INTRODUCTION
Disinfectants have a vital role in keeping acceptable health standards by significantly decreasing microbial loads and inactivating pathogens (Antonio et al., 2017). Quaternary ammonium compounds (QACs) are the main biocide (antiseptics and disinfectants) used in various fields such as health care and agriculture as well as in industry (Ebrahimi et al., 2016).
Benzalkonium chloride (BC) is the most important QACs used for surfaces disinfection in medical care applications because of their antibacterial activity (Antunes et al., 2016) in addition to its important use as a preservative in aqueous formulations to prevent several infections (Nones et al., 2017).
Pseudomonas aeruginosa is considered to be a frequent and severe cause of acute nosocomial infections, especially affecting intensive care unit (ICU) and immune-compromised patients (Cabot et al., 2016).Previous studies suggested that exposure to sub-inhibitory concentrations of BC leads to co- and cross-resistance to other antimicrobial agents (Rakic-Martinez et al., 2011).
Pseudomonas aeruginosa resists the lethal effects of BC by several mechanisms, such as increased efflux pump activity, mutations in GyrA and ParC genes that encode DNA gyrase and topoisomerase IV in addition to changes in the bacterial membrane (Ferreira et al., 2011).
While there are at least 12 structural genes for the resistance-nodulation-division efflux systems have been identified in P. aeruginosa genome, four of them are clinically-important (MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM) (Lister et al., 2009). Mex systems co-expression has been reported in P. aeruginosa clinical isolates where its variable effect on antibiotic susceptibility has been observed (Poonsuk and Chuanchuen, 2014).
Phenotypic and genetic tests used to identify acquired resistance due to efflux pump overexpression in P. aeruginosa isolates. Over-expression of efflux pumps could increase the MICs of many antimicrobials. Different compounds have been known as efflux pump inhibitors (EPIs) due to its ability to broadly inhibit several known multidrug EPs in P. aeruginosa (Adabi et al., 2015).The purpose of this study was the cross-resistance determination of the BC-adapted P. aeruginosa with other antimicrobial agents. In addition to phenotypic and genotypic evaluation of the resistance caused by efflux pump activity.
MATERIALS AND METHODS
Bacterial isolates
Totally, 88 non-duplicated P. aeruginosa isolates were collected from clinical settings at four hospitals, Kasr El-Aini Hospital, Ain-Shams University Hospital, Chest Hospital, and Al Salam Hospital in Cairo, Egypt along the period of January–March 2015. Isolates those were Gram-negative with greenish-yellow colonies, fluorescent-yellow colonies, and blue color colonies on Cetrimide Agar, Pseudomonas Agar F, and Pseudomonas Agar P, respectively. Oxidase, catalase, and nitrate reductase positive isolates were identified as P. aeruginosa (Mahon et al., 2014). Pseudomonas aeruginosa wild-type strain (ATCC 15442) was purchased from Microbiologica USA.
Benzalkonium chloride
BC was purchased from Scharlabs S. L., Spain.
Minimum inhibitory concentration (MIC) of benzalkonium chloride
The MIC of benzalkonium chloride was carried out against the 88 P. aeruginosa isolates and P. aeruginosa strain (ATCC 15442) using agar dilution method due to precipitation of BC at high concentrations, following Clinical and Laboratory Standards (CLSI) (formerly NCCLS) in document M100-S14 (NCCLS, 2004; Kawamura-Sato et al., 2010). One microliter of each bacterial suspension contained104 cfu ml−1 was inoculated on Mueller-Hinton agar (Oxoid) with different BC concentrations, then incubated at 37°C for 18 hours and the MIC was recorded. The BC adapted isolates were selected for further investigation.
Antibiotics susceptibility testing (disk diffusion method)
Antibiotic susceptibility pattern of BC-adapted isolates was determined by the single disk diffusion test (Kirby-Bauer method) (Bauer et al., 1966; CLSI, 2008). According to CLSI (2008), the following antibiotic discs (Oxoid) were used: Gentamicin—CN (10 µg), Piperacillin–tazobactam PIP/TAZ (100/10 µg), Ciprofloxacin—CIP (5 µg), Amikacin—AK (30 µg), Ceftazidime—CAZ (30 µg), Imipenem—IPM (10 µg), and Piperacillin—PRL (100 µg)
Determination of cross-resistance to other antimicrobials
The MIC of cetrimide (Hopkin & Williams, UK), iodine (El-Nile Company, Egypt), and ciprofloxacin (ADWIA, Egypt) was determined by the agar dilution method (Kawamura-Sato et al., 2010; NCCLS, 2004), but due to ethanol (Fisher Scientific, UK) evaporation, its MIC was determined by macro-dilution method (Mazzola et al., 2009).
Detection of efflux pump genes by PCR technique
PCR amplification of the efflux pump regulatory genes (MexR, NfxB, MexT, and MexZ) was performed using primers shown in Table 1 (Linares et al., 2005). The PCR mixture containing 100 ng of chromosomal DNA, 0.5 µM concentration of each primer, and Dream Taq™ Green PCR Master Mix (Thermo-Fisher Scientific, USA) was heated for 3 minutes at 94°C. This was followed by 32 cycles of 30 seconds at 94°C; 30 seconds at 53°C (NfxB), 55°C (MexT), 58°C (MexR), or 60°C (MexZ); and 60 seconds at 72°C, with final 10 minutes extension step at 72°C.
Phenotypic determination of efflux pumps activity
Ethidium bromide (EtBr)-agar cartwheel method (screening method)
Plates of Tryptic Soy Agar (TSA) containing EtBr concentrations (Sigma Aldrich) ranging from 0 to 5 mg/l were swabbed with bacterial cultures. Cartwheel pattern was made by dipping a swab into each culture and streaked them from the central circle to the margin of the plate.
EtBr-agar plates were incubated for 16–18 hours at 37°C. Each EtBr plate was examined under a UV light. The minimal concentration of EtBr which produced fluorescence of the swabbed culture was recorded for the wild-type strain and BC-adapted isolates. The higher the concentration of EtBr required for the appearance of fluorescence the greater the presumptive EP activity (Martins et al., 2013).
Sequencing of the efflux pump genes
PCR products of the selected isolate and P. aeruginosa ATCC 15442 strain were purified using the DNA purifying kit (Thermo Fischer, USA) following the manufacturer’s instruction. The samples were processed using DNA sequencing kit (Thermo Fischer, USA) and analyzed in an automatic DNA sequencer (Applied Biosystem—serial no. G: 43 A: 39 T: 30 C: 41) in order to analyze mutation in genes. The DNA sequences were aligned and homology searches were performed using BLAST program (www.ncbi.nlm.nih.gov/blast) and the translation to amino acids was performed using ExPASy translate https://web.expasy.org/translate/.
 | Table 1. Primer sequences used in efflux pump regulatory genes detection. [Click here to view] |
Efflux pumps inhibitors (EPIs) (confirmatory method)
Determination of MIC of EPIs
The MIC of EPIs (listed in Table 2) has been determined by microtiter method (Wiegand et al., 2008). A 100 µl of broth was added into the negative control well (column 12) and 50 µl into the positive control well (column 11). Fifty microliters of each EPIs dilution (twice the desired final conc.) was added into the respective well (columns 1–10) for each bacterial isolates. The bacterial suspension was adjusted to 1 × 108 1 ml−1 and diluted to 1:100. All wells except the negative control were inoculated with 50 µl of the bacterial inoculums with final inoculum concentration 5 × 105 cfu ml−1.
Determination of MIC of BC in combination with EPIs
The antimicrobial efficacy of BC in combination with EPIs against BC-adapted P. aeruginosa was evaluated using the agar dilution method. The concentrations tested typically ranged from 2 to 3 concentration below the MIC of BC and EPIs. Organisms and agar dilution plates were prepared as for the MIC determination with exception of the addition of BC and EPIs plus an addition of agar medium to reach the 20 ml of the total (NCCLS, 2004). The fractional inhibitory concentration (FIC) index was calculated to evaluate the effect of the combinations (Satish et al., 2005).
.png) | Table 2. Efflux pumps inhibitors used. [Click here to view] |
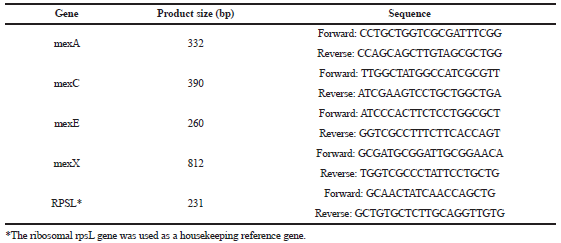 | Table 3. Primers used in RT-PCR. [Click here to view] |
Genotypic determination of efflux pumps activity
Gene expression analysis of MexA, MexC, MexE, and MexX by real-time PCR
RT–PCR was used to determine the expression of the genes encoding the four major P. aeruginosa efflux pumps MexA, MexC, MexE, and MexX of the selected isolate and P. aeruginosa wild-type strain ATCC 15442 (as a control).
Total RNA was extracted by the RNeasy Mini Kit (Qiagen, Germany) following its instructions and QIAGEN RNase-Free DNase Set (Qiagen, Germany) was used to remove the residual DNA. Finally, RNA was dissolved in 50 µl of RNase-free water. Synthesis of cDNA was performed by reverse transcription using QIAGEN OneStep RT-PCR Kit (Thermo Fisher). rpsL gene expression was measured as an internal control for confirmation that the amounts of RNA were equal in all of the RT-PCRs. The amplification of the selected genes was performed with specific primers (listed in Table 3). The reaction mixtures were incubated at 50°C for 30 minutes and 5 minutes at 94°C. Followed by 40 cycles of 15 seconds at 94°C; 30 seconds at 57°C (for MexA), 55°C (for MexC and MexE), 60°C (for MexX and rpsL); and 1 minute at 72°C, with final 7 minutes elongation at 72°C (Linares et al., 2005).
Analysis of the SYBR green RT-PCR results
Stratagene MX3005P software was used to determine the amplification curves and Ct values. The Ct of each sample was compared with that of the control group according to the “ΔΔCt” method to estimate the variation of gene expression on the RNA of the different samples (Yuan et al., 2006).
RESULTS AND DISCUSSION
Antibiotic resistance in bacterial pathogens has increased worldwide in the last decade leading to failures in the treatment of infectious diseases in human and animals (Gnanadhas et al., 2013).
Nosocomial infection is a worldwide problem and reported in 7%–8% of in-patients. Pseudomonas aeruginosa is responsible for 6%–25% of nosocomial infections. Treatment of infections caused by P. aeruginosa is very complicated because of their remarkable virulence characteristics and frequent antibiotic resistance (Yuruken et al., 2016).
Benzalkonium chloride (BC) is a QAC which have broad-spectrum antimicrobial activity (Ebrahimi et al., 2016; McCay et al., 2010). Low and higher level of adaptation to the BC has been observed in P. aeruginosa strains (Langsrud et al., 2003). Adaptation to BC in P. aeruginosa is mediated primarily by the efflux pumps activity, most notably the Mex systems (Lomovskaya et al., 1999). Vijaya et al. (2016) reported that about 22% of multidrug-resistant P. aeruginosa isolates showed insusceptibility to BC.
Determination of minimal inhibitory concentration (MIC) of Benzalkonium Chloride (BC) against Pseudomonas aeruginosa isolates
The MIC of the wild-type strain and P. aeruginosa isolates were shown in Table 4, which indicated that 20 isolates were 2,048 mg/l, 55 isolates were 512 mg/l, and 13 isolates, as well as wild-type strain was 256 mg/l. The 20 BC-adapted isolates were selected for the further testing.
Lambert and Pearson (2000) reported that the MIC of BC against P. aeruginosa isolates was 4,844, 1,462, 346, and 83.7 mg/l. Also, Walsh et al. (2003) found the MIC of BC against P. aeruginosa isolates was 500 mg/l.
Sources of the 20 BC-adapted isolate
The 20 BC-adapted isolates were collected from ICU, sputum, pus, urine, and wounds (Fig. 1).
Antibiotics susceptibility screening by Disk diffusion method
The antibiotics susceptibility of the 20 BC-adapted isolates against the selected antibiotics is shown in Figure 2, which indicates that all isolates were resistant to at least six antibiotics (multidrug resistant).
.png) | Table 4. MIC values of BC. [Click here to view] |
 | Figure 1. The sources of the BC-adapted isolates. [Click here to view] |
.png) | Figure 2. Antimicrobial susceptibility pattern of BC-adapted isolates: CAZ = ceftazidime; FEP = Cefepime; PRL = Piperacillin; CN = Gentamicin; AK = Amikacin; ATM = Aztreonam; CIP = Ciprofloxacin; IPM = Imipenem; TZP = Piperacillin-tazobactam. [Click here to view] |
 | Figure 3. Detection of Efflux pump regulatory genes: (A) MexR gene, (B) NfxB gene, (C) MexT gene, and (D) MexZ gene by garose gel electrophoresis of PCR-amplified products. Lane 1 represented DNA ladder; lane 2, the amplified products for wild-type strain (ATCC 15442); and lanes from 3 to 22 represent the amplified products for the selected 20 isolates (52, 87, 1, 7, 15, 28, 37, 51, 76, 86, 90, 108, 116, 117, 132, 134, 135, 149, 153, and 154, respectively). [Click here to view] |
Determination of cross-resistance to the other antimicrobial agents
The MIC values of the selected 20 BC-adapted isolates and the wild-type strain ATCC 15442 were compared with cetrimide (QAC), ethanol (Alcohol), iodine (halogens), and ciprofloxacin (quinolones) for determination of cross-resistance to the other antimicrobial agents. The obtained results showed that the 20 BC-adapted isolates were also adapted to cetrimide with MIC 7,000 mg/l and ciprofloxacin with MIC was 64 or 128 mg/l, but not adapted to ethanol and iodine. As reported by Buffet-Bataillon et al. (2016), it was found that adaptation to BC leads to cross-resistance with fluoroquinolones.
Detection of efflux pump genes (MexR, NfxB, MexT, and MexZ) by PCR technique
The amplification products of MexR, NfxB, MexT, and MexZ genes were identified by agarose gel electrophoresis with ethidium bromide staining. The amplified products were 637-bp fragment for MexR gene, 939-bp fragment for NfxB, 997-bp fragment for MexT gene, and 781-bp fragment for MexZ gene.
MexR gene was detected in 16 isolates, NfxB gene in 18 isolates, MexT gene in 10 isolates, and MexZ gene in 17 isolates (Fig. 3).
The four genes were found collectively in nine isolates (isolates no. 7, 15, 28, 37, 51, 52, 86, 87, and 154), these nine isolates were selected for the further investigation.
Evaluation of the efflux pump activity by Ethidium bromide (EtBr) cartwheel method
The presumptive identification of the highest EP activity in the nine isolates and the wild-type strain ATCC 15442 was carried out by using the EtBr-agar Cartwheel method. Isolates express efflux pumps by variable levels as shown in Figure 4. The ATCC strain was found to fluoresce at conc. 0.5 mg/l of EtBr, which means it has no efflux pump activity (negative control).
The minimum concentration at which the adapted isolates showed fluorescence was 1mg/l for isolates no. 15, 51, and 154, while 2.5 mg/l for isolates no. 28 and 37, but isolate no. 86 was 4.5 mg/l, isolates no. 7 and 52 were 5 mg/l, however, the isolate no. 87 was more than 5 mg/l. These results were agreed with El-Naggar et al. (2011), who reported that EtBr was effluxed in P. aeruginosa.
The previous results revealed that isolate no. 87 has a high efflux pump activity.
Sequencing
To identify mutations, sequences of the selected isolate no. 87 and wild-type strain ATCC 15442 were compared with P. aeruginosa PAO1 (Pseudomonas Genome Database, (http://www.pseudomonas.com). Figures 5–8 showed the amino acids alignment of efflux pump regulatory genes MexR, NfxB, MexT, and MexZ of wild-type strain PAO1, wild-type strain ATCC 15442, and isolate no. 87, respectively.
As shown in Figures 5–8, in the MexR regulatory gene, a novel mutation in codon number 126 was correlated to resistance to fluoroquinolone, which is substitution of amino acid Valine (V) to Glutamic acid (E), which was reported in previous investigations (Nguyen et al., 2018; Vaez et al., 2018). Results of several independent studies revealed different mutations, for example, Suman et al. (2006) reported 24 silent and four missense mutations in 14 clinical isolates of P. aeruginosa, and Savov et al. (2014) found another mutation in codon 44 in one strain, substitution of amino acid lysine to methionine.
No mutations were detected in NfxB in this study, Esquisabel et al. (2011) found several different amino acid substitutions, while the amino acid substitutions of Asparagine (N) to Glycine (G) in codon 68 and Arginine (R) to Histidine (H) in codon 33 were previously reported in clinical strains of P. aeruginosa (Livermore, 2002; Suman et al., 2006).
In MexT, two substitutions were found, Proline (P) to Alanine (A) in codon 81 and Phenylalanine (F) to Isoleucine (I) in codon 172. These mutations were also observed in wild-type strain ATCC 15442 so they considered as a non-significant mutation.
In MexZ, three substitutions were found. One substitution was Leucine (L) to Arginine (R) in codon 138. In addition to two conservative substitutions producing amino acids with similar physiochemical properties [Methionine (M) to Valine (V) in codon 1 and Aspartic acid (D) to Glutamic acid (E) in codon 155]. These results were as reported by Nemat-Gorgani (2009). Esquisabel et al. (2011) observed the same substitution, but in another codon (Leu-162-Arg).
 | Figure 4. Fluorescence of the isolates on agar plates containing increasing concentrations of EtBr. cultures were swabbed in TSA plates containing increasing concentrations of EtBr. Following overnight incubation at 37°C for 16 hours, fluorescence was detected under UV light. No. 1: ATCC 15442, from 2 to 10 refer to isolates no. 7, 15, 28, 37, 51, 52, 86, 87, and 154, respectively. [Click here to view] |
.png) | Figure 5. Alignment of amino acid sequences of the MexR gene. [Click here to view] |
.png) | Figure 6. Alignment of amino acid sequences of the NfxB gene. [Click here to view] |
 | Figure 7. Alignment of amino acid sequences of the MexT gene. [Click here to view] |
.png) | Figure 8. Alignment of amino acid sequences of the MexZ gene. [Click here to view] |
Efflux pumps inhibitors (EPIs) (confirmatory method)
The MIC values of carbonyl cyanide m-chlorophenylhydrazone (CCCP) was 128 mg/l and MIC value of sertraline was 1,000 mg/l for both isolates no. 87 and the wild-type strain ATCC 15442. But trimethoprim and epinephrine have no antimicrobial activity.
Efflux pump activity of isolate no. 87 was evaluated in the absence and presence of an efflux pump inhibitors (EPI) used.
According to the obtained results, the insusceptibility of the isolate 87 to BC decreased upon the combination of BC with the sertraline (2,048–256 mg/l BC). Sertraline had the most potent antimicrobial activity among the four EPIs used. It was found that the concentration 256 mg/l of BC + 800 mg/l of Ser. is the best concentrations which inhibit the growth of all bacterial isolates. While CCCP, TMP, and EP have no effect.
The obtained results revealed that efflux pumps in isolate 87 play an important role in adaptation development to BC.
Data presented in Table 5 revealed that the FIC index values stated a partial synergistic antimicrobial activity of BC-Ser against all tested bacterial isolates (FIC > 0.5 but < 1).
Bohnert et al. (2011) reported that resistance of the isolates to BC decreased in the presence of the EPI. The mutant also showed a 94-fold decrease in sensitivity to ciprofloxacin (McCay et al., 2010).
Real-time PCR
Changes in expression of the MexA, MexC, MexE, and MexX genes of BC-adapted isolate no. 87 were measured in the absence and presence of a combination of BC + Ser. (at sub-inhibitory conc.); expression was compared to that of wild-type strain ATCC 15442 as shown in Table 6.
For untreated BC-adapted isolate no. 87, MexA gene was expressed at a 9.189-fold, MexC 8.5-fold, MexE 5.5-fold, and MexX 4-fold higher than the control sample. These results are in agreement with previous studies which reported that 4- to 10-fold increase in the activity of a mexA (Dumas et al., 2005; Gomaa et al., 2016; Pasca et al., 2012; Srikumar et al., 2000).
In another study by Wolter et al. (2009), it was documented that the expression of mexA gene in PA431 was two-fold higher than PAO1, while genes mexC and mexE showed higher expression by about 340- to 375-fold, respectively, in PA30 than in PAO1, and increased three- to four-fold in strain PA431. In another hand, of mexX gene expression in PA431 was higher than PAO1 by 50-fold and lower in PA30 by two-fold.
McCay et al. (2010) observed that the expression of both mexA and mexC was increased in the BC-adapted isolate. Expression of mexE and mexX in BC-adapted isolate was half that of 10421 in absence of BC, although expression of mexX was increased in BC-adapted isolate than 10421 in presence of BC.
Decreased expression was noticed by the addition of a combination of BC + SER to the growth medium (256 mg/l BC + 700 mg/l SER); the expression of MexA, MexC, MexE, and MexX was decreased to 3.9-, 3-, 1.9-, and 1.4-fold, respectively. These results indicated that BC-adapted isolates overexpressed the four efflux pump genes; and by treatment with sertraline (EPI), the expression of these genes was down-regulated to quarter; so we investigated that the efflux pump was the mainly resistant mechanism to the BC.
.png) | Table 5. The FIC index of benzalkonium chloride and sertraline. [Click here to view] |
.png) | Table 6. Expression of MexA, MexC, MexE, and MexX of the BC-adapted isolate no. 87 (untreated and treated). [Click here to view] |