INTRODUCTION
Diabetic kidney disease (DKD) is one of the main causes of end-stage renal disease (ESRD) globally that necessitates renal replacement. It is a chronic microvascular consequence of type 2 diabetes mellitus (T2DM) that affects approximately 20%–30% of patients [1,2]. DKD is identified by albuminuria and/or an initially elevated glomerular filtration rate (GFR) that decreases in the middle to the end stage [3]. According to epidemiological studies conducted in different Asian countries, microalbuminuria prevalence rates range between 8% and 32% among T2DM patients [4]. Metabolic and hemodynamic processes are crucial in the pathophysiology of DKD with local Renin-Angiotensin-Aldosterone System (RAAS) activation being a part of the hemodynamic pathway [5].
One of the major enzymes in the RAAS is an angiotensin-converting enzyme (ACE) [6]. The ACE is encoded by the ACE gene, which is a 21 Kb long and consists of 26 exons and 25 introns. It is located on chromosome 17q23 [7]. The most common ACE gene polymorphism is the insertion (I)/deletion (D) polymorphism, which is defined by the absence or presence of a 287 bp fragment in intron 16 of the gene [8]. This polymorphism influences ACE enzyme activity in the blood with decreased (allele I) and increased (allele D) enzyme activity, respectively [9]. The I\D polymorphism of this gene has been well studied regarding its relationship with diabetic renal and cardiovascular complications. The plasma ACE was obtained mostly from tissue vascular endothelial cells, indicating that patients with the DD genotype may have greater tissue angiotensin (Ang) II levels. Because Ang II may influence smooth muscle cell development and result in myointimal hyperplasia following endothelial damage, treatment with ACE inhibitors (ACEIs) ameliorated myointimal proliferation after vascular injury [10]. A recent study suggests that increasing serum ACE levels in DKD patients may predict DKD, and persistently rising ACE indicates DKD progression. Furthermore, elevated ACE levels may hint at retinal impairment in people with early-stage DKD [11].
Angiotensin-converting enzyme 2 (ACE2) is the only known ACE homolog with enzymatic activity; it is a monocarboxypeptidase that degrades Ang II to Ang 1–7. It is abundant in the kidneys. It was demonstrated that ACE2 activity is altered in DKD, hypertensive renal disease, and several models of kidney damage [12]. Furthermore, urinary ACE2 is associated positively with the degree of albuminuria and proteinuria in individuals with T2DM, indicating elevated ACE2 protein and activity in diabetic animals’ urine and serum [13]. Angiotensin type 1 receptor (AGT1R) is found in many tissues, including vessel walls, the lung, and the kidney. When activated, it can modulate renal function, and its expression pattern is associated with nephropathy. Meta-analysis has proven the significant association between the AGT1R A1166C polymorphisms and DKD, especially in Asians and the T2DM population [14]. The presence of the C allele may be linked to increased AGT1R gene expression or a greater affinity of the receptor for Ang II. The increased Ang II activity makes the kidney more susceptible to the adverse hyperglycemia effect, either by changes in systemic or renal hemodynamics or by affecting renal cell function [15]. A variety of drugs that block the RAAS, such as ACEIs and angiotensin receptor blockers (ARBs), are frequently administered for treating hypertension; also, these drugs are known to reduce proteinuria in DKD patients, either alone or in combination [16]. However, not all patients respond uniformly to ACEI treatment in terms of renoprotection. The causes of the inconsistent antiproteinuric response to these drugs remain unknown. Individuals with the DD genotype, or D allele, had higher circulation and tissue ACE activity than individuals with the I allele. This might result in interindividual diversity in antiproteinuric responses to RAAS inhibition with either ACEIs or ARBs [17]. The impact of ACEIs on proteinuria is varied, with reductions in proteinuria ranging from 20% to 80% in a variety of renal diseases [10]. Additionally, previous study have shown that the percentage reduction in proteinuria for the DD genotype was significantly higher than for the ID and II genotypes, indicating that the ACE DD genotype is more susceptible to the antiproteinuric effect of ACEIs than the ACE II and ID genotypes [18]. Another study discovered that ACEI medication reduced mortality and renal end points (death due to renal failure, dialysis, eGFR of <15 ml/minute/1.73 m2 or more than 50% loss of eGFR) more efficiently in I allele carriers than in DD [19]. Another research found that having an ACE II genotype and a cumulative genetic risk score of <1 was linked with a greater renoprotective response to ACEIs in people with T2DM who had normoalbuminuria. However, there was no significant difference in renoprotective efficacy with 3-year ACEIs treatment depending on ACE I/D genotypes in T2DM with nephropathy [6]. While regarding the association of AGT1RA1166C and response to ACEIs, a study on diabetic patients suggests that genotypes AC + CC are associated with an increased response to captopril in people with T2DM as compared to genotype AA [20].
The significance of this study results from the fact that the prevalence of DKD in Iraq is high and most diabetic patients develop nephropathy [4], which was silent in the earlier stages. Also to our knowledge, there is no genetic study on Iraqi patients with DKD that investigates the effect of genetic polymorphism on the antiproteinuric response of ACEIs.
The objective of this study was to investigate the role of genetic variations of the ACE I\D and AGT1R A1166C gene polymorphisms in the antiproteinuric effect of ACEIs therapy T2DM patients.
MATERIALS AND METHODS
Study design and setting
This cross-sectional study was carried out on already-diagnosed T2DM patients who visited the Diabetes and Endocrinology Center at Merjan Medical City in Babylon, Iraq, from March 2022 to January 2023.
Participants
Simple convenience sampling method was used to gathered patients (because of the inclusion and exclusion criteria, especially those chosen on the basis of taking a particular treatment and avoiding another treatment, make the selection of patients difficult and time-consuming. Also, the study took several months). However, the numbers of patients in the present study was more than the number of patients in the previous similar study conducted by Felehgari et al. [21] in Iran which include 45 (both normalbuminuric and macroalbuminuric patients) who are captopril users. The present study included 76 hypertensive T2DM patients with DM duration ≥5 years taking ACEI (43 patients taking captopril dose range (25–100 mg) and 33 patients taking lisinopril dose range (5–20 mg) divided into two groups according to urinary albumin-to-creatinine ratio (ACR).
1. Group 1: includes 31 T2DM patients who are ACEI users and without DKD.
2. Group 2: includes 45 T2DM patients who are ACEI users and have DKD.
The exclusion criteria included alcoholics, patients with renal illness prior to the onset of diabetes, patients whose blood pressure is extremely high and uncontrolled (>160/100 mm Hg), and those who were treated with calcium channel blockers, in addition to those patients treated with sodium glucose co-transporter 2 inhibitors (such as cangaliflozin or dapagliflozin). Other exclusion criteria were the presence of any sign or symptom of an autoimmune illness or an inflammatory kidney condition, and non-compliance with therapy according to the eight-item Morisky Medication Adherence Scale [Scores <6, which are considered low] [22]. Finally, patients with secondary causes of albuminuria, such as obstructive renal disease, renal stone disease, and acute urinary tract infection, or patients with drug-induced nephrotoxic injury were also excluded from the present study.
A standardized protocol was used to gather medical histories and data from study participants. DKD was defined as T2DM with an ACR ≥30 mg/g. This test was performed directly on each patient by taking a random spot of urine and measuring urine creatinine using a colorimetric reaction (Jaffe reaction) using a Biolab kit [23], while urine albumin was measured using an Abnova BCG Albumin kit [24]. ACR (mg\g) is then computed in g/l; the principle of these tests are described in details [25].
Collection and analysis of blood samples
After an overnight fast, venous blood samples were taken from each patient in a sitting position using disposable syringes. After using a tourniquet, 10 ml of blood were drawn from each person, 2 ml were placed in ethylenediamine tetra acetic acid tubes, 1 ml was used directly for the detection of HbA1c by using Cobas e411 apparatus and, the other 1 ml was stored at −80°C for genetic analysis and 8 ml were slowly pushed into a gel and clot activator disposable tube without anticoagulant and left to coagulate for 10–15 minutes before being centrifuged to obtain serum for testing. Part of the obtained serum was used directly for the measurement of fasting blood sugar (FBS), creatinine, and lipid profile using a fully automated Kromo Linear apparatus by the staff in the Diabetic and Endocrinology Center. While the remaining serum was stored in two separate eppendorf tubes at −80°C until the time for measuring cystatin C (CysC), kidney injury molecule 1 (KIM1) levels, ACE1 activity, and ACE2 levels in serum by ELISA technique (BT Laboratory/China kit), which are plate-based assays for detecting and quantifying a specific protein in a complex mixture. Briefly, the plate was pre-coated with Human CST3 antibody for CysC detection, Human HAVCR1 antibody for KIM-1 detection, Human ACE antibody for ACE1 detection, and Human ACE2 antibody for ACE2 detection. The target protein present in the sample is added and binds to antibodies coated on the wells, and then biotinylated Human CST3, HAVCR1 antibody, and biotinylated Human ACE1 or ACE2 were added and bound to the target protein in the sample. Then Streptavidin-HRP was added and bound to the biotinylated antibody. After incubation, unbound Streptavidin-HRP was washed away during a washing step. Substrate solution was then added, and color developed in proportion to the amount of human protein in the sample. The reaction was terminated by the addition of an acidic stop solution and absorbance was measured at 450 nm. To define CysC, KIM-1, ACE1, or ACE2 values, a standard curve was plotted [26–29]. The eGFR was calculated using the CKD-EPI Creatinine-Cystatin Equation (2021) online calculator [30].
GENETIC ANALYSIS
The Favor Prep Blood Genomic DNA Extraction Mini Kit was used to extract genomic DNA from peripheral blood leukocytes from frozen venous blood for genetic evaluation. ACE I\D polymorphisms were detected by high-resolution melting real time (HRM-RT) [31] using a Rotor-GeneQ(Qiagen®) thermal cycler. Using three primers:
• Primer1 ACE1 CATCCTTTCTCCCATTTCTC
• Primer2 ACE2 TCGGATTACAGCCCTGATACAG
• Primer3 ACE3 ATTTCAGAGCTGGAATAAAATT
The polymerase chain reaction (PCR) was conducted in a 0.1-ml microtube with 50 μl of final volume, including 20 μl SYBR Green PCR Master Mix, 3 μl of primers, 2 μl genomic DNA, 1 μl Mgcl2, and 24 μl DNAse-free water. The cycling conditions included denaturation at 95°C for 5 minutes, 35 cycles of denaturation at 95°C for 20 seconds, and annealing and elongation at 55°C for 60 seconds. After DNA amplification, the PCR products were submitted to the dissociation curve. The samples were cooled to 60°C and gradually raised to 85°C. The intensity of fluorescence (F) was recorded during temperature rise, and the signal was plotted as a function of temperature to provide the dissociation curve for each sample. The dissociation peaks were subsequently created by charting the negative derivative of fluorescence as a function of temperature (−dF/dT vs. T). The dissociation curve was then visually inspected for genotyping, and the results of some of the current study samples are demonstrated in Figures 1 and 2.
The AGT1R A1166C polymorphism was detected by the PCR and restriction fragment length polymorphism (PCR–RFLP) methods [32]. The primers were designed using the NCBI-primer BLAST online software [33], which gives a product length of 316 bp and includes:
• Forward primer ATGAGCACGCTTTCCTACCG
• Reverse primer TTCTTCGAGCAGCCGTCATC
 | Figure 1. HRM-RT amplification and melting curve raw data. [Click here to view] |
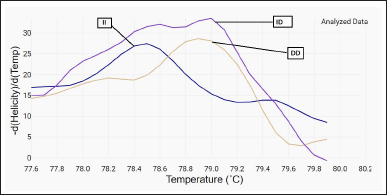 | Figure 2. HRM-RT analyzed data showed three different genotype according to their melting temperature (II, ID, DD). [Click here to view] |
The PCR amplification has been carried out in a final volume of 20 μl, including 2 μl of genomic DNA, 1 μl of each primer, 8 μl master mix, 0.5 μl MgCl2, and 7.5 μl DNAse-free water. The optimal conditions were selected, including denaturation at 94°C for 5 minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, elongation at 72°C for 30 seconds and final extension at 94°C for 5 minutes. Through 2% agarose gel electrophoresis with ethidium bromide (EtBr) staining and direct visualization in UV light, the PCR products were examined. Then PCR products were taken for restriction by an appropriate restricted enzyme, which was selected with the aid of SnapGene viewer software (V6.0.5). The restriction reaction is carried out by using a mixture of 3 μl of PCR product, 0.25 μl of the selected restriction enzyme (DdeI), restriction buffer 1.5 μl. The reaction mixture was then completed to 13 μl by 8.5 μl DNAse-free water. Vaseline oil was added to reduce evaporation since the reaction mixture was incubated overnight at 60°C in a water bath. Then, utilizing 2% agarose gel electrophoresis, the length of the PCR product, the specificity of the PCR reaction, and the restricted PCR product were examined, and the results of the gel electrophoresis image from the current study samples are shown in Figure 3.
Statistical analysis
The Statistical Package for the Social Sciences software for Windows version 26.0 (IBM Corp., Armonk, NY) was implemented for the statistical analysis. The Shapiro-Wilk test was used to test the normality of the continuous variables. For normally distributed data, the continuous variables are given as mean ± SD, and for skewed data, as median (IQR). An independent T test, or Mann-Whitney test, and a Kruskal–Wallis H test were used for analyzing the data. The categorical variables are expressed as numbers and frequencies and were analyzed using the chi-square test. Binary logistic regression was used for the prediction correlation between the ACE1 level and both genes after controlling for (age, gender, and DKD state). Statistical significance was defined as a p-value of <0.05.
RESULTS
The demographic, clinical, genetic, and laboratory data of the study groups are expressed in Table 1. Which reveals significant differences only for retinopathy, urinary albumin, urinary creatinine, and ACR. Also, significant differences were seen in serum KIM1, with higher levels in the DKD patient group compared to normalbuminuric patients group.
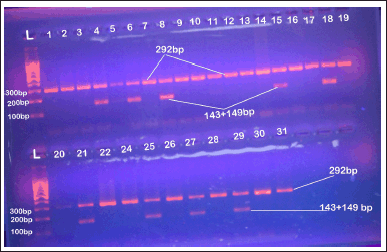 | Figure 3. Agaroes gel electrophoresis image show RFLP-PCR product analysis for AGT1RA1166C gene stained with EtBr in 2% agarose gel at 70 volt for 1 hour, 1,000 bp DNA ladder, lanes L 10 0bp step DNA ladder; lanes 4,6,8,15,18,21,25,27and 29 AC genotype; other lanes AA genotype. [Click here to view] |
Genetic analysis shows no significant differences between study groups for both genes, with a higher prevalence for DD followed by ID, and II genotypes for ACE I/D polymorphism. for the AGT1R and A1166C, the AA genotype has the highest prevalence, followed by the AC genotype, and the mutant CC isn’t seen in the present study.
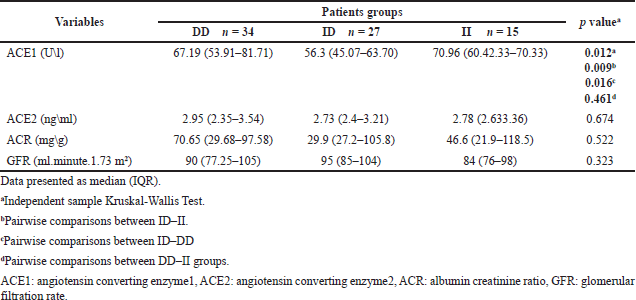
Table 2 displays the differences in biomarker levels that evaluate the antiproteinuric effect of ACEIs according to ACE I\D genotypes, which show a significant difference between groups (p = 0.012). A pairwise comparison of groups revealed that T2DM patients carrying the ID genotype have significantly lower ACE1 levels compared to DD and II carriers. ACE2 levels were higher in DD carriers, followed by II carriers, and ID carriers had the lowest ACE2. In addition, ID carriers had a lower ACR and a higher GFR compared to other genotypes, but these differences were not significant.
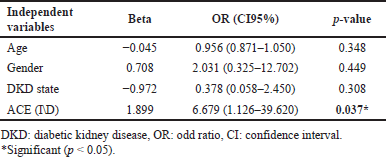
The result of binary logistic regression for S.ACE1 levels as a dependent variable after dividing in to two categories (normal level ≤40 U\l, abnormal level >40 U\l) in Table 3 revealed ACE (I\D) genotypes as a significant predictor of S.ACE1 level after controlling age, gender, and DKD state. This means that ACE (I\D) genotypes affect response to ACEIs treatment.
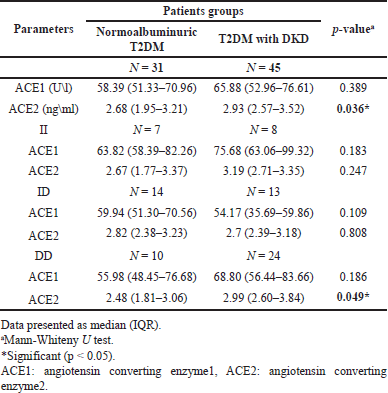
Serum ACE1 and ACE2 levels were higher in DKD patients compared to normalbuminuric patients, which is significant only for ACE2. Serum ACE1 and ACE2 were higher in nephropathic patients with II and DD genotypes and lower for carriers of ID genotype compared to normalbuminuric patients; however, No significant differences were observed for ACE1 in all genotypes, while for ACE2, the differences were only significant in carriers of DD genotype (p = 0.049) between the nephropathic and normalbuminuric groups, as demonstrated in Table 4.
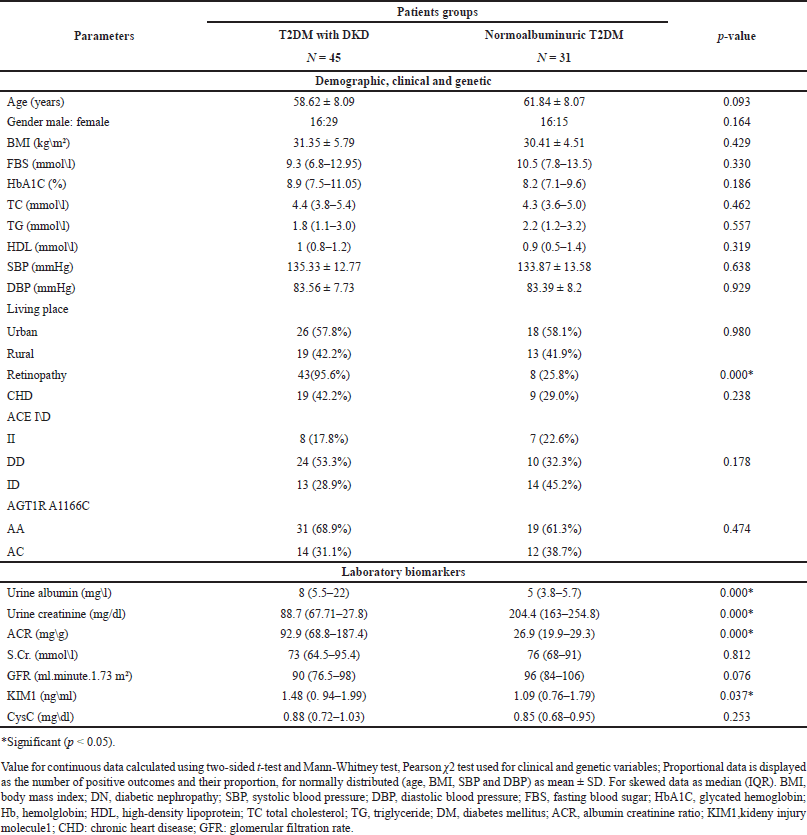 | Table 1. Demographic, clinical, genetic and laboratory data of the study groups. [Click here to view] |
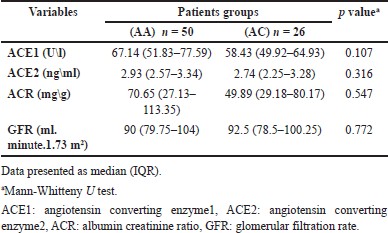
Table 5 displays the differences in biomarker levels that evaluate the antiproteinuric effect of ACEIs in AGT1R (A1166C) genotypes. There were no significant differences between the wild-type AA and heterozygote AC genotypes in all measured parameters. The AC genotype had lower levels of ACE1, ACE2, and ACR and a higher level of GFR.
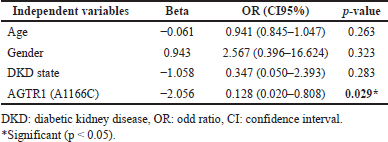
The result of binary logistic regression to determine the effect of AGTR1 (A1166C) genotypes on the prediction of S.ACE1 levels as a dependent variable in Table 6 demonstrated that after adjusting for age, gender, and DKD status, AGTR1 (A1166C) genotypes are a significant predictor of S.ACE1 level, which means that AGTR1 (A1166C) genotypes affect response to ACEIs treatment.
 | Table 2. Differences in biomarkers levels according to ACE I\D genotypes. [Click here to view] |
 | Table 3. Binary logistic regression of ACE1 in T2DM who are ACEIs user. [Click here to view] |
 | Table 4. The ACE1 activity and ACE2 concentration in various ACE genotypes in the presence of ACEI therapy between DKD and normalbuminuric patients. [Click here to view] |
 | Table 5. Differences in biomarkers levels according to AGT1R (A1166C) genotypes. [Click here to view] |
 | Table 6. Binary logistic regression of ACE1 in T2DM who are ACEIs user. [Click here to view] |
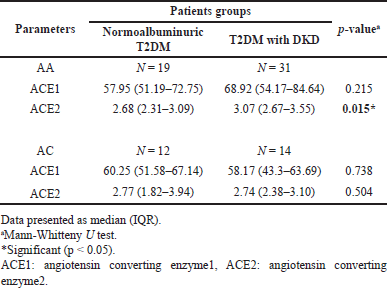
Table 7 shows that ACE1 levels were higher in DKD patients, especially for AA genotype carriers, compared to normalbuminuric patients, but still not significant. In addition, ACE2 levels were significantly higher in DKD patients who carry the AA genotype (p = 0.015), while for the AC genotype, even the levels were higher in the DKD group but did not significantly differ from normalbuminuric patients.
 | Table 7. The ACE1 activity and ACE2 concentration in various AGT1R genotypes in the presence of ACEI therapy between DKD and normalbuminuric patients. [Click here to view] |
DISCUSSION
DKD is a silent illness that goes undetected for lengthy periods of time [34]. Both albuminuria and GFR are significantly better predictors of ESRD; however, urine ACR has a bigger influence on clinical practice than GFR. As a result, the presence of albuminuria suggests glomerular and tubular damage, whereas a GFR problem only shows glomerular damage [35]. In the present study, when dividing patients according to ACR, significant differences were seen between DKD and normalbuminuric patients for retinopathy, urine albumin, urine creatinine, ACR, and KIM1, with higher levels in nephropathic patients.
The ACE I/D polymorphism is responsible for more than 40% of interindividual variability in serum or tissue ACE activity, and persons with the DD genotype have greater levels of tissue and plasma ACE than persons with the II genotype; also, its association with DKD has been extensively studied [6,10,21]. The mechanism underlying ACEIs’ antiproteinuric action is not well understood. However, it is believed that ACEIs produce efferent arteriolar vasodilation of the glomerulus and so reduce intraglomerular hypertension, resulting in an antiproteinuric effect. Also, it has been demonstrated that ACEIs improve glomerular membrane size-selective dysfunction, resulting in an anti-proteinuric effect [36]. The present study analyzed the antiproteinuric effect of ACEIs by comparing levels of both ACE1 and ACE2 and renal markers (ACR and GFR) in different genotypes. Results revealed that all diabetic patients show significant differences between II, DD, and ID genotypes for serum ACE1 activity, with a lower level seen in ID carriers. Additionally, ID carriers had non-significantly lower ACE2, ACR, and a higher GFR, suggesting that patients with the ID genotype have a better response to ACEIs. When controlling covariates (age, gender, and DKD state), the ACE I\D polymorphism is a significant predictor of ACE1 level. Then when the patients were grouped based on their ACR, significant differences were observed only in ACE2, with higher levels of both ACE1 and ACE2 in DKD groups. When further subdividing according to ACE genotypes, there were no inter-genotype differences except for ACE2 in DD carriers; additionally, ID carriers have lower levels of both markers in DKD groups compared to II and DD genotypes, which also means ID carriers have the best ACEIs antiproteinuric response. Previous follow-up studies about the association between ACE I\D polymorphism and ACEIs renoprotective effects in T2DM showed conflicting results: Ha et al. [18] found that the DD genotype had a considerably larger percentage of reductions in urine protein and albumin excretion in comparison to ID and II genotypes following ACEIs treatment for 3 months, while So et al. [6] discovered that use of RAAS inhibitors for approximately 3.7 years was related to better survival and renoprotection, with these beneficial impacts being most noticeable among the II and ID carriers. Whereas Cheema et al. [19] found that following ACEIs treatment for 36 months, II genotype had a better renoprotective response to ACE1s in normalbuminuric T2DM, while for DKD patients, no significant difference in renoprotective effect among different ACE I\D genotypes. Aggarwal et al. [36] revealed that the reduction in albuminuria in response to 6 months of ACEIs was independent of the ACE I\D polymorphism. While Felehgari et al. [21] is the only cross-sectional study on T2DM that shows patients with normalbuminuria carrying the DD genotype on captopril were more effective at lowering serum ACE activity than other genotypes, in DKD, the II genotype had the lower serum ACE activity. The differences in these results may be due to the different ethnic groups included in each study; the Felehgari et al. [21] study included only Kurd ethnicity, while the current study was performed in a region with predominantly Arab ethnicity.
The AGT1R A1166C variation has long been hypothesized to be an Ang II sensitivity modulator. This SNP, which is situated in the 3’UTR of the AGT1R gene, may have an effect on AGT1R expression through modulating microRNA binding. The miR155 selectively downregulates the A allele [37]. Therefore, the apparent increased Ang II sensitivity associated with the C allele would be the consequence of increased receptor expression caused by an abrogation of miR155 control [38]. The present study revealed that AC carriers have lower ACE1, ACE2, and ACR and higher GFR, but the differences were not significant, and after controlling age, gender, and DKD state, the result of binary logistic regression revealed that AGTR1 (A1166C) was a significant predictor of ACE1 level. Also, when subdivided according to albuminuria state, DKD patients in both AA and AC genotypes have higher ACE1 and ACE2 compared to normalbuminuric patients, and this was significant only for ACE2 in AA carriers. In addition, the AC carriers in DKD groups still have a lower level of markers that mean a better response to ACEIs compared to the AA genotype. The cause behind the better response may be related to the fact that C allele carriers are associated with an increase in ACE activity and are augmented in Ang II, which makes the response to RAAS blockers higher in C allele carriers, as seen in this study [39]. There has been no previous research on the relationship between AGT1R A1166C and response to ACEIs in DN.
The main limitations of this study are the design of the study, since prospective cohort study is better for evaluating response to treatment, and association of genetic polymorphisms with treatment response. Also, small number of patients included in the study, as well as the including two ACEIs and using different doses of each drug could result in variations in treatment response. Additionally this study mainly focuses on ACE1 and ACE2 levels and ACR as outcomes, additional parameters could be considered in forthcoming research.
CONCLUSION
This study revealed that both genes [ACE (I\D) and AGT1RA1166C] are significant predictors of ACE1 level after controlling age, gender, and DKD state. Also, heterozygote ID carriers had lower ACE1 and ACE2 levels compared to other ACE I\D genotypes even after grouping the patients according to albuminuric state, identifying a better ACEI response in ID carriers, and this was confirmed by having a lower ACR and a higher GFR. While for the AGT1R (A1166C) genotypes, AC genotypes had lower ACE1 and ACE2 levels with a lower ACR and higher GFR compared to wild-type AA, which means AC carriers had a better ACEI response. The present findings may be validated to ensure their reliability and generalizability by long-term follow-up of larger, more diverse populations on ACEI medications. Also, future research could benefit from a broader array of clinical endpoints, such as long-term renal function, to provide a more comprehensive view of the treatment response.
ACKNOWLEDGMENT
We’d like to gratitude all participants in this study.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
This study received no external financial support.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
Verbal informed consent from patients included in the study was obtained before specimens were taken. Prior to recruiting individuals for this study, ethical committee approval was gained from the Research Ethics Committee of the University of Baghdad–College of Pharmacy (approval number: RECAUBCP4102021A on 4/10/2021), and also from the medical institutions that study protocol, the subject information, and the consent form were reviewed and approved by the local ethical committee from the Diabetes and Endocrinology Center as well as from the Research Unit of Center for the Training and Human Development of the Babylon Health Directorate in Babylon province on March 8, 2022, according to document number 36.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Jasim HS, Altaie AF, Saloum WT, Ali AH. Comparative study of new biomarkers in Iraqi DM2 with and without complications. Rep Biochem Mol Biol. 2022;11(3):400–4. CrossRef
2. Akram W. Initial recognition and prophecy of diabetic nephropathy in type I diabetes in a sample of Iraqi patients. AL-Kindy Col Med J. 2012;8:94–102.
3. Hamid GS, Allawi AA, Ghudhaib KK. Correlation of pentosidine with kidney diseases in Iraqi patients with diabetic nephropathy. Iraqi J Sci. 2021;62(10):3436–42. Available from: https://ijs.uobaghdad.edu.iq/index.php/eijs/article/view/3999
4. Ali AA, Al Lami FH. Prevalence and determinants of microalbuminurea among type 2 diabetes mellitus patients, Baghdad, Iraq, 2013. Saudi J Kidney Dis Transpl. 2016;27(2):348–55. CrossRef
5. Salih BH, Ali SH, Allehibi KI. Serum aldosterone levels in patients with diabetic nephropathy in relation to vascular calcification. Iraqi J Pharm Sci. 2019;28:53–63.
6. So WY, Ma RC, Ozaki R, Tong PC, Ng MC, Ho CS, et al. Angiotensin-converting enzyme (ACE) inhibition in type 2, diabetic patients—interaction with ACE insertion/deletion polymorphism. Kidney Int. 2006;69(8):1438–43. CrossRef
7. Al-Awadi SJ, Ghareeb MA, Oleiwi AA, Salo WH, Moner AI. Genotype distribution of angiotensin i- converting enzyme in iraqi arab population. Iraqi J Cancer Med Genet. 2011;4(2):24–9.
8. Al-Radeef MY, Fawzi HA, Allawi AA. ACE gene polymorphism and its association with serum erythropoietin and hemoglobin in Iraqi hemodialysis patients. Appl Clin Genet. 2019;12:107–12. CrossRef
9. Reis AAdS, Silva EGd, Santos KdF, Anjos LRBd, Santos RdS, Freiria-Oliveira AH. Do ACE and ACE2 polymorphisms influence in the pathogenesis of diabetic nephropathy?. Res Sq. 2021. CrossRef
10. Ha SK. ACE insertion/deletion polymorphism and diabetic nephropathy: clinical implications of genetic information. J Diabetes Res. 2014;2014:846068. CrossRef
11. Huang K, Liang Y, Wang K, Ma Y, Wu J, Luo H, et al. Elevated ACE levels indicate diabetic nephropathy progression or companied retina impaired. Front Clin Diabetes Healthc. 2022;3:1. CrossRef
12. Soler MJ, Wysocki J, Batlle D. ACE2 alterations in kidney disease. Nephrol Dial Transplant. 2013;28(11):2687–97. CrossRef
13. Varagic J, Ahmad S, Nagata S, Ferrario CM. ACE2: angiotensin II/angiotensin-(1-7) balance in cardiac and renal injury. Curr Hypertens Rep. 2014;16(3):420. CrossRef
14. Zhuang Y, Niu F, Liu D, Sun J, Zhang X, Zhang J, et al. Association between AGTR1 A1166C polymorphism and the susceptibility to diabetic nephropathy: evidence from a meta-analysis. Medicine (Baltimore). 2018;97(41):e07689. CrossRef
15. Ribeiro-Oliveira A Jr, Nogueira AI, Pereira RM, Boas WW, Dos Santos RA, Simões e Silva AC. The renin-angiotensin system and diabetes: an update. Vasc Health Risk Manag. 2008;4(4):787–803.
16. Toblli JE, Bevione P, Di Gennaro F, Madalena L, Cao G, Angerosa M. Understanding the mechanisms of proteinuria: therapeutic implications. Int J Nephrol. 2012;2012:546039. CrossRef
17. Moriyama T, Kitamura H, Ochi S, Izumi M, Yokoyama K, Yamauchi A, et al. Association of angiotensin I-converting enzyme gene polymorphism with susceptibility to antiproteinuric effect of angiotensin I-converting enzyme inhibitors in patients with proteinuria. J Am Soc Nephrol. 1995;6(6):1676–8. CrossRef
18. Ha SK, Yong Lee S, Su Park H, Ho Shin J, Jung Kim S, Hun Kim D, et al. ACE DD genotype is more susceptible than ACE II and ID genotypes to the antiproteinuric effect of ACE inhibitors in patients with proteinuric non-insulin-dependent diabetes mellitus. Nephrol Dial Transplant. 2000;15(10):1617–23. CrossRef
19. Cheema BS, Kohli HS, Sharma R, Bhansali A, Khullar M. Endothelial nitric oxide synthase gene polymorphisms and renal responsiveness to RAS inhibition therapy in type 2 diabetic Asian Indians. Diabetes Res Clin Pract. 2013;99(3):335–42. CrossRef
20. Volkan-Salanci B, Dagdelen S, Alikasifoglu M, Erbas T, Hayran M, Erbas B. Impact of renin-angiotensin system polymorphisms on renal haemodynamic responsiveness to acute angiotensin-converting enzyme inhibition in type 2 diabetes mellitus. J Renin Angiotensin Aldosterone Syst. 2009;10(1):41–50. CrossRef
21. Felehgari V, Rahimi Z, Mozafari H, Vaisi-Raygani A. ACE gene polymorphism and serum ACE activity in Iranians type II diabetic patients with macroalbuminuria. Mol Cell Biochem. 2011;346(1–2):23–30. CrossRef
22. Al-Tukmagi HF, AL-Auqbi TF. Assessment of morisky medication adherence scale (8-mmas) in a sample of iraqi type 2 diabetic patients. IRAQI J Community Med. 2015;28(4):212–5.
23. Biolab. Creatinine kinetic method. Available from: https://www.biolabo.fr/pdfs/noticesE/biochimieE/AT-80107.pdf
24. Abnova. BCG albumin assay kit. Available from: https://www.abnova.com/products/products_detail.asp?catalog_id=KA1612
25. Yahya A, Kadhim D, Abdalhadi N. Kidney injury molecule-1 and cystatin C as early biomarkers for renal dysfunction in Iraqi type 2 diabetes mellitus patients. J Adv Biotechnol Exp Ther. 2023;6(3):673–85.
26. Human Cystatin C, CYS-C ELISA Kit, Jiaxing, China.
27. Human Kidney Injury Molecule 1, KIM-1 ELISA Kit, Jiaxing, China.
28. Human Angiotensin Converting Enzyme, ACE ELISA Kit, Jiaxing, China.
29. Human Angiotensin Converting Enzyme 2, ACE2 ELISA Kit, Jiaxing, China.
30. Inker LA, Eneanya ND, Coresh J, Tighiouart H, Wang D, Sang Y, et al. New creatinine-and cystatin c–based equations to estimate gfr without race. N Engl J Med. 2021;385:1737–49.
31. Alp E, Menevse S. Comparison of conventional and real time pcr methods to determine of the ace I/D and angiotensinogen m235t polymorphisms. Marmara Med J. 2009;22:27–33.
32. Halder K, Purkait P. Association of angiotensin II type I receptor (agtr1) gene polymorphism and type2 diabetes & nephropathy among the Eastern Indian Bengali patients. Diabetes Obes Int J Diabetes Obes Int J. 2020;5:1–13.
33. Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. CrossRef
34. Yamamoto Y, Hanai K, Mori T, Yokoyama Y, Yoshida N, Murata H, et al. Kidney outcomes and all-cause mortality in people with type 2 diabetes exhibiting non-albuminuric kidney insufficiency. Diabetologia. 2022;65(1):234–45. CrossRef
35. Ibrahim RK, Ghudhaib KK, Allawi AA. Determining ACE-2 level and some relevant biochemical parameters and studying the effect of gender in Iraqi diabetic patients with glomeruli and renal tubules fibrosis as early prediction marker. Baghdad Sci J. 2023;20(6):2256–64.
36. Aggarwal N, Kare PK, Varshney P, Kalra OP, Madhu SV, Banerjee BD, et al. Role of angiotensin converting enzyme and angiotensinogen gene polymorphisms in angiotensin converting enzyme inhibitor-mediated antiproteinuric action in type 2 diabetic nephropathy patients. World J Diabetes. 2017;8(3):112–9. CrossRef
37. de Denus S, Dubé MP, Fouodjio R, Huynh T, LeBlanc MH, Lepage S, et al. A prospective study of the impact of AGTR1 A1166C on the effects of candesartan in patients with heart failure. Pharmacogenomics. 2018;19(7):599–612. CrossRef
38. Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, et al. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3’ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81(2):405–13. CrossRef
39. Miller JA, Thai K, Scholey JW. Angiotensin II type 1 receptor gene polymorphism predicts response to losartan and angiotensin II. Kidney Int. 1999;56(6):2173–80. CrossRef