INTRODUCTION
The use of antibiotics has provided defense against potentially fatal bacterial infections for over a century. However, widespread and inappropriate antibiotic use combined with the evolution of organisms has led to the rise of multidrug-resistant organisms that are now resistant to most, if not all, antibiotic classes. These organisms are usually called extensively drug-resistant or pan-resistant. There are differences in antibiotic usage patterns worldwide based on factors such as healthcare practices, prescribing habits, accessibility to healthcare, and regulatory policies [1–4]. The emergence of antibiotic resistance also threatens life-saving medical advances such as chemotherapy, organ transplants, and various surgeries. It is primarily because antibiotic-resistant bacteria increase the risk of serious and life-threatening sepsis [5]. In addition, bacterial biofilm communities also contribute to antibiotic resistance. A biofilm is a complex community of bacteria adhered to surfaces and containing a variety of bacterial colonies and single-cell types. It contains a matrix of extracellular polymeric substances, including environmental deoxyribonucleic acid (DNA), proteins, and polysaccharides. As a result of this complex structure, biofilms are highly resistant to antibiotics, making them a major cause of persistent infections, especially in healthcare settings involving indwelling medical devices [6]. The bacteria within biofilms display reduced susceptibility to antimicrobial treatments [7]. They also play a crucial role in developing and persisting antibiotic resistance in various multi-drug resistant (MDR) pathogens, including the Acinetobacter baumannii. It contributes to its resistance phenotype by providing a physical barrier that hinders the penetration and efficacy of antibiotics. Interestingly, A. baumannii has the highest biofilm formation rate among pathogens, reaching approximately 90% [8–10]. Besides these, other pathogen factors, mainly the outer membrane protein A (ompA), biofilm-associated protein (bap), chaperon-usher pilus (csu) or others, contribute to the adherence, communication, invasion, cytotoxicity, and biofilm production [11–13]. Most antibiotics introduced in recent decades are modified versions of those discovered in the 1980s. To respond to the growing problem of antimicrobial resistance, one of the forward-looking strategies involves revitalizing the effectiveness of existing antibiotics against bacterial pathogens [14–16]. There is a growing body of research on using natural compounds to reverse antibiotic resistance. By combining antibiotics with natural products, it is possible not only to overcome resistance but also to reduce the required dosage, thereby minimizing associated side effects [17–20]. Subsequently, there is a need for novel treatment approaches to address infections associated with biofilms.
In recent years, there has been a notable increase in research focusing on exploring the therapeutic potential of traditional medicinal herbs and their bioactive compounds to tackle these challenges [21]. Studies exploring the antibiofilm effects of plant phenolics have unveiled promising findings. These natural compounds derived from plants have demonstrated the capacity to disrupt and modulate bacterial regulatory mechanisms, thereby impeding the formation and development of biofilms [22]. Curcumin, derived from the plant Curcuma longa, is a bioactive compound renowned for its antimicrobial characteristics. Curcumin exhibits a wide-ranging antibacterial effect, encompassing various bacteria, including those that have developed resistance to conventional antibiotics. Nevertheless, most research studies have primarily focused on examining the antibacterial effects of curcumin against standard strains of bacteria; however, reports on its efficacy against resistant bacterial biofilms are limited [23]. Given this context, the present study uses molecular docking techniques to explore the potential of curcumin and/or its novel derivatives against various biofilm-related pathogens, namely selected protein structures bacteria, fungi, and human pathogens. The efficacy against resistant biofilm using selected protein structures of Staphylococcus aureus, A. baumannii, Escherichia coli, Candida albicans, and P. aeruginosa as receptors have been studied using curcumin as ligand.
Lakadong turmeric (LKD), grown in Meghalaya (India), is one of the highest curcumin-yielding turmerics in the world. Despite claims of high yield, there needs to be more scientific reports on curcumin isolated from LKD. Moreover, the clinical application of curcumin as an antibacterial is limited due to its poor solubility. In our earlier study, we isolated the curcumin from the LKD rhizome by successive extraction followed by isolation of the Chloroform extract by column chromatography. This isolated curcumin was purified, characterized by fourier-transform infrared spectroscopy (FTIR), nuclear magnetic resonance (NMR), differential scanning calorimeter (DSC), high-performance thin layer chromatography (HPTLC), MS, and high-performance liquid chromatography (HPLC), and was loaded into poly (lactic-co-glycolic acid) (PLGA) nanosuspensions-based carbopol Nanogel. These Cur-NS Nanogel were evaluated for their potential anti-inflammatory and wound-healing activity in rats [24]. In the present study, therefore, the anti-bacterial and biofilm formation in Gram-positive and negative bacteria have been studied for these Lakadong-derived cur-nanogels through in-vitro assay.
METHODOLOGY
Selection and preparation of protein
The crystal structure of the selected targets was based on the association with bacteria, fungi, and human pathogens. The website research collaboratory for structural bioinformatics [Protein Data Bank(PDB); https://www.rcsb.org/] was utilized to access this valuable resource and retrieve the structural information of the receptor molecule from the database. The selected protein structures with the PDB ID were 7C7U (S. aureus; biofilm associated protein-BSP domain), 4QGG (S. aureus subsp. aureus MRSA252; Thymidylate kinase); 6FJY (A. baumannii; CsuC); 3TD4 (A. baumannii; Outer membrane protein omp38); 6B8Y (Homo sapiens; TGF-beta receptor type-1); 6F86 (E. coli O157:H7; DNA gyrase subunit B); 6CJS (C. albicans SC5314; Heat shock protein 90 homolog); 5OE3 (P. aeruginosa PAO1; Anthranilate--CoA ligase), and 4HZ5 (Enterococcus faecalis V583; DNA topoisomerase IV, B subunit), respectively. Discovery Studio 2022 was used to remove the water molecules and heteroatoms in the crystal structure to prevent potential interference during docking. Also, polar hydrogen was added and saved in .pdb format.
Ligand preparation and structure
Curcumin was selected as a ligand for the docking study and was extracted using the PubChem database (https://pubchem.ncbi.nlm.nih.gov) (Fig. 1). The PubChem database contains information about chemical substances and their biological activities. The two-dimensional structure of a specific compound was retrieved and saved as an structure data file (SDF). Then, the structure was minimized using Marvin Sketch (ChemAxon) software. The SDF format was converted to the PDB format using the Open Babel software’s graphical user interface (GUI), ensuring the compounds’ preparation for docking prediction.
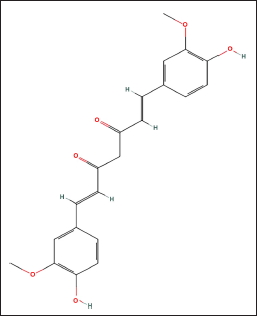 | Figure 1. The 2-D structure of Curcumin (https://pubchem.ncbi.nlm.nih.gov). [Click here to view] |
Molecular docking
In the present work, the docking of the prepared ligand with the selected proteins was conducted using Biovia Discovery Studio 2022 (DS 2022) software. The docking process employed the CDOCKER technique, renowned for its precision in predicting ligand-protein interactions. The binding energy for the ligand-protein complex was determined following the docking study. Subsequently, an in situ ligand minimization procedure was executed to enhance data accuracy and refine the ligand structure. Utilizing a nonbond list radius of 14.0 enabled considering nonbonded interactions in estimating binding energy. An examination of the binding posture and ligand orientation within the active site was performed to comprehend the exact interactions and mechanisms governing ligand-protein binding [25].
Preparation and characterization of LKD-Cur nanogel
Cur-NS Nanogel from the isolated curcumin was prepared by the method discussed in our earlier study [24]. Briefly, the isolated curcumin was loaded into PLGA nanosuspensions by nanoprecipitation method followed by conversion to carbopol-based Nanogel. The nanogels were characterized physicochemically in terms of pH, content, spreadability, gel strength, viscosity, and drug permeation. The optimized nanogel was then evaluated for its antibacterial and antibiofilm activity. The isolated curcumin was obtained by successive extraction by soxhlet using chloroform, hexane, ethyl acetate, and methanol, followed by isolation and purification of the Chloroform extract using column chromatography. This was then characterized by FTIR, NMR, DSC, HPTLC, and MS, and its purity was analyzed by HPLC [24].
Chemicals and bacterial culture
LKD curcumin isolated from our earlier study with high purity (established from the NMR, MS, FTIR, XRD, HPTLC, and HPLC studies) was used in the study [24]. The positive control Ciprofloxacin (2 mg/ml) was procured from SRL Chem-78079. The Microbial Type Culture Collection and Gene Bank (MTCC): P. aeruginosa (MTCC3541) and S. aureus (MTCC96) were used as bacterial cultures. All other chemicals or reagents used for the study were obtained from verified suppliers for carrying out the in-vitro assays.
Preliminary antimicrobial activity (anti-bacterial-zone inhibition test)
The preliminary antimicrobial activity of curcumin was determined using the Kirby-Bauer method, also known as the zone inhibition method. The Mueller-Hinton Agar (MHA) plates were prepared and inoculated with 100 μl of bacterial culture (P. aeruginosa and S. aureus), adjusted to a 0.5 McFarland Unit, followed by placing the discs containing 10 μl of different concentrations (ranging from 0 to 100 mg/ml). The solvent disc served as the vehicle control, while the Ciprofloxacin disc (10 μg) served as the positive control. The plates were then incubated for 24 hours at 37°C. Finally, the antibacterial activity of the discs was assessed by measuring the clarity of the inhibition zones around them. All the experiments were performed in triplicate.
Biofilm formation assay using scanning confocal microscope
An analysis of biofilm morphology investigated the antibiofilm property of LKD-Cur Nanogel via an Inverted laser scanning confocal microscope [ILSCM (ZEISS, LSM 900)] at 63× magnification. Briefly, the logarithmic growth phase culture of the microbes [gram-positive (S. aureus) and gram-negative (P. aeruginosa)] was adjusted to 0.5 McFarland standard. From this, 1 ml of a 10-times diluted culture with a 0.5 McFarland standard was placed onto a coated coverslip within a 50 mm petri plate. A control plate and a treated plate were prepared separately.10 μl of this culture was introduced onto a coated glass slide with 100 μl of medium. Afterward, the slide was incubated for 24 to 48 hours at 35°C. Following incubation, the slide was gently swirled to remove the medium, leaving the biofilm behind. It was then rinsed with water and air dried once more for 15 minutes, after which the coverslip with the biofilm was allowed to air dry. The dried coverslip containing the biofilm was then mounted on a slide using an antifade mountant containing DAPI (a fluorescent dye), and photos were captured.
Potential curcumin derivatives
A series of potential curcumin derivatives labeled as CC(1–5) and depicted in Figure 2 were designed through modifications made to the core structure of curcumin. These derivatives were then subjected to an assessment of their antibacterial activity against specific protein structures. These protein structures were identified by their corresponding PDB IDs: 7C7U, 4QGG, 6FJY, 3TD4, 6B8Y, 6F86, 6CJS, 5OE3, and 4HZ5.
The subsequent step involved conducting docking evaluations for these curcumin derivatives using the Biovia Discovery Studio 2022 (DS 2022) software. After performing the docking simulations, the best binding poses for each ligand with the respective protein were identified. These binding poses were evaluated in terms of their binding affinities, which were quantified in units of Kcal/mol. The binding affinity indicates how strong the interaction is between the ligand and the protein, with lower values representing stronger binding.
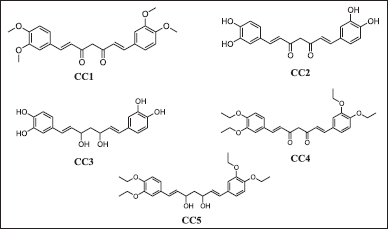 | Figure 2. The 2-D structure of curcumin derivatives (CC1-5). [Click here to view] |
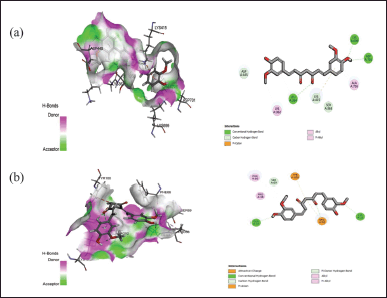 | Figure 3. Interaction of curcumin with the active binding site of protein (a) 7C7U and (b) 4QGG showing the 2-D diagram. [Click here to view] |
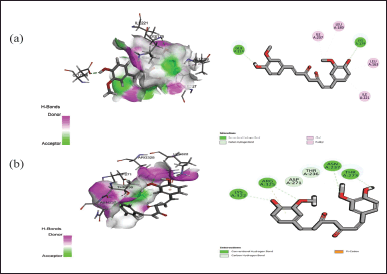 | Figure 4. Interaction of curcumin with the active binding site of protein (a) 6FJY and (b) 3TD4 showing the 2-D diagram. [Click here to view] |
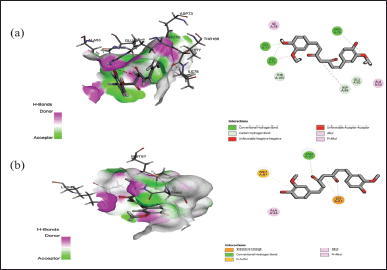 | Figure 5. Interaction of curcumin with the active binding site of protein (a) 6F86 and (b) 6CJS showing the 2-D diagram. [Click here to view] |
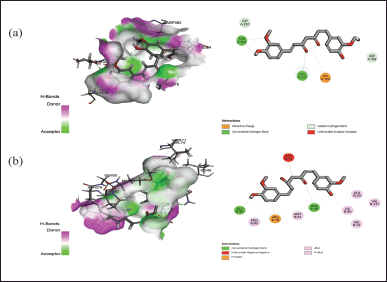 | Figure 6. Interaction of curcumin with the active binding site of protein (a) 5OE3 and (b) 4HZ5 showing the 2-D diagram. [Click here to view] |
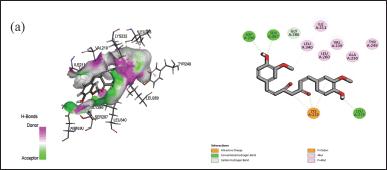 | Figure 7. Interaction of curcumin with the active binding site of protein (a) 6B8Y showing the 2-D diagram. [Click here to view] |
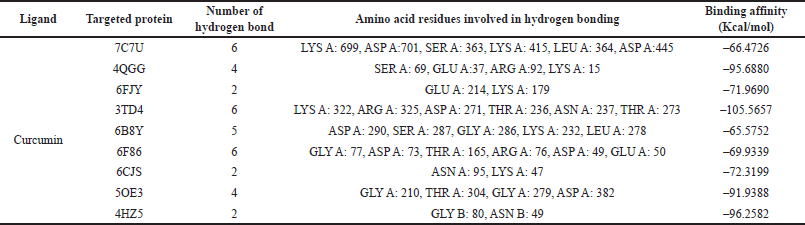 | Table 1. Docking energy values (binding energy) of curcumin expressed in Kcal/mol. [Click here to view] |
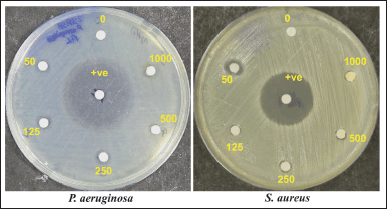 | Figure 8. Anti-bacterial activity of curcumin against P. aeruginosa (MTCC 3541) and S. aureus (MTCC 96). The amount present per well is in μg. +ve: Positive control (Ciprofloxacin). [Click here to view] |
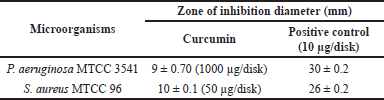 | Table 2. Anti-bacterial potential of curcumin against selected human pathogens. [Click here to view] |
RESULTS
Molecular docking study
The docking assessment was conducted to investigate the binding interaction between the selected protein structures (namely with the PDB IDs: 7C7U, 4QGG, 6FJY, 3TD4, 6B8Y, 6F86, 6CJS, 5OE3, 4HZ5), and with the ligand molecule, Curcumin. As shown in Figures 3–7, the curcumin molecule successfully binds within the active binding pocket of the selected protein with strong van der Waals forces. Among the selected proteins, curcumin showed the highest binding affinity (–105.5657 Kcal/mol) with the targeted protein 3TD4, whereas the protein 7C7U showed the least affinity (–66.4726 Kcal/mol) (Table 1). Furthermore, on looking at the hydrogen bond interaction, 3TD4 and 6F86 scored the highest number of hydrogen bond interactions toward curcumin via six bonds, whereas 6FJY, 6CJS, and 4HZ5 showed the least number of hydrogen bond interactions. The 2D results of the amino-acid residues for the selected protein-curcumin interactions showing the number of hydrogen bond interactions are tabulated in Table 1.
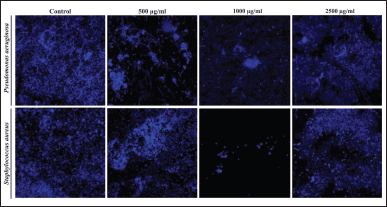 | Figure 9. Visualization of the biofilm inhibition of control and treated (Curcumin; 500, 1,000, and 2500 μg/ml) against P. aeruginosa and S. aureus. [Click here to view] |
Preparation and characterization of LKD-Cur-nanogels
Homogenous Cur-nanogel was obtained from nanosuspensions with size 150 ± 50 nm and PDI 0.2 ± 0.1. The Cur-NS-nanogel showed satisfactory physical properties (in terms of pH, gel strength, viscosity, and spreadability), drug loading, and stability. Ex vivo permeation studies in rats showed controlled permeation with steady-state flux of 21.28 ± 0.23 μg/cm2/hr in 24 hours [24].
Anti-bacterial-zone inhibition test
The anti-microbial activity test revealed that LKD-Cur Nanogel exhibits significant anti-bacterial activity against P. aeruginosaMTCC3541 with a mean zone inhibition of 9 ± 0.70 mm [Mean ± standard deviation (SD)] at an amount of 1,000 μg/disk and 10 ± 0.1 (50 μg/disk) against S. aureus (Fig. 8; Table 2).
Biofilm formation assay
The biofilm formation assay depicted that LKD-Cur Nanogel disrupted the established bacterial (both for P. aeruginosa and S. aureus) biofilms (formed on microtiter plates) at 1,000 μg/ml concentration compared with the control. However, further curcumin concentration increase at 2,500 μg/ml failed to eliminate the biofilms (Fig. 9) effectively.
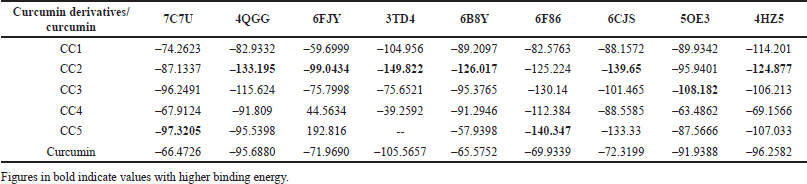 | Table 3. Docking energy values (binding energy) of curcumin derivatives and curcumin expressed in Kcal/mol against different targeted proteins. [Click here to view] |
Potential curcumin derivatives
The results of the modified curcumin derivatives [CC(1–5)] revealed a successful binding within the active binding pocket of the selected proteins with strong van der Waals forces. All the curcumin derivatives showed a strong binding affinity with all the selected proteins compared to the curcumin parent molecule. Among all the curcumin derivatives, CC2 showed an excellent binding affinity with the highest number of the targeted proteins 4QGG, 6FJY, 3TD4, 6B8Y, 6CJS, 4HZ5, followed by CC5 (7C7U and 6F86) and CC3 (5OE3) (Table 3).
DISCUSSION
The data from docking studies aid in developing novel multi-targeted medications that can be further explored in pre-clinical and clinical investigations [26]. It has long been recognized that plants contain bioactive compounds that can be used in biomedicine to treat various illnesses. The World Health Organization (WHO) has identified antimicrobial resistance as a global health threat of major concern. Consequently, the WHO prioritized antimicrobial resistance as a critical research area and called for effective strategies to address this global challenge [27]. Accordingly, this study aimed to screen Curcumin and/or their novel derivatives and docked against various biofilm-related pathogens, namely bacteria, fungi, and human pathogens. Furthermore, the antimicrobial and antibiofilm efficacy of LKD-Cur Nanogel prepared from earlier studies was studied.
Herbal plant extracts may prevent pathogens from attaching to biofilms, preventing the formation of biofilms. This extract may help treat biofilm-related infections and might be included in drug formulations to fight bacterial infections caused by biofilms. Several plant extracts containing phenolic compounds effectively counter biofilms’ effects [28,29]. The findings of our in-silico study shed light on the potential therapeutic value of Curcumin, a bioactive phenolic compound derived from C. longa. Our investigation revealed that Curcumin exhibited high selectivity toward a range of targeted proteins, including 7C7U, 4QGG, 6FJY, 3TD4, 6B8Y, 6F86, 6CJS, 5OE3, and 4HZ5. Importantly, the docking simulations demonstrated strong binding interactions and acceptable docking energy between Curcumin and these proteins, primarily driven by hydrogen bond interactions. Therefore, the confirmation of robust binding interactions validates their functional activities and provides insights into potential mechanisms of action. Bacterial biofilms, consisting of communities of bacteria, form and envelop themselves within an extracellular matrix commonly composed of proteins. Infectious bacteria can modulate these proteins, vital in developing bacterial biofilms [30]. In response to environmental factors, acidic conditions, and low calcium levels, the S. aureus Bap protein undergoes self-assembly, forming functional amyloid aggregates that contribute to constructing the biofilm matrix [31]. Another plausible mechanism involves the utilization of thymidylate kinase (PDB ID: 4QGG) due to its antibacterial properties. The Bap protein (PDB ID: 7C7U) forms functional amyloid structures and biofilm production. These enzymes are crucial in various biological processes, including bacterial DNA replication, resilience against antibiotics, and the immune system [32,33]. The bacterial enzymes Gyrase B (PDB ID: 6F86) and Topoisomerase IV subunit B (PDB ID: 4HZ5) are responsible for modulating the DNA’s topological structure during replication [34]. Furthermore, A. baumannii (PDB ID: 6FJY, 3TD4), a major cause of healthcare-associated infections, forms robust biofilms aided by CsuC pili and outer membrane protein omp38. These revealed insights into bacterial attachments to nonbiological surfaces via hydrophobic interactions [35,36]. Considering C. albicans (PDB ID: 6CJS) growing drug resistance, this makes heat shock protein 90 (Hsp90) homolog an attractive target for treating invasive fungal infections [37]. Another opportunistic gram-negative bacteria, P. aeruginosa (PDB ID: 5OE3), is implicated in acute and chronic nosocomial infections and often leads to patient mortality due to its ability to form a biofilm, thereby boosting drug resistance and increasing virulence [38]. Hence, blocking these proteins could stop biofilm growth, and one such approach can be achieved through the active principles present in the plants for their anti-bacterial effects.
The anti-microbial activity test of LKD-Cur Nanogel was carried out using the Kirby-Bauer method (zone inhibition method). Our findings showed that curcumin has substantial antimicrobial potential (zone of inhibition) against P. aeruginosa and S. aureus. The results demonstrate curcumin’s ability to inhibit P. aeruginosa growth, an opportunistic pathogen associated with various infections, especially among immunocompromised patients. Similarly, S. aureus, a common pathogen, causes a wide variety of infections, including infection of the skin and respiratory system. The result of the study aligns with the attributes of S. aureus in combating infections. These are consistent with previous reports highlighting the ability of curcumin’s antimicrobial action on various pathogens [39,40]. This could be attributed to its mechanisms of action, which include interfering with the essential cellular processes, disrupting the cell membranes, and modulating bacterial gene expression [41].
Forming a biofilm is one of the mechanisms by which bacteria reduce their ability to resist antibiotics. The scanning electron microscopy analysis of LKD-Cur nanogel-treated bacterial cells revealed disruption of the established bacterial biofilms (both for P. aeruginosa and S. aureus) at 1,000 μg/ml concentration compared with the control. However, further curcumin concentration increases at 2500 μg/ml showed inconsistent results and failed to eliminate the biofilms effectively. This indicated that curcumin had an inhibitory effect on the biofilm formation of P. aeruginosa and S. aureus. This might be due to disruption in bacterial attachments or interference with the regulation of extracellular polysaccharides (EPS) and extracellular DNA (eDNA), leading to biofilm production cessation [42,43].
The results of the modified curcumin derivatives [CC(1–5)] suggest excellent potency against different targeted proteins with a strong binding affinity. These modified curcumin ligands are well-settled inside the protein cavity. They might be suitable candidates for treating infections caused by pathogenic bacterial biofilms, thereby showing better anti-biofilm efficacy than the parent molecule of curcumin. However, it is important to acknowledge our study’s limitations, particularly in validating these findings concerning biofilm-associated pathogens through in-vitro assays.
CONCLUSION
In conclusion, the in-silico and in-vitro study demonstrates the promising therapeutic potential of Curcumin, a bioactive phenolic compound derived from C. longa, in targeting biofilm-related pathogens. Through strong binding interactions and acceptable docking energy, curcumin exhibited high selectivity toward several key proteins involved in biofilm formation, including those responsible for bacterial DNA replication and modulation of DNA topological structure. The LKD-derived Curcumin nanogels showed good antibacterial activity and effectiveness against biofilms formed by S. aureus and P. aeruginosa. Moreover, the modified novel curcumin derivatives exhibit robust binding and potential efficacy against targeted proteins. Hence, LKD-derived curcumin and its modified novel derivatives could be a new source of antibacterial agents against different biofilm-related pathogens. LKD-Cur Nanogel can be a promising approach for antibacterial and antibiofilm therapy.
ACKNOWLEDGEMENT
The authors would like to acknowledge the in-silico facilities provided by the DST-FIST laboratory [Department of Science and Technology-Fund for Improvement of S&T Infrastructure (DST-FIST), Government of India (GoI)], to Girijananda Chowdhury Institute of Pharmaceutical Science (GIPS), GCU, Guwahati, Assam.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
DST-FIST Grant Sponsored by the Department of Science and Technology (DST), Government of India. Reference No. SR/FST/COLLEGE-/2021/1172 for “FIST Program-2021” (TPN-69342).
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Al-Tawfiq JA, Momattin H, Al-Ali AY, Eljaaly K, Tirupathi R, Haradwala MB, et al. Antibiotics in the pipeline: a literature review (2017–2020). Infection. 2022;50(3):553–64.
2. Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645.
3. Al-Tawfiq JA, Stephens G, Memish ZA. Inappropriate antimicrobial use and potential solutions: a middle eastern perspective. Expert Rev Anti Infect Ther. 2010;8:765–74.
4. Septimus EJ. Antimicrobial Resistance: an antimicrobial/diagnostic stewardship and infection prevention approach. Med Clin North Am. 2018;102(5):819–29.
5. Cardile S, Del Chierico F, Candusso M, Reddel S, Bernaschi P, Pietrobattista A, et al. Impact of two antibiotic therapies on clinical outcome and gut microbiota profile in liver transplant paediatric candidates colonized by carbapenem-resistant Klebsiella pneumoniae CR-KP. Front Cell Infect Microbiol. 2021;11:730904.
6. Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019;8:76.
7. Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2(2):114–22.
8. Qi L, Li H, Zhang C, Liang B, Li J, Wang L, et al. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front Microbiol. 2016;7:483.
9. Sung JY. Molecular characterization and antimicrobial susceptibility of biofilm-forming Acinetobacter baumannii clinical isolates from Daejeon, Korea. Korean J Clin Lab Sci. 2018;50:100–109.
10. Salmani A, Shakerimoghaddam A, Pirouzi A, Delkhosh Y, Eshraghi M. Correlation between biofilm formation and antibiotic susceptibility pattern in Acinetobacter baumannii MDR isolates retrieved from burn patients. Gene Rep. 2020;21:100816.
11. Colquhoun JM, Rather PN. Insights into mechanisms of biofilm formation in Acinetobacter baumannii and implications foruropathogenesis. Front Cell Infect Microbiol. 2020;10:253.
12. Chapartegui-González I, Lázaro-Díez M, Bravo Z, Navas J, Icardo JM, Ramos-Vivas J. Acinetobacter baumannii maintains its virulence after long-time starvation. PLoS One 2018;13:e0201961.
13. Uppalapati SR, Sett A, Pathania R. The outer membrane proteins OmpA, CarO, and OprD of Acinetobacter baumannii confer a two-pronged defense in facilitating its success as a potent human pathogen. Front Microbiol. 2020;11:589234.
14. Sheu CC, Chang YT, Lin SY, Chen YH, Hsueh PR. Infections caused by Carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Front Microbiol. 2019;10:80.
15. Nasr T, Bondock S, Ibrahim TM, Fayad W, Ibrahim AB, AbdelAziz NA, et al. New acrylamide-sulfisoxazole conjugates as dihydropteroate synthase inhibitors. Bioorg Med Chem. 2020;28(9):115444.
16. Kumar M, Sarma DK, Shubham S, Kumawat M, Verma V, Nina PB, et al. Futuristic non-antibiotic therapies to combat antibiotic resistance: a review. Front Microbiol. 2021;12:609459.
17. Alvarez-Martinez FJ, Barrajon-Catalan E, Micol V. Tackling antibiotic resistance with compounds of natural origin: a comprehensive review. Biomedicines. 2020;8(10):405.
18. Su T, Qiu Y, Hua X, Ye B, Luo H, Liu D, et al. Novel opportunity to reverse antibiotic resistance: to explore traditional Chinese medicine with potential activity against antibiotics-resistance bacteria. Front Microbiol. 2020;11:610070.
19. Elhawary EA, Mostafa NM, Labib RM, Singab AN. Metabolomic profiles of essential oils from selected Rosa varieties and their antimicrobial activities. Plants (Basel). 2021;10(8):1721.
20. Cheesman MJ, Ilanko A, Blonk B, Cock IE. Developing new antimicrobial therapies: Are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacogn Rev. 2017;11(22):57–72.
21. Barbieri R, Coppo E, Marchese A, Daglia M, Sobarzo-Sanchez E, Nabavi SF, et al. Phytochemicals for human disease: an update on plant-derived compounds antibacterial activity. Microbiol Res. 2017;196:44–68.
22. Silva LN, Zimmer KR, Macedo AJ,Trentin DS. Plant natural products targeting bacterial virulence factors. Chem Rev. 2016;116:9162–236.
23. Hussain Y, Alam W, Ullah H, Dacrema M, Daglia M, Khan H, et al. Antimicrobial potential of curcumin: therapeutic potential and challenges to clinical applications. Antibiotics (Basel). 2022;11(3):322.
24. Sarma SK, Dutta U, Bharali A, Kumar S, Baruah S, Sarma H, et al. Isolation of curcumin from Lakadong turmeric of Meghalaya and development of its PLGA-Cur-NS loaded nanogel for potential anti-inflammatory and cutaneous wound healing activity in rats. Future J Pharm Sci. 2023;9(1):85.
25. Johari S, Sharma A, Sinha S, Das A. Integrating pharmacophore mapping, virtual screening, density functional theory, molecular simulation towards the discovery of novel apolipoprotein (apoE ε4) inhibitors. Comput Biol Chem. 2019;79:83–90.
26. Patwardhan B. Editorial: the new pharmacognosy. Comb Chem High Throughput Screen. 2014;17:97.
27. Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–18.
28. Kannappan A, Santhakumari S, Srinivasan R, Pandian SK, Ravi AV. Hemidesmus indicus, a traditional medicinal plant, targets the adherence of multidrug-resistant pathogens to form biofilms. Biocatal Agric Biotechnol. 2019;21:101338.
29. Stefanovic OD, Tesic JD, Comic LR. Melilotus albus and Dorycnium herbaceum extracts as source of phenolic compounds and their antimicrobial, antibiofilm, and antioxidant potentials. J Food Drug Anal. 2015;23(3):417–24.
30. Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A, Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol, 2020;27(12):1202–208.
31. Taglialegna A, Navarro S, Ventura S, Garnett JA, Matthews S, Penades JR, et al. Staphylococcal bap proteins build amyloid scaffold biofilm matrices in response to environmental signals. PLoSPathog. 2016;12(6):e1005711.
32. Kawatkar SP, Keating TA, Olivier NB, Breen JN, Green OM, Guler SY, et al. Antibacterial inhibitors of gram-positive thymidylate kinase: structure-activity relationships and chiral preference of a new hydrophobic binding region. J Med Chem. 2014;57:4584–97.
33. Ma J, Cheng X, Xu Z, Zhang Y, Valle J, Fan S, et al. Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch. EMBO J. 2021;40(14):e107500.
34. Tari LW, Trzoss M, Bensen DC, Li X, Chen Z, Lam T, et al. Pyrrolo pyrimidine inhibitors of DNA Gyrase B (GyrB) and Topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. Bioorganic Med Chem Lett. 2013;23:1529–36.
35. Pakharukova N, Tuittila M, Paavilainen S, Malmi H, Parilova O, Teneberg S, et al. Structural basis for Acinetobacter baumannii biofilm formation. Proc Natl Acad Sci USA. 2018;115(21):5558–63.
36. Park JS, Lee WC, Yeo KJ, Ryu KS, Kumarasiri M, Hesek D, et al. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J. 2012;26(1):219–28.
37. Yuan R, Tu J, Sheng C, Chen X, Liu N. Effects of Hsp90 Inhibitor ganetespib on inhibition of azole-resistant Candida albicans. Front Microbiol. 2021;12:680382.
38. Shahab M, Danial M, Khan T, Liang C, Duan X, Wang D, et al. In-silico identification of lead compounds for Pseudomonas aeruginosa PqsA enzyme: computational study to block biofilm formation. Biomedicines. 2023;11(3):961.
39. Di Salle A, Viscusi G, Di Cristo F, Valentino A, Gorrasi G, Lamberti E, et al. Antimicrobial and antibiofilm activity of curcumin-loaded electrospun nanofibers for the prevention of the biofilm-associated infections. Molecules. 2021;26(16):4866.
40. Dai C, Lin J, Li H, Shen Z, Wang Y, Velkov T, et al. The natural product curcumin as an antibacterial agent: current achievements and problems. Antioxidants (Basel). 2022;11(3):459.
41. Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482–501.
42. Rasamiravaka T, Labtani Q, Duez P, El Jaziri M. The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int. 2015;2015:1–17.
43. Ganesh PS, Rai VR. Attenuation of quorum-sensing-dependent virulence factors and biofilm formation by medicinal plants against antibiotic-resistant Pseudomonas aeruginosa. J Tradit Complement Med. 2018;8(1):170–77.