INTRODUCTION
Chemometrics is the application of a mathematical, statistical, and logical method to construct a technique or experiment and acquire the most chemical information from the findings produced. It aids in the processing of chemical data as well as the selection of an appropriate source of herbal drugs; for example, in cases where a manufacturer is investing in a company to manufacture herbal medicines from herbal sources such as Austria and Egypt and wants to know which particular source will provide the manufacturer with the maximum amount of active ingredient from the herbal drug. Here is an application that uses chemometric data processing studies to group compounds with highly active ingredients utilizing mathematical analysis and statistical representation [1]. By fusing the terms “metri” for measurement and “kemo” for chemistry, Svante Wold created the term “kemometri” (chemometrics in English) in 1972, and it is broadly divided into two categories: Unsupervised and supervised pattern recognition techniques, when taking a qualitative assessment into account multivariate calibration for quantitative analysis is the second [2].
The issue of poor-quality pharmaceuticals in the market can be attributed to two primary factors: inadequate production standards, leading to substandard medicines, and fraudulent activities [3]. Counterfeit drugs may involve various forms of fraud and adulteration, such as lacking the declared active pharmaceutical ingredient (API), containing a different API, or having a lower API strength. Various methodologies, particularly those employing spectroscopic techniques and chemometric methods, have been proposed to detect substandard and counterfeit pharmaceuticals. Spectroscopy, especially near-infrared (NIR), when combined with exploratory data analysis (EDA), classification, and regression methods, plays a significant role in developing effective, high-performance, rapid, non-destructive, and sometimes online approaches to assess pharmaceutical quality and compliance with production and pharmacopeia standards [4]. However, the abundance of available chemometric tools applicable to handling spectroscopic data, among others introduces the potential for misapplication [5].
Samples are categorized according to particular characteristics that are described by their measurements. The term “pattern recognition” refers to the process of finding similarities and regularities in the data based on the measurements made on the samples that are being analyzed [6]. A “pattern” is the set of measurements that characterize each sample in a given dataset. “Recognition” is the process of determining that property of interest by classifying a sample into its appropriate category [7]. The data might consist of peak regions and retention times (RTTs) for chromatograms, transmittance or absorbance for spectroscopic methods, and so on. These data are plotted in the same number of dimensions as the variables to appear as variables (multivariate data). However, this review describes the various aspects regarding the use of current trends included in chemometric software which will consist of significant parameters associated with the data variables used for the identification, analysis, interpretation, and evaluation of herbal medicines [8].
CURRENT METHODS OR TECHNIQUES USED FOR TRENDS AND INNOVATIONS IN CHEMOMETRICS
The classification of chemometric methods is as follows:
Pattern recognition techniques
Over the years, the application of pattern recognition methods has been extensively utilized to categorize samples analyzed through high-performance thin-layer chromatography (HPTLC) and high-performance liquid chromatography (HPLC) with evaporative light scattering detection. Chemometrics assesses the pretreatment data obtained from HPLC fingerprints and HPTLC fluorescence pictures for the identification of patterns and similarities [9].
Unsupervised pattern recognition
Numerous approaches most notably cluster analysis (CA) are used in unsupervised pattern techniques to categorize samples (or objects) according to chemical measurements. Analyses based on similarity and exploratory factors are the two categories of unsupervised methods. The initial stage is to find commonalities among items [10]. Similarity analysis (SA) uses the correlation coefficient R and is the most often used and straightforward method. In addition to the whole fingerprint SA, distinctive peak relative peak area and RTT can be employed. Connecting the item or EDA is the next stage. Hierarchical cluster technique (HCA), is the most often used method. Building qualitative branching structures, or dendrograms, that allow for visualization is the core of the hierarchical clustering technique [11].
Similarity analysis
A simple method for making an initial assessment for data analysis is to use SA. Statistical computations are used to assess if two fingerprint samples, one as the test sample and the other as the reference or standard sample, are identical or different [12]. The assessment of herbal medications entails determining similarity or dissimilarity using distance or similarity measures. Analyzing the similarity of peak height, peak area, and peak-to-peak ratio in fingerprints has shown to be an efficient way of evaluating the quality of medicinal plants [13].
Exploratory data analysis
EDA seeks to find hidden patterns in data that may not be obvious based on current knowledge. There are several tools available for data exploration, including principal component analysis (PCA), Factor Analysis, and Projection Pursuit. These strategies create latent variables, which are not immediately observable, by linearly merging original variables according to a preset criterion [14]. They explain the contrasts between items. Exploratory analysis is used to collect data on cluster propensity groupings, factor correlations, and sample-variable associations. HCA is frequently used for the exploratory investigation of chemical fingerprints [15].
Hierarchical clustering techniques
A dendrogram is frequently used to display the findings of HCA, a method that seeks to establish a cluster hierarchy by comparing fingerprint data for similarity [16]. HAC classifies items according to their spatial similarities, using a qualitative technique to demonstrate sample correlations and clusters. Two main approaches, agglomerative and divisive, compare samples by progressively merging or separating observations as they ascend the hierarchy [17]. In the divisive technique, as one goes down the hierarchy, each cluster’s samples are split from the start [18]. To find the best timing for merging or dividing clusters, linkage criteria specifying sample dissimilarity in HCA is essential. HCA tries to arrange data into naturally occurring clusters and includes a metric that indicates the dissimilarity of individual samples. The main objective of HCA is to unveil patterns in two dimensions by visually representing these clusters through a dendrogram. The HCA dendrogram operates on the assumption that samples with similar numerical values are positioned close to each other, suggesting that the most similar samples exhibit the highest proximity [19]. CA calculates distances between clusters by employing both Euclidean and Mahalanobis distances. The determination of similarity depends on the observed separation among individual data points within the clusters [20].
Supervised pattern recognition technique
Supervised pattern recognition techniques have been applied extensively to dataset categorization. Building a classification model uses calibration or training sets using pre-known data. Before evaluating the model on unidentified data, the predictive qualities are first confirmed by analyzing it using an independent sample set with prior known information [21]. The prevalent techniques employed in categorizing herbal products include linear discriminant analysis (LDA), artificial neural networks (ANNs), soft independent modeling of class analogy (SIMCA), and orthogonal projections to latent structures–discriminant analysis (PLS-DA) [22].
Classification analysis
Multiclass issues were the subject of discriminant analysis, sometimes referred to as “hard modeling.” The differentiation of each geographical origin, growth year, botanical origin, and manufacturing batch, for instance, as well as the differentiation of various processing levels or procedures in the final goods, are some of the primary issues. PLS-DA, k-nearest neighbors, and Journal Pre-proof 21-dimensional procedures were the most often used discriminant approaches, such as unfolded-PLS-DA, multi-way PLS-DA (N-PLS-DA), and others [23,24].
Class-modeling analysis, often known as “soft modeling,” is concerned primarily with the target class and is concentrated on the single-class problem. The primary class-modeling approaches include unequal distributed classes, SIMCAs, multivariate statistical process control (MSPC), and so on [25].
Regression analysis
In regression analysis of HM fingerprint, the multivariate calibration algorithm aimed to model a link between fingerprint data and a corresponding continuous property, i.e., the content of chemical compounds or water, adulteration ratio, pharmacological or toxicological activity, and so on. The commonly used regression techniques were PLSR, ANN, support vector regression, and so on. Generally, regression analysis is carried out for different applications depending on the properties of fingerprint data [26].
Multivariate calibration methods
Multivariate calibration techniques often involve using various factors such as responses at different potentials or wavelengths, or across a full range of data points to determine concentrations. This approach offers several advantages, including the reduction of noise and the elimination of interference [27]. Examining the entire dataset, rather than focusing on a single point, can make it easier to detect and remove noise. Additionally, interferences should be taken into account if their measurement characteristics differ significantly from those of the target substance [28]. Generally, multivariate methods tend to be more effective than univariate methods. It is possible to always create a univariate model from a multivariate model, which allows for gathering more information without losing any important data variable [29].
Partial least squares (PLS)
Reducing complexity in the feature space aids in creating a concise representation while retaining maximum diversity. Simplifying the features is less computationally intensive compared to other methods and is not only easy to understand but also effectively depicts the relationships within spectral data by analyzing variance levels [30]. Regression techniques like PLS are employed to identify connections between input and output variables assuming they originate from a common set of underlying factors [26].
Cluster analysis (CA)
An exploratory pattern recognition technique called CA seeks to organize things into clusters so that related objects are grouped together. Although CA is an effective exploratory technique for spotting patterns and trends, it does not offer quantitative information about them. PCA, SA, and HCA can successfully categorize and differentiate medicinal plant fingerprints. These methods seem particularly well-suited for quality assurance applications [31]. A multivariate method called CA groups elements according to their shared traits. It groups parts according to how similar they are to one another in space. Because of this, the cluster shows both high intergroup homogeneity and significant intergroup heterogeneity [32].
Linear discriminant analysis (LDA)
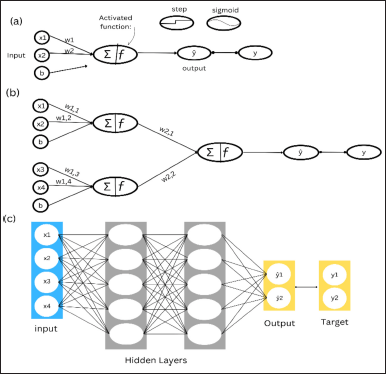
As the name implies, LDA is a linear model for classification and dimensionality reduction (Fig. 1). Most typically used in pattern classification tasks for feature extraction [33]. LDA is a technique for supervised pattern recognition. It focuses on determining the best boundaries across classes. It finds a multivariate linear function of the variable that optimizes the ratio of both variances to within-group variance. Because multidimensional data occur when the number of variables exceeds the number of observations where we cannot apply LDA directly [34].
 | Figure 1. Linear discriminant analysis. [Click here to view] |
Artificial neural networks (ANNs)
ANNs have a significant influence on chemometrics. In chemometrics, ANNs are extremely effective for modeling complicated interactions within datasets, particularly in spectroscopy, chromatography, and sensor data processing. As part of a greater variety of multivariate analytic techniques, ANNs are better renowned for their capacity to identify nonlinear patterns and connections seen in complicated chemical datasets [35]. ANNs are increasingly used in chemometrics for various purposes, including calibration, classification, and prediction. ANNs show versatility in identifying complicated patterns in calibration data, making them particularly effective in reproducing hard chemical processes [36].
Calibration is the act of building a link between instrumental measurements and certain properties that enables precise quantifications [37]. ANNs are versatile and adaptive, making them suited for varied applications in chemometrics, resulting in advancements in analytical chemistry, process optimization, and quality assurance within the chemical industry [38]. ANNs often consist of multiple parameters that must be calculated. To reduce model overfitting, training ANN with an experimental dataset that is at least three to five times larger than the number of parameters is recommended. After estimating the parameters, utilize a separate dataset to evaluate the proposed model [39].
Principal component analysis (PCA)
PCA is commonly employed to handle multivariate data in the absence of prior information about the studied samples. The fundamental concept behind PCA is to reduce the dimensionality of a dataset with numerous interrelated variables, aiming to preserve the utmost variance in the dataset [40]. PCA is a multivariate approach used to identify the primary cause of variability in datasets. It detects cluster formatting and establishes the association between object and variable [41]. When utilized in conjunction with chemometrics, the described HPLC-DAD method has demonstrated considerable efficacy in identifying resources of Ziziphus jujuba var. inermis and can be applied in the chemotaxonomic characterization of the same [42]. Plant medications have been shown to include heavy metals, pesticide residue, mycotoxin, and synthetic prescription or non-prescription pharmaceuticals, raising concerns about their safety [43]. It is essential to identify these elements to ensure the safety of herbal medicines. Recently, there has been extensive research on trace elements like iron, copper, zinc, and manganese, as they are known to play crucial roles in biological systems and may be associated with the specific geographical origin and therapeutic effectiveness of the plants. Data analysis, employing PCA and HCA, was employed to establish connections between these elements and to classify the herbal samples [44].
SOFTWARES USED FOR THE EVALUATION OF HERBAL DRUGS IN THE FIELD OF CHEMOMETRICS
PLS_Toolbox
PLS Toolbox could be a collection of fundamental and progressed chemometric schedules that work inside the MATLAB® computational environment. It contains the tools that chemical engineers, chemists, and other researchers need to study their data and build predictive models [45]. Indeed less experienced clients can perform capable examinations with interactive state-of-the-craftsmanship instruments. PLS_Toolbox incorporates the foremost progressed suite of multi-way instruments accessible within the world as well as a tremendous cluster of other capacities [46].
PLS_Toolbox offers an integrated visual interface and an extensive collection of over 300 tools designed for diverse specialized fields. Named after the PLS regression technique, a widely adopted calibration method in various applications, the toolbox goes beyond this standard, encompassing all the computational tools necessary for chemical engineers, analytical chemists, and other data scientists to effectively analyze their data and construct predictive models [47].
Unscrambler X
The Unscrambler X stands as a commercial software solution dedicated to multivariate data analysis. Its primary function lies in the calibration of multivariate data, often applied to analytical data sources like Raman spectroscopy and NIR spectroscopy. This calibration process aims to construct predictive models, facilitating real-time spectroscopic analysis of materials. Originally developed by Harald Martens in 1986, the program later came under the management of computer-aided modeling (Fig. 2).
 | Figure 2. Flowchart describing process of PCA. [Click here to view] |
Applications
(a) Product development: Creating innovative products is a costly and time-intensive endeavor. It is also dangerous. Unscrambler helps to speed product development, minimize the time it takes to bring items to market, and improve current products. The use of design of experiments skills enables the creation of product designs that are less susceptible to fluctuations in raw material quality or qualities.
(b) Production optimization.
(c) Process control: Using Unscrambler improves the ability to keep batch and continuous processes operating smoothly and effectively. It provides an easy multivariate control chart with early defect identification and process deviation notifications. This will assist operators to avoid quality and consistency issues in continuous production operations [48].
Soft independent modeling of class analogy
SIMCA-online is meant to work with process data where a process is observed over time, whereas SIMCA-offline can be used to model and analyze nearly any data. SIMCA-online SIMCA caters to more than just “data scientists.” You do not need an advanced degree in statistics or computer science to execute projects involving “data mining,” multivariate calibration, or predictive modeling with SIMCA. This software simplifies the technical aspects of “data science,” enabling R&D and quality engineers to leverage multivariate methods, data visualizations, and process intelligence without requiring an extensive technical background [49].
SIMCA, when used to evaluate multivariate data, gives an overview of the signals and converts them into information regarding sample and/or process quality. Fast and non-invasive spectroscopic sensors are an important aspect of the process analytical technology (PAT) application because they may provide process understanding and, ultimately, build the groundwork for a control strategy to be used in the manufacturing process [50].
This software solution offers easy post-batch interpretation and analysis of massive process datasets, provides a summary of all forms of process information, major trends, correlations, and patterns all in one handy data model, and allows for speedier troubleshooting, reducing the risk of costly downtime. The newly improved program features an easy graphical interface as well as the ability to manage complicated data such as rewriting, dividing and merging, and more. SIMCA projects can be instantly uploaded to an accessible SIMCA-online server for real-time data display of the process [51].
APPLICATIONS OF CHEMOMETRICS IN QUALITY EVALUATION
After outlining the inception and evolution of chemometrics and detailing its frequently utilized techniques for managing data from hybrid analytical devices, we will elucidate the utilization of chemometrics in assessing quality. This will be illustrated through specific instances in the assessment of the authenticity, effectiveness, uniformity, and safety of medicinal plants.
Authenticity
Each therapeutic plant harbors distinct components, and, as a result, scrutinizing constituents along with their specific chemical proportions allows for the differentiation and identification of authentic medicinal plants from counterfeit ones. In this context, a fingerprint analysis of Cassia seeds was conducted utilizing HPLC-UV at two wavelengths, and chemometrics techniques were applied for the assessment [52]. PCA analysis was employed to group samples into four clusters based on their plant sources and preparation procedures. The categories of the four distinct samples in the test set were accurately predicted using PLSs, back propagation ANN, and radial basis function ANN [53].
Safety
The safety concerns associated with herbal remedies include the presence of heavy metals, residual pesticides, mycotoxins, and both prescribed and non-prescribed synthetic drugs. These contaminants may arise from mineral components, contamination, or adulteration. It is imperative to accurately identify and quantify these elements to guarantee the safety of herbal medicines [54]. Certain trace elements like iron, copper, zinc, and manganese have significant functions in biological systems and are currently under extensive study due to their potential connections with the specific growth environment and therapeutic effectiveness of plants. Conversely, elements like lead and cadmium, even in trace quantities, are known to be toxic. Consequently, it is crucial to determine the levels of these toxic elements, emphasizing their central importance [55].
Application of chemometrics in other fields
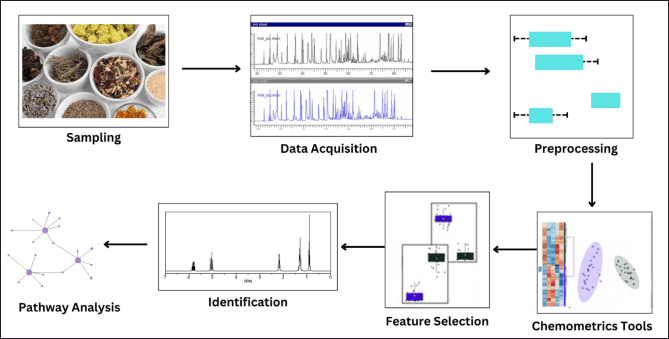
The definition of chemometrics suggests that the process focuses exclusively on managing and analyzing chemical data. However, in the biological sciences, numerous analyses now rely on “rapid” or alternative methods that ascertain chemical or physical criteria. Chemometrics has found extensive use in key areas :
Taxonomic studies of food-associated organisms, particularly species or strains of food-borne pathogens like Salmonella [67]. Studies on food spoilage, where chemical changes during storage are assessed in relation to microbial growth [68]. The application of bioassays for the identification of food contaminants, such as residues of veterinary antibiotics, is common. Numerous other studies in the realm of microbiology, particularly microbial taxonomics, are also prevalent [69]. When it comes to monitoring instances of foodborne diseases or spoilage outbreaks, having a thorough comprehension of the precise causative organism is frequently essential. While conventional diagnostic techniques are satisfactory for discerning microorganisms at the levels of genus, species, and strain, there is often an epidemiological necessity for more conclusive and swift identification. We have additionally summarized applications of chemometrics and software used in Table 1 below.
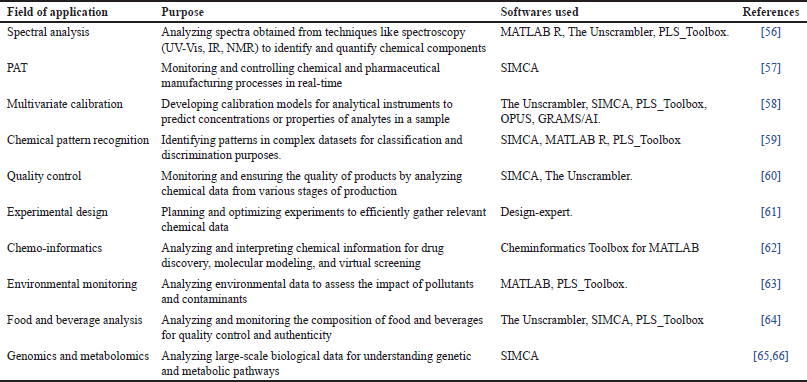 | Table 1. Application of chemometrics and softwares used in other fields. [Click here to view] |
FUTURE PROSPECTS OF CHEMOMETRICS SOFTWARES
Chemometrics finds significant application in the field of process analytical chemistry. Periodic reviews on this subject are available in Analytical Chemistry every 2 years. The primary objective of research in this domain is the utilization of measurements obtained from chemical analyzer systems and chemometrics to supervise, enhance, and manage chemical processes [70]. These applications primarily focus on improving process yield, enhancing product quality, minimizing waste, reducing processing time, and enhancing safety. Chemical analyzer systems encompass various technologies, including temperature, pressure, and flow sensors, gas or liquid chromatography, flow-injection analysis, electrochemistry, x-ray spectrometry, nuclear magnetic resonance (NMR) and microwave spectroscopy, mass spectrometry, ultrasonic methods, as well as spectroscopic methods like UV/Visible, NIR, mid-infrared, and Raman spectroscopy [71]. Common chemometrics approaches for process analytical applications involve multivariate calibration methods such as time-series analysis, PLS, and MSPC [72].
Chemometrics and multivariate techniques are progressively employed in the food and feed chemistry sectors to conduct in-depth analyses of extensive datasets [73].
PCA is both objective and intuitive. Discriminant function analysis is capable of assigning each sample to its respective group based on the constructed model. Meanwhile, SIMCA can ascertain whether a given sample conforms to the established model category. Another study examines the use of chemometrics in conjunction with low-resolution time-domain NMR measurements, specifically focusing on relaxation profiles, to characterize various food products. Other reviews within this field explore the application-specific use of chemometrics and spectroscopic techniques in the food industry, covering aspects such as the postharvest evaluation of fruit and vegetable quality, the characterization of essential oils, and an in-depth examination of the history and methodologies of chemometrics in the wine industry [1].
The analysis of intricate genomics-based data in systems biology necessitates the application of multivariate data analysis techniques to derive valuable insights [74]. Simultaneous variations across different levels, such as organism-specific differences and temporal variations, are observed in these datasets [75]. Conventional two-way methods like PCA tend to mathematically confound the distinct types of variation present in these datasets. A novel method, referred to as multilevel component analysis, has been recently proposed to untangle the various types of variation [76].
CONCLUSION
Modern software-supported chemometrics has significantly altered the pattern of carrying out analytical chemistry and data analysis. The advanced algorithms could be used to discern the relevant trends in extended data analysis in chemical studies and for interpreting experimental results obtained. These, in turn, improve the speed and accuracy of processing data while improving the reliability of the analytical output obtained. Other applications include multivariate analysis and equipment calibration; this is due to improved research output emanating from the fields of study concerned. The complexity of the chemical composition is one of the challenges in herbal medicine. Spectroscopy, chromatography, and hybrid chemometric methods are some of the techniques that are vital in guaranteeing the authenticity and quality of herbal products. These techniques enable the researchers to explain the complex interactions observed in herbal systems. Current chemometric software is becoming more user-friendly and specialized, and this makes us anticipate its increased accessibility and improvement in data analysis and interpretation. Automation, better data visualization, and interdisciplinary collaboration are going to become ubiquitous in the future. With artificial intelligence and machine learning integrated and tested out, the predictive capabilities of chemometric software are becoming advanced, thereby increasing their efficacy. All of these enhancements may bring new and important implications for herbal medicine in terms of plants’ chemical composition and interaction, quality assurance, and identification of new therapeutic agents. As such, this enables the scientist to surmount beyond prevailing boundaries toward scientific breakthroughs in fields.
LIST OF ABBREVIATIONS
ANN, artificial neural networks; API, active pharmaceutical ingredient; BP-ANN, back propagation ANN; CA, cluster analysis; CAMO, computer aided modeling; EDA, exploratory data analysis; HCA, Hierarchical cluster technique; HPLC, high-performance liquid chromatography; HPTLC, high-performance thin-layer chromatography; LDA, linear discriminant analysis; MSPC, multivariate statistical process control; NIR, near-infrared; NMR, nuclear magnetic resonance; PAT, process analytical technology; PCA, principal component analysis; PLS, partial least square; PLS-DA, projections to latent structures–discriminant analysis; RTTs, retention times; SA, similarity analysis; SIMCA, soft independent modeling of class analogy.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
No new data were created or analyzed in this review. Data sharing is not applicable to this article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Rebiai A, Hemmami H, Zeghoud S, Ben Seghir B, Kouadri I, Eddine LS, et al. Current application of chemometrics analysis in authentication of natural products: a review. Comb Chem High Throughput Screen. 2022;25:945–72.
2. Takamura A, Ozawa T. Recent advances of vibrational spectroscopy and chemometrics for forensic biological analysis. Analyst. 2021;146:7431–49.
3. Sarraguça MC, Lopes JA. Quality control of pharmaceuticals with NIR: from lab to process line. Vib Spectrosc. 2009;49:204–10.
4. Liu X, Bai B, Rogers KM, Wu D, Qian Q, Qi F, et al. Determining the geographical origin and cultivation methods of Shanghai special rice using NIR and IRMS. Food Chem. 2022;394:133425.
5. Robotti E, Marengo E. Chemometric multivariate tools for candidate biomarker identification: LDA, PLS-DA, SIMCA, ranking-PCA. Methods Mol Biol. 2016;1384:237–67.
6. Cao X, You G, Li H, Li D, Wang M, Ren X. Comparative investigation for rotten xylem (kuqin) and strip types (tiaoqin) of scutellaria baicalensis georgi based on fingerprinting and chemical pattern recognition. Molecules. 2019;24:2431.
7. Lavine B, Workman J. Chemometrics. Anal Chem. 2008;80:4519–31.
8. Acevska J, Stefkov G, Cvetkovikj I, Petkovska R, Kulevanova S, Cho J, et al. Fingerprinting of morphine using chromatographic purity profiling and multivariate data analysis. J Pharm Biomed Anal. 2015;109:18–27.
9. Huang Q, Yang D, Jiang L, Zhang H, Liu H, Kotani K. A novel unsupervised adaptive learning method for long-term electromyography (EMG) pattern recognition. Sensors. 2017;17:1370.
10. Sensinger JW, Lock BA, Kuiken TA. Adaptive pattern recognition of myoelectric signals: exploration of conceptual framework and practical algorithms. IEEE Trans Neural Syst Rehabil Eng. 2009;17:270–8.
11. Zhang X, Mei X, Wang Z, Wu J, Liu G, Hu H, et al. Chemical fingerprint and quantitative analysis for the quality evaluation of docynia dcne leaves by high-performance liquid chromatography coupled with chemometrics analysis. J Chromatogr Sci. 2018;56:575–81.
12. Xie J-P, Ni H-G. Chromatographic fingerprint similarity analysis for pollutant source identification. Environ Pollut. 2015;207:341–4.
13. Schulze AE, De Beer D, Mazibuko SE, Muller CJF, Roux C, Willenburg EL, et al. Assessing similarity analysis of chromatographic fingerprints of Cyclopia subternata extracts as potential screening tool for in vitro glucose utilisation. Anal Bioanal Chem. 2016;408:639–49.
14. Souza M, José Comin J, Moresco R, Maraschin M, Kurtz C, Emílio Lovato P, et al. Exploratory and discriminant analysis of plant phenolic profiles obtained by UV–vis scanning spectroscopy. J Integr Bioinform. 2021;18:20190056.
15. Silva EFR, da Silva Santos BR, Minho LAC, Brandão GC, de Jesus Silva M, Silva MVL, et al. Characterization of the chemical composition (mineral, lead and centesimal) in pine nut (Araucaria angustifolia (Bertol.) Kuntze) using exploratory data analysis. Food Chem. 2022;369:130672.
16. G?d?k B. Antioxidant, antimicrobial activities and fatty acid compositions of wild Berberis spp. by different techniques combined with chemometrics (PCA and HCA). Molecules. 2021;26:7448.
17. Al-Dabooni S, Wunsch D. Model order reduction based on agglomerative hierarchical clustering. IEEE Trans Neural Netw Learn Syst. 2019;30:1881–95.
18. McLachlan GJ, Bean RW, Ng S-K. Clustering. Methods Mol Biol. 2008;453:423–39.
19. Sharma A, López Y, Tsunoda T. Divisive hierarchical maximum likelihood clustering. BMC Bioinformatics. 2017;18:546.
20. Aeria G, Claes P, Vandermeulen D, Clement JG. Targeting specific facial variation for different identification tasks. Forensic Sci Int. 2010;201:118–24.
21. Oliveira S, Douglas de Sousa Fernandes D, Véras G. Overview of analytical techniques associated with pattern recognition methods in sugarcane spirits samples. Crit Rev Anal Chem. 2019;49:477–87.
22. Shamsudin S, Selamat J, Sanny M, A R SB, Jambari NN, Khatib A. A comparative characterization of physicochemical and antioxidants properties of processed Heterotrigona itama honey from different origins and classification by chemometrics analysis. Molecules. 2019;24:3898.
23. Mohamad Asri MN, Verma R, Mahat NA, Nor NAM, Mat Desa WNS, Ismail D. Discrimination and source correspondence of black gel inks using Raman spectroscopy and chemometric analysis with UMAP and PLS-DA. Chemom Intell Lab Syst. 2022;225:104557.
24. Afy-Shararah M, Salonitis K. Integrated modeling of “soft” and “hard” variables in manufacturing. Int J Adv Manuf Technol. 2022;122:4259–65.
25. Harnly J. Botanical authentication using one-class modeling. J AOAC Int. 2023;106:1077–86.
26. Salehi M, Zare A, Taheri A. Artificial neural networks (ANNs) and partial least squares (PLS) regression in the quantitative analysis of respirable crystalline silica by Fourier-transform infrared spectroscopy (FTIR). Ann Work Expo Health. 2021;65:346–57.
27. Ahmad I, Sheraz MA, Ahmed S, Anwar Z. Multicomponent spectrometric analysis of drugs and their preparations. Profiles Drug Subst Excip Relat Methodol. 2019;44:379–413.
28. Akbari Lakeh M, Abdollahi H. Known-value constraint in multivariate curve resolution. Anal Chim Acta. 2018;1030:42–51.
29. Ferreira RJ, Rosa TR, Ribeiro J, Barthus RC. Simultaneous metal determination in artisanal cachaça by using voltammetry and multivariate calibration. Food Chem. 2020;314:126126.
30. Chong J, Wishart DS, Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics. 2019;68:e86.
31. Li Q, Qi L, Zhao K, Ke W, Li T, Xia L. Integrative quantitative and qualitative analysis for the quality evaluation and monitoring of Danshen medicines from different sources using HPLC-DAD and NIR combined with chemometrics. Front Plant Sci. 2022;13:932855.
32. Charles AL, Alamsjah MA. Application of chemometric techniques: an innovative approach to discriminate two seaweed cultivars by physico-functional properties. Food Chem. 2019;289:269–77.
33. Oravec M, Beganovi? A, Gál L, ?eppan M, Huck CW. Forensic classification of black inkjet prints using Fourier transform near-infrared spectroscopy and linear discriminant analysis. Forensic Sci Int. 2019;299:128–34.
34. Liu J, Xu Y, Liu S, Yu S, Yu Z, Low SS. Application and progress of chemometrics in voltammetric biosensing. Biosensors. 2022;12:494.
35. Beke ÁK, Gyürkés M, Nagy ZK, Marosi G, Farkas A. Digital twin of low dosage continuous powder blending – Artificial neural networks and residence time distribution models. Eur J Pharm Biopharm. 2021;169:64–77.
36. Brinson RG, Elliott KW, Arbogast LW, Sheen DA, Giddens JP, Marino JP, et al. Principal component analysis for automated classification of 2D spectra and interferograms of protein therapeutics: influence of noise, reconstruction details, and data preparation. J Biomol NMR. 2020;74:643–56.
37. Liu H-L, Zeng Y-T, Zhao X, Ye Y-L, Wang B, Tong H-R. Monitoring the authenticity of pu’er tea via chemometric analysis of multielements and stable isotopes. Food Res Int. 2020;136:109483.
38. Cruz-Tirado JP, Lima Brasil Y, Freitas Lima A, Alva Pretel H, Teixeira Godoy H, Barbin D, et al. Rapid and non-destructive cinnamon authentication by NIR-hyperspectral imaging and classification chemometrics tools. Spectrochim Acta A Mol Biomol Spectrosc. 2023;289:122226.
39. Bevilacqua M, Nescatelli R, Bucci R, Magrì AD, Magrì AL, Marini F. Chemometric classification techniques as a tool for solving problems in analytical chemistry. J AOAC Int. 2014;97:19–28.
40. Dastjerdi AG, Akhgari M, Kamali A, Mousavi Z. Principal component analysis of synthetic adulterants in herbal supplements advertised as weight loss drugs. Complement Ther Clin Pract. 2018;31:236–41.
41. Du W, Li X, Yang Y, Yue X, Jiang D, Ge W, et al. Quantitative determination, principal component analysis and discriminant analysis of eight marker compounds in crude and sweated Dipsaci Radix by HPLC-DAD. Pharm Biol. 2017;55:2129–35.
42. Khan MN, Ul Haq F, Rahman S, Ali A, Musharraf SG. Metabolite distribution and correlation studies of Ziziphus jujuba and Ziziphus nummularia using LC-ESI-MS/MS. J Pharm Biomed Anal. 2020;178:112918.
43. Nie Q-X, Zhang Q-X, Zhu Y-J, Wu P-J, He Y-L, Wang J-Y. Relationship between the UPLC fingerprints of citrus reticulata “Chachi” leaves and their antioxidant activities. Int J Anal Chem. 2022;2022:1–11.
44. Li H, Feng T, Wen Y, Li L, Liu Y, Ren X, et al. Comparative investigation for raw and processed products of euodiae fructus based on high-performance liquid chromatography fingerprints and chemical pattern recognition. Chem Biodivers. 2021;18:e2100281.
45. Bastien P, Bertrand F, Meyer N, Maumy-Bertrand M. Deviance residuals-based sparse PLS and sparse kernel PLS regression for censored data. Bioinformatics. 2015;31:397–404.
46. Manley SC, Hair JF, Williams RI, McDowell WC. Essential new PLS-SEM analysis methods for your entrepreneurship analytical toolbox. Int Entrep Manag J. 2021;17:1805–25.
47. Sarstedt M, Hair JF, Pick M, Liengaard BD, Radomir L, Ringle CM. Progress in partial least squares structural equation modeling use in marketing research in the last decade. Psychol Mark. 2022;39:1035–64.
48. Biancolillo A, Marini F. Chemometric methods for spectroscopy-based pharmaceutical analysis. Front Chem. 2018;6:576.
49. Saleh AA, Hegazy M, Abbas S, Elkosasy A. Development of distribution maps of spectrally similar degradation products by Raman chemical imaging microscope coupled with a new variable selection technique and SIMCA classifier. Spectrochim Acta A Mol Biomol Spectrosc. 2022;268:120654.
50. Fabijani? I, ?avuži? D, Mandac Zubak A. Meningococcal polysaccharides identification by NIR spectroscopy and chemometrics. Carbohydr Polym. 2019;216:36–44.
51. Marengo E, Robotti E, Bobba M, Righetti PG. Evaluation of the variables characterized by significant discriminating power in the application of SIMCA classification method to proteomic studies. J Proteome Res. 2008;7:2789–96.
52. Pomerantsev AL, Rodionova OY. Multiclass partial least squares discriminant analysis: taking the right way—A critical tutorial. J Chemom. 2018;32:e3030.
53. Sainani KL. Introduction to principal components analysis. PM&R. 2014;6:275–8.
54. Giuliani A. The application of principal component analysis to drug discovery and biomedical data. Drug Discov Today. 2017;22:1069–76.
55. Belmonte-Sánchez E, Romero-González R, Garrido Frenich A. Applicability of high-resolution NMR in combination with chemometrics for the compositional analysis and quality control of spices and plant-derived condiments. J Sci Food Agric. 2021;101:3541–50.
56. Yang J, Xu J, Zhang X, Wu C, Lin T, Ying Y. Deep learning for vibrational spectral analysis: recent progress and a practical guide. Anal Chim Acta. 2019;1081:6–17.
57. Challa S, Potumarthi R. Chemometrics-based process analytical technology (PAT) tools: applications and adaptation in pharmaceutical and biopharmaceutical industries. Appl Biochem Biotechnol. 2013;169:66–76.
58. Abdolmaleki A, Ghasemi J, Shiri F, Pirhadi S. Application of multivariate linear and nonlinear calibration and classification methods in drug design. Comb Chem High Throughput Screen. 2015;18:795–808.
59. Cuypers W, Lieberzeit PA. Combining two selection principles: sensor arrays based on both biomimetic recognition and chemometrics. Front Chem. 2018;6:268.
60. Arvanitoyannis IS, Vlachos A. Maize authentication: quality control methods and multivariate analysis (Chemometrics). Crit Rev Food Sci Nutr. 2009;49:501–37.
61. Brereton RG, Jansen J, Lopes J, Marini F, Pomerantsev A, Rodionova O, et al. Chemometrics in analytical chemistry—part I: history, experimental design and data analysis tools. Anal Bioanal Chem. 2017;409:5891–9.
62. Xu J, Hagler A. Chemoinformatics and drug discovery. Molecules. 2002;7:566–600.
63. Dupont MF, Elbourne A, Cozzolino D, Chapman J, Truong VK, Crawford RJ, et al. Chemometrics for environmental monitoring: a review. Analytical Methods. 2020;12:4597–620.
64. Efenberger-Szmechtyk M, Nowak A, Kregiel D. Implementation of chemometrics in quality evaluation of food and beverages. Crit Rev Food Sci Nutr. 2018;58:1747–66.
65. Beger RD, Sun J, Schnackenberg LK. Metabolomics approaches for discovering biomarkers of drug-induced hepatotoxicity and nephrotoxicity. Toxicol Appl Pharmacol. 2010;243:154–66.
66. Eriksson L, Antti H, Gottfries J, Holmes E, Johansson E, Lindgren F, et al. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (GPM). Anal Bioanal Chem. 2004;380:419–29.
67. Brereton RG, Jansen J, Lopes J, Marini F, Pomerantsev A, Rodionova O, et al. Chemometrics in analytical chemistry—part II: modeling, validation, and applications. Anal Bioanal Chem. 2018;410:6691–704.
68. Segelke T, Schelm S, Ahlers C, Fischer M. Food authentication: truffle (Tuber spp.) species differentiation by FT-NIR and chemometrics. Foods. 2020;9:922.
69. Biancolillo A, D’Archivio AA, Marini F, Vitale R. Editorial: novel applications of chemometrics in analytical chemistry and chemical process industry. Front Chem. 2022;10:926309.
70. Workman J, Koch M, Veltkamp D. Process analytical chemistry. Anal Chem. 2005;77:3789–806.
71. De Zan MM, Teglia CM, Robles JC, Goicoechea HC. A novel ion-pairing chromatographic method for the simultaneous determination of both nicarbazin components in feed additives: chemometric tools for improving the optimization and validation. Talanta. 2011;85:142–50.
72. Timmerman ME. Multilevel component analysis. Br J Math Stat Psychol. 2006;59:301–20.
73. Di C, Crainiceanu CM, Jank WS. Multilevel sparse functional principal component analysis. Stat. 2014;3:126–43.
74. Madsen R, Lundstedt T, Trygg J. Chemometrics in metabolomics—a review in human disease diagnosis. Anal Chim Acta. 2010;659:23–33.
75. Du, Zhang, Hou, Zhao, Li. Combined 2D-QSAR, principal component analysis and sensitivity analysis studies on fluoroquinolones’ genotoxicity. Int J Environ Res Public Health. 2019;16:4156.
76. Hendriks MMWB, Eeuwijk FA van, Jellema RH, Westerhuis JA, Reijmers TH, Hoefsloot HCJ, et al. Data-processing strategies for metabolomics studies. TrAC Trends Anal Chem. 2011;30:1685–98.