INTRODUCTION
Herpes simplex viruses (HSV) is a pathogen that has given rise to concerns at a worldwide level. Approximately 67% of people are affected with HSV 1 and 11.3% are affected with HSV 2 infection around the globe [1,2]. In the population aged between 0 to above 45 years, HSV-1 seroprevalence among male adults in India ranged from 78.5%–93.6% and among female adults from 75.5% to 97.8%, in 2016. The seroprevalence of HSV-2 was found to be between 3.2% to 0.7% in males and 6.4% to 26.5% in females in India [3]. Herpes infection causes recurrent oral and genital ulcerations however some more serious and rare complications may include meningitis, encephalitis, neonatal infection, and keratitis [4]. HSV-2 infection is hugely responsible for the HIV epidemic [5].
Several drugs have been developed against these HSVs, like acyclovir and its analogues that act upon the viral DNA polymerase or thymidine kinase (TK) which are considered as the gold standard drug against HSV. However, in addition to the low bioavailability of acyclovir, one of the major drawbacks is that the small molecule targets proteins with high variability due to the mutation of the gene sequences of DNA polymerase or TK [6–8]. The emergence of resistance also contributed to the mismanagement of the antiherpetic drugs [9]. The appearance of resistant varieties of viruses that are unresponsive to multiple drugs is another matter of concern [10]. The absence of any marketable vaccines for humans against the herpes viruses creates an urgent need to develop new techniques and identify new targets against HSV [11,12].
These concerns necessitate the need for novel targets and the use of advanced therapeutic technologies. The ul15 gene is highly conserved across the Herpesviridae family and can therefore act as a viable target [8]. The protein encoded by ul15 gene is the large subunit for the terminase complex required for viral DNA encapsidation [13–15]. The importance of UL15 protein in viral DNA processing and packaging has been demonstrated by many research groups using null mutants and recombinant viruses. The conserved nature of the protein and its significance in the viral life cycle indicates that it can act as a good target in herpesvirus infection therapy [8,16].
The RNA interference (RNAi) strategy provides an exciting avenue to design highly specific antiviral agents. The siRNA revolution has provided a huge impetus to the field of therapeutics leading to the release of 7 FDA-approved siRNA-based therapeutics [17]. The synthetic double-stranded (ds) siRNA is a 21-nucleotide long molecule that is introduced in the host cell. The biogenesis of the siRNA takes place in the cell cytoplasm. The Dicer complex cleaves the long ds RNA into shorter ds siRNA molecules. One of these strands in the ds siRNA molecule has the sequence complementarily with the target mRNA, while the other strand is called the passenger strand. The RNA-induced silencing complex (RISC) associates with the guide strand of the siRNA, while degrading the passenger strand. The mechanism of action of gene inhibition by the siRNA contributes to the specific binding of the guide strand with the target gene; the consequence of this binding is RNA degradation and loss of gene expression [18].
The advantage of using the siRNA technique over other techniques in the treatment of HSV 1 and 2 is that (1) there will be no off-target effects experienced due to the high specificity of the well-designed siRNA, (2) a very low concentration of siRNA for targeting specific genes at the transcriptional level (3) capacity to target undruggable targets [19].
Therefore, the current study provides an effective strategy for designing siRNA molecules using in silico tools and in vitro tools to validate the designed siRNAs. The study establishes the use of siRNA targeting ul15 gene product alone and in combination with low acyclovir concentration as a therapeutic agent for Herpesvirus infection. The problem of resistance occurs due to the development of small molecule drugs that target proteins produced by highly variable genes. The siRNA therapeutic that targets a conserved gene with low variability like ul15 will help in overcoming the problem of resistance and in targeting multiple strains of HSV.
MATERIALS AND METHODS
In silico design and analysis of siRNA ul15 gene sequence retrieval
The UL15 coding gene sequences from the strains of HSV1 were retrieved from the BLAST output using the HSV1 (NC_001806.2) gene sequence as a query. This allowed access to a complete compendium of the ul15 gene sequences across different strains.
Homology analysis for conserved area
Multiple sequence alignment is required to be carried out for the different viral strains to determine the areas showing sequence conservation. MEGAX software (version 10.2.6) was used to carry out multiple sequence alignment of the ul15 gene to determine the conserved areas of the ul15 gene across the different strains of HSV1 [20].
Target site identification for siRNA designing
siRNAs were designed using the siPred [21], siRNA Pred [22] and IDT SciTools siRNA Design Tool [23]. Rules of Ui-Tei [24], Amarzguioui and Prydz [25], and Reynolds et al. [26] were applied to designing the siRNA. Inhibition efficiency was restricted to >70% and cross-reacting human transcripts were checked.
Off-target similarity search using BLAST
The selection of suitable siRNA was based on the results of the BLAST search which was performed against the human genome and transcriptome [27] to identify the possible off-target matches.
Secondary structure prediction
The GC content and self-complementarity were checked using OligoCalc [28] and MaxExpect program was used to determine the secondary structure of the siRNA [29].
Determining the interaction of target mRNA with siRNA
DuplexFold program [30] of the RNA structure web server [29] was used to determine the thermodynamic interaction of the targeted mRNA strand and the siRNA guide strand. A stronger interaction between them is indicative of better efficacy of the designed siRNA. The interaction between the siRNA and target mRNA was further ascertained through the HDOCK webserver [31].
In vitro testing of designed siRNA
Cells and virus
The viral passage and antiviral screening were carried out using Vero cells (African green monkey kidney cells) in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) obtained from Gibco. The virus was passaged using a maintenance medium containing DMEM in 1% FBS. HSV1 was maintained at −80OC until use.
Determination of virus titer
Viral dilutions were prepared from 10−1 to 10−10 to determine the TCID50 (Tissue Culture Infectivity Dose50) for each virus. Each dilution (100 μl) was added to a column of the 96-well plate and the plate was observed microscopically. The dilution that showed cytopathic effect (CPE) in 50% of the wells was selected and used for further assays [32].
Cytotoxicity of designed siRNA
Acyclovir was procured from Cayman Chemical Company (Ann Arbor, MI) and siRNAs were procured from Sigma Aldrich (St. Louis, MO). Cytotoxicity of the siRNA molecules and siRNA in combination with acyclovir was analyzed. siRNA molecules were administered at 50 nM while acyclovir was administered at 18 μM. Acyclovir concentration is known to be non-cytotoxic to Vero cells at concentrations ≤20 μM [33]. The designed siRNAs were transfected into Vero cells cultured in 96-well plates using X-tremeGENE (Roche) according to the manufacturers’ instructions. The viable cells were determined after 5 days by following the 2-(2,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay. 100 μl of 0.2 g/ml MTT was added to the cells and the viable cells were determined by checking the formation of formazan crystals. The formazan crystals were dissolved in 50 μl of DMSO and the absorbance was checked at 570 nm in a microplate reader (ELx800, BioTek Instruments, Inc., Winooski, VT) [34].
Antiviral activity
The antiviral activity of the siRNA and siRNA acyclovir combination was determined by infecting confluent monolayers of Vero cells (2 × 104 cells/well) in 96-well microtiter plates for 2 hours. Vero cells were infected with HSV1 at 10TCID50 for 2 hours. siRNA transfection was carried out on Vero cells post infection. The initial antiviral activity was analyzed at concentrations of 0.1, 0.5, 10, and 50. For the combination studies, acyclovir (18 μM) was administered with the siRNA treatment (50 nM) to the Vero cells and incubated at 37°C for 5 days in 5% CO2 incubator. The viable cells were determined after 5 days by following the same MTT assay mentioned above.
Analysis of ul15 gene expression using real time-polymerase chain reaction (RT-PCR)
A monolayer of Vero cells was cultured in a six-well microtitre plate which was infected for 2 hours with 10TCID50 dose of HSV1. The siRNA and siRNA+acyclovir treatments were introduced to the Vero cells and complete RNA was extracted after 72 hours using TRIzol reagent (Invitrogen). The extracted total RNA was converted to cDNA using iScript cDNA Synthesis Kit (Bio-Rad). RT-PCR was conducted using TB green® Premix Ex Taq™ II (TII RnaseH Plus) (TaKaRa, Kusatsu-shi, Japan) in Applied Biosystems® QuantStudio® 5 Real-Time PCR System (Thermo Fisher, Waltham, MA). The primers used for amplifying the ul15 gene were: Forward primer: 5’TTGTCGACGAGGCCAACTTT3’; Reverse primer: 5’AAAGCTCGTACTGGCCTTCC3’; while the amplification of the housekeeping gene (Huel) was done using the primers: Forward primer: 5’TCAGACGACGAAGTCCCCATGAAG3’; Reverse primer: 5’TCCTTACGCAATTTTTTCTCTCTGGC3’ [35]. Duplicates of Ct values were averaged and normalized using the housekeeping genes. They were compared with the viral untreated samples using the Livak method of fold change in the target gene mRNA expression which was expressed as 2–ΔΔCT [36].
Statistical analysis
The experiments for each condition were repeated at least three times and the data is expressed as mean ± SD. ANOVA (one-way) followed by the Dunnett post hoc test was performed on the data.
RESULTS AND DISCUSSION
In silico design and analysis of siRNA
Design of siRNA
The online freeware siRNA Pred, siPRED, and IDT software were used to design siRNA, and the sequences are generated as per the efficiency expected for these siRNA sequences against the target mRNA. The software generated multiple sequences as outcomes out of which two sequences were taken for further analysis. The selection of the two siRNA molecules was based on their high inhibition capacity and lack of cross-reacting species (Table 1). This selection was done also to satisfy the rules for siRNA design and conservation across HSV strains [24–26]. The selected siRNA1 was designed using siPred where it showed an inhibition efficacy of 78.94% and siRNA2 was selected by using the IDT software as it showed no cross-reacting transcripts indicating no off-target effects. The off-target effect of the selected siRNA was further tested to check any unintentional matches with the human genome and transcriptome using the standalone BLAST package [27]. No significant similarity was observed and therefore, the sequences were used for further analysis. Therefore, the selected siRNA molecules are expected to have a silencing effect on the ul15 gene only, with no off-target effects on the host.
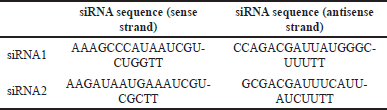 | Table 1. The siRNA sequences designed using the online freewares siRNA Pred, siPred, and IDT software. [Click here to view] |
Conservation of the selected siRNA sequences across different strains
The ul15 gene is known to be one of the most conserved genes in the Herpesvirus family [8]. However, to target multiple strains of the virus, it is important that the gene should be conserved. Therefore, the multiple sequence alignment was carried out for ul15 gene in HSV1 using in MEGAX software which showed high sequence conservation (Fig. 1). The area shown in the image depicts the zones that were eventually used for designing the siRNA. Therefore, the siRNA molecules were designed to be highly specific to the viral target and to be capable of inhibiting the other strains of the virus. This is highly advantageous as it effectively overcomes the necessity of designing multiple therapeutics for the different strains belonging to the same virus family.
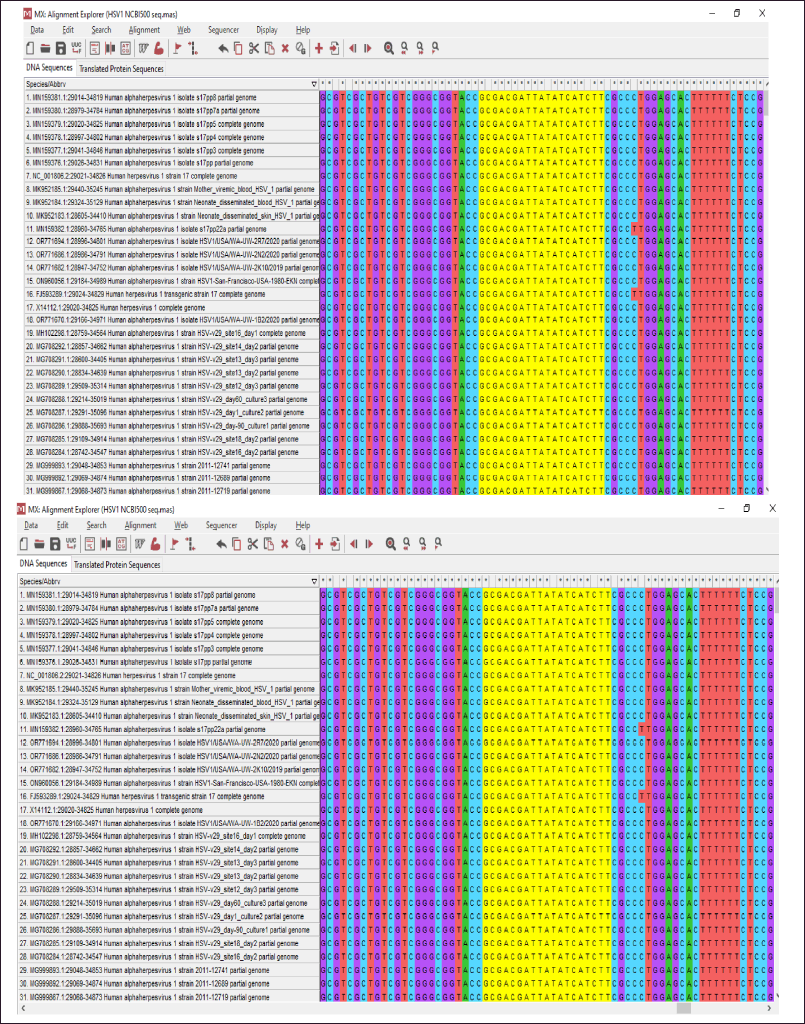 | Figure 1. The gene sequences encoding UL15 protein from different strains of HSV1 was aligned using MEGAX. Multiple sequence alignment of HSV1 UL15 gene sequences was done to analyse conserved areas and to determine areas that are suitable for siRNA design. (A) siRNA 1 and (B) siRNA 2 was designed using the highlighted area. [Click here to view] |
GC content calculation and secondary structure formation
The GC content of the siRNA molecules was maintained between 30% to 60% since a low value causes poor and off-target binding, while a high value blocks the RISC complex and helicase from unzipping the siRNA duplex [37]. GC content analysis of the predicted siRNA1 was 43% and for siRNA2 was 33%. Here, recommended level of GC content is between 30%–64% [38]. The secondary structure for the siRNA was also analyzed using MaxExpect (Fig. 2). The free energy for the formation of secondary structures for the siRNA1 was 1.7 kcal/mol and for siRNA2 was 1.4 kcal/mol. The positive and high values indicate no spontaneous secondary structure formation. Therefore, the designed siRNA molecules have an optimum value of GC content which will allow stable binding with RISC, with lower chances of off-target effects.
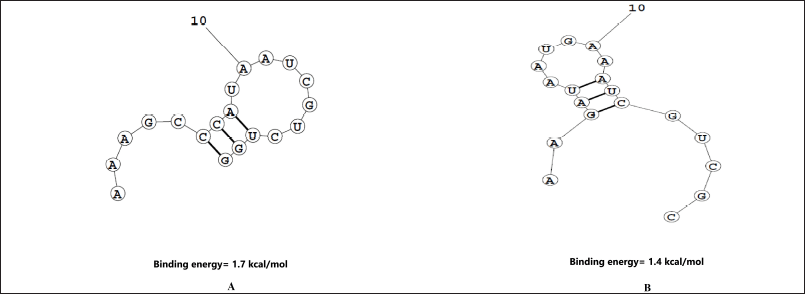 | Figure 2. The secondary structures for two designed siRNA predicted using MaxExpect webserver. (A) siRNA1; (B) siRNA2. The energy required for the secondary structure formation is indicative of the potential of siRNA strand folding. The higher the energy value, the lower the probability of folding. [Click here to view] |
Thermodynamics of target-guide strand interaction
Free energy of binding between the target and guide strand was calculated using DuplexFold (Fig. 3). The value was −32.9 for siRNA1 and the two binding patterns for siRNA2 with the target site had the binding energies of −17.9 and −17.4 kcal/mol. The structures were further visualized after docking in HNADOCK and the results are visible in Figure 4. The lower binding energies indicate a higher probability of binding of the siRNA with the target mRNA indicating the better probability of inhibition [39].
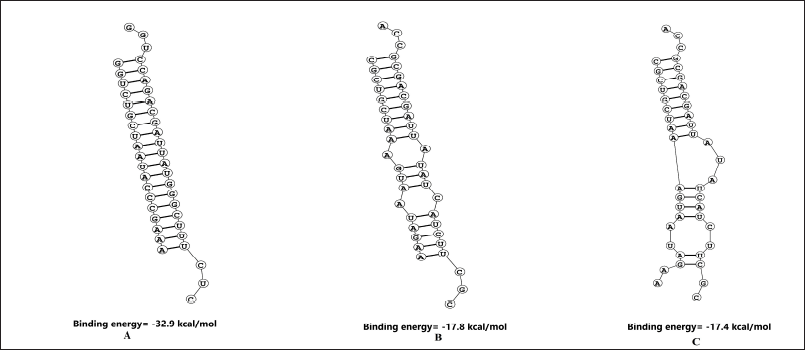 | Figure 3. The probability of siRNA and target mRNA site binding predicted using Duplexfold webserver. (A) siRNA1; (B) and (c) siRNA2. The low energy values indicate the high binding probability between the target mRNA and the designed siRNA. [Click here to view] |
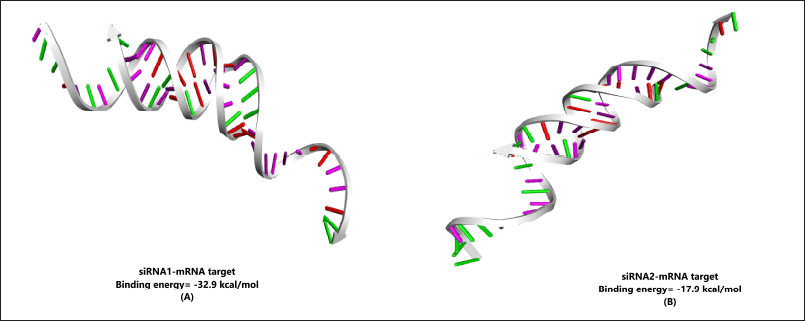 | Figure 4. HNADOCK results for docking between (A) siRNA1; and (B) siRNA2 with their respective target mRNA. The image depicts the stable formation of the double helical structure between the two designed siRNA strands over the targeted zone in the mRNA sequence. [Click here to view] |
In vitro testing of designed siRNA
Cytotoxicity of the designed siRNA
Cytotoxicity of the siRNA was tested at 50 nM and at 18 μM for acyclovir as this is the concentration at which the subsequent assays will be carried out. It was observed that both the siRNA molecules did not display any cytotoxicity on Vero cells at 50nM (Fig. 5). The combination of siRNA (50 nM) with acyclovir at 18 μM was also found to be non-cytotoxic. Acyclovir has an IC50 value of 4 μM; therefore, the selected concentration was expected to have a slightly higher antiviral activity on the infected cells [40]. These concentrations of siRNA and acyclovir were further used for the antiviral activity assay.
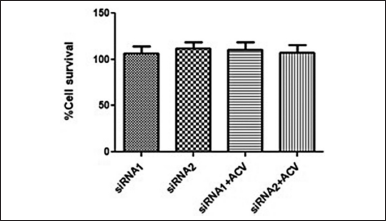 | Figure 5. The cytotoxicity of the designed siRNA and the siRNA+ acyclovir combinations was tested on Vero cells. The siRNA concentration was maintained at 50nM and that of acyclovir was at 18 μM. The cells were able to withstand the treatments and survived 48 hours of the treatment. [Click here to view] |
Antiviral activity of the designed siRNA
HSV1 viral load in the infected cells treated with specific siRNAs were reduced when compared to HSV1-infected control cells. The screening of antiHSV effect of siRNA molecules was initially tested at 0.5, 10, and 50 nM. The 50 nM concentration shows close to 50% antiHSV activity for both siRNA molecules and therefore, this concentration was used for further testing of the siRNA molecules. The antiviral effects of the siRNA molecules (50 nM) were tested individually and in combination with acyclovir (18 μM). The siRNA 1 and 2 showed the capacity to control HSV1 infection at 50 nM by showing percent antiviral activity of 50% and 25%, respectively. The synergistic activity of siRNA and acyclovir was observed (Fig. 6) when the siRNA 1 and siRNA2 at a concentration of 50 nM were co-administered with acyclovir at 18 μM.
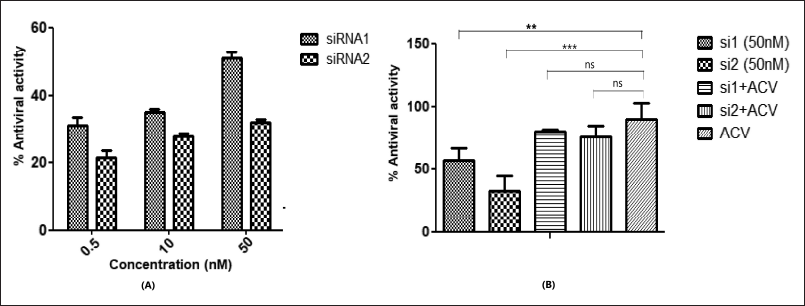 | Figure 6. Antiviral activity of siRNA and acyclovir as anti-HSV1 agent. (A) siRNAs 1 and 2 were administered at a range of concentrations to determine the IC50. (B) The concentration of 50 nM was selected for siRNA antiviral activity while the acyclovir concentration was set at 25 μM as a standard for comparison. Percentage antiviral activity is representative of the results of three independent experiments and expressed as the means ± SDs. For the siRNA and acyclovir combinations, statistical significance was determined by one-way ANOVA, with Dunnett’s multiple comparison test compared to the acyclovir-treated samples. (p < 0.05 given as ***). Data were plotted using Graph Pad Prism software version 8.0.2. [Click here to view] |
The results show the higher activity of siRNA 1 as compared to siRNA2, at the same concentration. This may be attributed to the sequence complementarity at the seed regions of the targeted gene [41]. The seed region for siRNA 1 has complete complementarity with the target whereas siRNA 2 has few mismatches with the seed region. These mismatches were incorporated to account for siRNA design guidelines and off-target effects. However, there should be a balance established between the guidelines to be followed and the antiviral effect, as seen in the results of the current study.
Analysis of ul15 gene expression using RT-PCR
The results of RT-PCR analysis are depicted in Figure 7. There was a significant reduction in viral gene expression when compared with the untreated viral control. The fold change analysis revealed that siRNA1 reduced the ul15 gene expression to 1.7%, while siRNA2 reduced the gene expression to 2%. On combination with acyclovir (18 μM), viral gene expression reduced to 0.1% for siRNA1 and 0.6% for siRNA2. The acyclovir treatment was able to limit the gene expression to 0.8% at 18 μM. Therefore, the synergistic activity of siRNA molecules with acyclovir was demonstrated as a reduction in viral gene expression which was enhanced by adding the acyclovir in combination with siRNA as a treatment strategy.
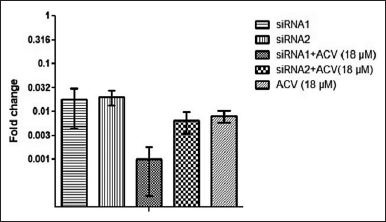 | Figure 7. Fold change in HSV1 ul15 gene expression in infected Vero cells treated with siRNA1 and 2 (50 nM) expressed as 2-??Ct value. [Click here to view] |
CONCLUSION
The current study provides a novel approach towards the development of antiHSV therapeutics. It highlights the use of the gold standard drugs like acyclovir in combination therapy to demonstrate a synergistic effect with RNAi agents. These gold standard drugs and the related nucleoside analogues have been in use since decades, however the emergence of resistant strains necessitates the evolution of new therapeutic approaches. In present study ul15 gene was targeted using siRNA approach has not been explored earlier. The results of RT-PCR demonstrated that the siRNA molecules designed in the current study had the capacity to reduce the u115 gene expression to ~2%. The study also demonstrated the capacity of designed siRNA molecules to reduce the infectivity of the HSV1 via the CPE assay post treatment. The advantage of targeting ul15 gene lies in the potential to target different strains of HSV1 due to the non-variable nature of the target gene.
ACKNOWLEDGMENT
The authors thank Manipal Center for Infectious Diseases (MAC-ID), Prasanna School of Public Health, Manipal Academy of Higher Education Manipal, India for the seed grant to complete the work. We also thank the Department of Pharmaceutical Biotechnology, Manipal College of Pharmaceutical Sciences and Department of Biotechnology, Manipal Institute of Technology, Manipal, for providing the necessary laboratory resources and support to conduct the research.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
The study has received financial support from Manipal Center for Infectious Diseases (MAC-ID); Ref No. MAC ID/SGA/2021/86, Prasanna School of Public Health, Manipal Academy of Higher Education Manipal, India.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used AI-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Looker KJ, Magaret AS, May MT, Turner KM, Vickerman P, Gottlieb SL, et al. Global and regional estimates of prevalent and incident herpes simplex virus type 1 infections in 2012. PLoS One 2015;10(10):1–17. CrossRef
2. Looker KJ, Magaret AS, Turner KM, Vickerman P, Gottlieb SL, Newman LM. Global estimates of prevalent and incident herpes simplex virus type 2 infections in 2012. PLoS One 2015;10(5):e114989. CrossRef
3. Cowan FM, French RS, Mayaud P, Gopal R, Robinson NJ, de Oliveira SA, et al. Seroepidemiological study of herpes simplex virus types 1 and 2 in Brazil, Estonia, India, Morocco, and Sri Lanka. Sex Transm Infect. 2003;79(4):286–90. CrossRef
4. Suazo PA, Tognarelli EI, Kalergis AM, González PA. Herpes simplex virus 2 infection: molecular association with HIV and novel microbicides to prevent disease. Med Microbiol Immunol. 2015;204:161–76. CrossRef
5. Wald A, Link K. Risk of human immunodeficiency virus infection in herpes simplex virus type 2 – seropositive persons: a meta-analysis. J Infect Dis. 2017;185(1):45–52. CrossRef
6. Sauerbrei A. Diagnosis, antiviral therapy, and prophylaxis of varicella-zoster virus infections. Eur J Clin Microbiol Infect Dis. 2016;35(5):723–34. CrossRef
7. Christophers J, Clayton J, Craske J, Ward R, Collins P, Trowbridge M, et al. Survey of resistance of herpes simplex virus to acyclovir in Northwest England. Antimicrob Agents Chemother. 1998;42(4):868–72. CrossRef
8. Karamitros T, Harrison I, Piorkowska R, Katzourakis A. De novo assembly of human herpes virus type 1 (HHV-1) genome, mining of non-canonical structures and detection of novel drug-resistance mutations using short-and long-read next generation sequencing technologies. PLoS One 2016;11(6):1–19. CrossRef
9. Bacon TH, Boon RJ, Schultz M, Hodges-Savola C. Surveillance for antiviral-agent-resistant herpes simplex virus in the general population with recurrent herpes labialis. Antimicrob Agents Chemother. 2002;46(9):3042–4. CrossRef
10. Boon RJ, Bacon TH, Robey HL, Coleman TJ, Connolly A, Crosson P, et al. Antiviral susceptibilities of herpes simplex virus from immunocompetent subjects with recurrent herpes labialie: a UK-based survey. J Antimicrob Chemother. 2000;46(2):324–5. CrossRef
11. Awasthi S, Hook LM, Shaw CE, Pahar B, Stagray JA, Liu D, et al. An HSV-2 trivalent vaccine is immunogenic in rhesus macaques and highly efficacious in guinea pigs. PLoS Pathogens 2017;13(1):e1006141. CrossRef
12. Awasthi S, Belshe RB, Friedman HM. Better neutralization of herpes simplex virus type 1 (HSV-1) than HSV-2 by antibody from recipients of GlaxoSmithKline HSV-2 glycoprotein D2 subunit vaccine. J Infect Dis. 2014;210(4):571–5. CrossRef
13. Yang K, Wills EG, Baines JD. A mutation in U L 15 of herpes simplex virus 1 that reduces packaging of cleaved genomes. J Virol. 2011;85(22):11972–80. CrossRef
14. Yu D, Weller SK. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 1998;243(1):32–44. CrossRef
15. Yu D, Sheaffer AK, Tenney DJ, Weller SK. Characterization of ICP6 :: lacZ insertion mutants of the UL15 gene of herpes simplex vrus type 1 reveals the translation of two proteins. J Virol. 1997;71(4):2656–65. CrossRef
16. Pi F, Zhao Z, Chelikani V, Yoder K, Kvaratskhelia M. Development of potent antiviral drugs inspired by viral hexameric DNA-packaging motors with revolving mechanism. J Virol. 2016;90(18):8036–46. CrossRef
17. Saju AF, Mukundan A, Divyashree MS, Chandrashekhar R, Mahadev Rao A. RNA diagnostics and therapeutics: a comprehensive review. RNA Biol. 2025;22(1):1–11. CrossRef
18. Qureshi A, Ahangar AG. A review on current status of antiviral siRNA. Rev Med Virol. 2018;28(4):1–11. CrossRef
19. Campeau E, Gobeil S. RNA interference in mammals: behind the screen. Brief Funct Genomics. 2011;10(4):215–26. CrossRef
20. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. CrossRef
21. Naito Y, Yoshimura J, Morishita S, Ui-Tei K. siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinform. 2009;10:1–8. CrossRef
22. Kumar M, Lata S, Raghava GPS. siRNApred: SVM based method for predicting efficacy value of siRNA. In: Proceedings of the first international conference on Open Source for Computer Aided Drug Discovery (OSCADD); Chandigarh: CSIR-IMTECH; 2009.
23. RNAi Design Tool™ program. Coralville, IA: IDT.
24. Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, et al. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32(3):936–48. CrossRef
25. Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochem Biophys Res Commun. 2004;316(4):1050–8. CrossRef
26. Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22(3):326–30. CrossRef
27. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinform. 2009;10:1–9. CrossRef
28. Kibbe WA. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35(suppl_2):W43–6. CrossRef
29. Bellaousov S, Reuter JS, Seetin MG, Mathews DH. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013;41(W1):W471–4. CrossRef
30. Piekna-Przybylska D, DiChiacchio L, Mathews DH, Bambara RA. A sequence similar to tRNA3Lys gene is embedded in HIV-1 U3–R and promotes minus-strand transfer. Nat Struct Mol Biol. 2010;17(1):83–9. CrossRef
31. Yan Y, Zhang D, Zhou P, Li B, Huang SY. HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365–73. CrossRef
32. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938;27(3):493–7. CrossRef
33. Brandi G, Schiavano GF, Balestra E, Tavazzi B, Perno CF, Magnani M. The potency of acyclovir can be markedly different in different cell types. Life Sci. 2001;69(11):1285–90. CrossRef
34. Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983;65(1–2):55–63. CrossRef
35. Chey S, Claus C, Liebert UG. Validation and application of normalization factors for gene expression studies in rubella virus-infected cell lines with quantitative real-time PCR. J Cell Biochem. 2010;110(1):118–28. CrossRef
36. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25(4):402–8. CrossRef
37. Friedrich M, Aigner A. Therapeutic SiRNA: state-of-the-art and future perspectives. BioDrugs 2022;36(5):549–71. CrossRef
38. Birmingham A, Anderson E, Sullivan K, Reynolds A, Boese Q, Leake D, et al. A protocol for designing siRNAs with high functionality and specificity. Nat Prot. 2007;2(9):2068–78. CrossRef
39. Chowdhury UF, Shohan MU, Hoque KI, Beg MA, Siam MK, Moni MA. A computational approach to design potential siRNA molecules as a prospective tool for silencing nucleocapsid phosphoprotein and surface glycoprotein gene of SARS-CoV-2. Genomics 2021;113(1):331–43. CrossRef
40. Grit GF, Märtson AG, Knoester M, Toren-Wielema ML, Touw DJ. Shedding a light on acyclovir pharmacodynamics: a retrospective analysis on pharmacokinetic/pharmacodynamic modelling of acyclovir for the treatment of varicella zoster virus infection in immunocompromised patients: a pilot study. Pharmaceutics 2022;14(11):2311. CrossRef
41. Angart P, Vocelle D, Chan C, Walton SP. Design of siRNA therapeutics from the molecular scale. Pharmaceuticals 2013;6(4):440–68. CrossRef