to 1 (indicates the ideal model). When the GH score exceeds 0.7, the model is very good to identify the active compounds (Niu et al., 2013; 2014). The result was used to screen the pharmacophores of the phytoconstituents in Morus sp. and A. flava (Fig. 3).
 | Figure 1. Hydrogen bond interaction between AT2 (left) and SEITU (right) with Glu377 and Tyr347 in human iNOS. Black dashed lines indicate hydrogen bonds, salt bridges, and metal interactions. Green solid line shows the hydrophobic interactions. Visualization by poseview. [Click here to view] |
Validation of the docking program
To validate the docking program, the co-crystallized inhibitors (AT2 and SEITU) were separated from the protein and were re-docked into their origin position. The re-docked pose of both inhibitors was superimposed with the origins, and the root mean square deviation (RMSD) was calculated (Fig. 4).
Molecular docking
Furthermore, a molecular docking of the 29 phytoconstituents of Morus sp. and A. flava into human iNOS binding pocket was performed. The compounds were generated by using ChemDraw Professional 15.0, the energy was optimized by employing MMFF94. Parameters observed were as follows: (1) the protein-ligand interaction; (2) the docking score in kcal/mol; and (3) inhibition constant (Ki).
RESULTS AND DISCUSSION
The crystal structure of human iNOS in complex with AT2 (PDB code: 3E7G; resolution 2.2 Å; R-value free 0.213) deposited by Garcin et al. (2008) and SEITU (PDB code: 4NOS; resolution 2.25 Å; R-value free 0.289) deposited by Fischmann et al. (1999), were selected as our protein targets. The interaction between the inhibitors and the enzyme was studied by employing JSmol (an open-source Java viewer for chemical structures in 3D: http://www.jmol.org) and PoseView (developed at the Center for Bioinformatics Hamburg and jointly provided with BioSolveIT as a community service at the PDB) (Fig. 1).
Three hydrogen bonds are observed in the interaction between AT2 and human iNOS. The nitrogen atom in the pyridine ring acts as hydrogen bond acceptor (HBA) to Glu377, while the amino acts as hydrogen bond donor (HBD). The carboxylic group attached to the piperidine ring plays as HBA to Tyr347. Both oxygens of the acid’s carboxylic group act as HBAs as shown in Fig. 1 (left). Four HBs are observed in the interaction between SEITU and human iNOS. All hydrogens attached to the thiourea group act as HBD to Glu377 and Trp372 as shown in Fig. 1 (right).
 | Figure 2. Pharmacophore features in the interaction between AT2 (left) and SEITU (right) with human iNOS created by using LigandScout. [Click here to view] |
 | Figure 3. Decoy-set method to validate the pharmacophore features. AUC100% of ROC curves are >0.50, revealed that the two models could identify the active compounds, hence the pharmacophore features are categorized as valid. [Click here to view] |
 | Figure 4. Superimposition of the origin and re-docked AT2 molecules (RMSD = 0.41). [Click here to view] |
Based on the interaction between AT2 and SEITU with human iNOS, pharmacophore features were generated by using LigandScout and comprised three features: one aromatic ring, HBDs, and one HBA. These pharmacophore features are located in the arginine-binding site of human iNOS (Fig. 2). An internal database was developed using 8307 compounds containing 234 active structures collected from DUD-E (http://dude.docking.org/targets/nos1). The calculated enrichment factor (Table 1) indicates that the pharmacophore features generated from both AT2 and SEITU, are efficient for database screening. The GH score ranges from 0 (indicates the null model) to 1 (indicates the ideal model). Our GH scores are > 0.70 and AUC100% of ROC curve = 0.87, revealed that this model could identify the active compounds and was categorized as valid (Table 1 and Fig. 3).
Validation was performed by re-docking the co-crystallized AT2 and SEITU into its origin position in the arginine-binding site of human iNOS. The result indicated a similar binding mode with that of the origin complex. Both AT2 and SEITU molecules interact with Glu377. The validity of the docking program was further confirmed by superimposing the re-docked pose of AT2 with the origin, which resulted in RMSD value of 0.41. This value is defined as valid (Fig. 4).
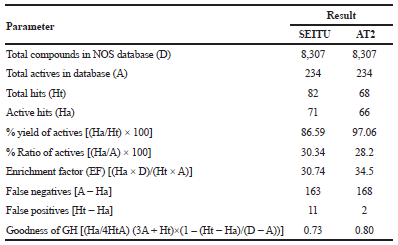 | Table 1. Statistical parameters of GH score validation. [Click here to view] |
Pharmacophore screening of the phytoconstituents of Morus sp. revealed that there are four best hits (kaempferol, steppogenin, quercetin, and rutin) that fitted the features, and therefore these compounds were predicted to be active in inhibiting iNOS (Table 2).
Kaempferole shows the best-fit score based on SEITU pharmacophore features (Table 2), which indicates its pharmacophore similarity with SEITU. This compound was reported could decrease NOS-2 mRNA level in LPS-stimulated J774.2 cells, moreover, an infusion of quercetin, kaempferole, and apigenin injected into the coronary circulation of guinea pig for 1 minute resulted in immediate vasodilatation. Quercetin (30 µM) showed the most potent vasodilator (Olszanecki et al., 2002).
Furthermore, 29 phytoconstituents contained in A. flava and M. alba were generated by ChemDraw Professional 15.0 and their energy was minimized by using MMFF94. The docking result is presented in Table 3.
The docking revealed that 16 phytoconstituents interacted with Glu377, an important amino acid residue that plays an important role in producing L-citrullin and NO in iNOS binding pocket.
This interaction is similar to that showed by AT2 and SEITU, both known inhibitors of iNOS. These two inhibitors are hydrogen-bonded to a conserved Glu377 in a narrow cleft within the larger active-site cavity containing heme and tetrahydrobiopterin (Fischmann et al., 1999; Garcin et al., 2008). However, the iNOS substrate L-arginine binds at the pterin site (Crane et al., 1998; Kadish et al., 2000).
The best docking score that belongs to sanggenon F is shown in Figure 5.
Sanggenon F shows the best affinity with the enzyme (docking score −9.88 kcal/mol; inhibition constant 0.57 μM), better than SEITU (docking score −5.48 kcal/mol; inhibition constant 96.8 μM). Sanggenon F could be potential as iNOS inhibitor and might compete with L-arginine in interacting with Glu377. This result was compared with that of other workers. Treatment with sanggenon C, a benzopyrone flavonoid isolated from the stem bark of Morus cathayana, could increase the level of intracellular Ca2+ and ATP and inhibited iNOS expression (Chen et al., 2017). Prenylated flavonoids, i.e., kuwanon C and sanggenon, were reported could down-regulate the iNOS expression in LPS-induced RAW 264.7 cells (Cheon et al., 2000). It has been reported that the C-2,3- double bond in flavonoid molecular structure is important. In vivo activity also depends on the patterns and numbers of hydroxylation/methoxylation. Especially, the A-ring 5,7-dihydroxyl and B-ring 3’,4’-catechol groups are important (Kim et al., 2004).
 | Table 2. Pharmacophore screening and modeling. [Click here to view] |
 | Table 3. Docking score of phytoconstituents contained in A. flava and M. alba with human iNOS. [Click here to view] |
 | Figure 5. HB interaction of sanggenon F with Glu377 (an important amino acid residue in human iNOS binding pocket) and with three other amino acid residues. [Click here to view] |
CONCLUSION
Phytoconstituents in Morus sp. and A. flava fit the pharmacophore features generated from AT2 or SEITU complex with iNOS, therefore, they might be potential as iNOS inhibitors. Of 29 phytoconstituents, sanggenon F in Morus sp. indicates the best the docking score. Its score is better than SEITU, a known iNOS inhibitor.
REFERENCES
Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem J, 2001; 357:593–615.
Chen LD, Liu ZH, Zhang LF, Yao JN, Wang CF. Sanggenon C induces apoptosis of colon cancer cells via inhibition of NO production, iNOS expression and ROS activation of the mitochondrial pathway. Oncol Rep, 2017; 38(4):2123–31.
Chen PF, Tsai AL, Berka V, Wu KK. Mutation of Glu-361 in human endothelial nitric-oxide synthase selectively abolishes L-arginine binding without perturbing the behavior of heme and other redox centers. J Biol Chem 1997; 272:6114–8.
Cheon BS, Kim YH, Son KH, Chang HW, Kang SS, Kim HP. Effects of prenylated flavonoids and biflavonoids on lipopolysaccharide-induced nitric oxide production from the mouse macrophage cell line, RAW 264.7. Planta Med, 2000; 66:596–600.
Choi EM, Hwang JK. Effects of Morus alba leaf extract on the production of nitric oxide, prostaglandin E2 and cytokines in RAW264.7 macrophages. Fitoterapia, 2005; 76:608–13.
Crane BR, Arvai AS, Ghosh DK, Wu C, Getzoff ED, Stuehr DJ, Tainer JA. Structure of nitric oxide synthase oxygenase dimer with pterin and substrate. Science, 1998; 279:2121–6.
Fischmann TO, Hruza A, Niu XD, Fossetta JD, Lunn CA, Dolphin E, Prongay AJ, Reichert P, Lundell DJ, Narula SK, Weber PC. Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat Struct Mol Biol, 1999; 6:233–42.
Gachhui R, Ghosh DK, Wu C, Parkinson J, Crane BR, Stuehr DJ. Mutagenesis of acidic residues in the oxygenase domain of inducible nitricoxide synthase identifies a glutamate involved in arginine binding. Biochemistry, 1997; 36:5097–103.
Garcin ED, Arvai AS, Rosenfeld RJ, Kroeger MD, Crane BR, Andersson G, Andrews G, Hamley PJ, Mallinder PR, Nicholls DJ, St-Gallay SA, Tinker AC, Gensmantel NP, Mete A, Cheshire DR, Connolly S, Stueh DJ, Aberg A, Wallace AV, Tainer JA, Getzoff ED. Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nat Chem Biol, 2008; 4:700–7.
Hong CH, Hur SK, Oh OJ, Kim SS, Nam KA, Lee SK. Evaluation of natural products on inhibition of inducible cyclooxygenase (COX-2) and nitric oxide synthase (iNOS) in cultured mouse macrophage cells. J Ethnopharmacol, 2002; 83:153–9.
Hošek J, Bartos M, Chudik S, DallAcua S, Innocenti G, Kartal M, Kokoska L, Kollar P, Kutil Z, Landa P, Marek R, Zavalova V, Zemlicka M, Smejkal K. Natural compound cudraflavone B shows promising anti-inflammatory propertiesin vitro. J Nat Prod, 2011; 74:614–9.
Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer, 2007; 121:2373–80.
Kadish KM, Smith KM, Guilard R. The porphyrin handbook. Biochemistry and binding: activation of small molecules, Elsevier, pp 313–4, 2000.
Keawpradub N, Dej-adisai S, Yuenyongsawad S. Antioxidant and cytotoxic activities of Thai medicinal plants named Khaminkhruea: Arcangelisia flava, Coscinium blumeanum and Fibraurea tinctoria. Songklanakarin J Sci Technol, 2005; 27(Suppl. 2):455–67.
Kim HP, Son KH, Chang HW, Kang SS. Anti-inflammatory plant flavonoids and cellular action mechanisms. J Pharmacol Sci, 2004; 96(3):229–45.
Marletta MA. Nitric oxide synthase: aspects concerning structure and catalysis. Cell, 1994; 78:927–30.
Niu MM, Dong FG, Tang S, Fida G, Qin JY, Qiu J, Liu K, Gao W, Gu Y. Pharmacophore modeling and virtual screening for the discovery of new type 4 camp phosphodiesterase (PDE4) inhibitors. PLoS One, 2013; 8(12):1–15.
Niu MM, Qin JY, Tian CP, Yan XF, Dong FG, Cheng ZQ, Fida G, Yang M, Chen H, Gu YQ. Tubulin inhibitors: pharmacophore modeling, virtual screening and molecular docking. Acta Pharmacol Sin, 2014; 35:967–79.
Olszanecki R, Gêbska A, Kozlovski VI, Gryglewski RJ. Flavonoids and nitric oxide synthase. J Physiol Pharmacol, 2002; 53(4):571–84.
Tiara A, Arief RH, Sudarsono. The antidepressant effects of (Arcangelisia flava (L.) Merr) water-soluble extract in Balb-C mice reviewed from immobility time by forced. Biol Med Nat Product Chem, 2014; 3(2):65–7.
Tierney DL, Martasek P, Doan PE, Masters BSS, Hoffman BM. Location of guanidino nitrogen of L-arginine substrate bound to neuronal nitric oxide synthase (nNOS): determination by Q-band pulsed ENDOR spectroscopy. J Am Chem Soc, 1998; 120:2983–4.