INTRODUCTION
Infectious diseases are major health concerns worldwide. These infections are commonly treated with a single antibiotic or a combination of some antibiotics [1]. However, improper, and excessive use of antibiotics may cause bacteria to develop various resistance mechanisms, hindering the treatment of patients with infections caused by resistant strains and possibly promoting the risk of mortality [2]. Currently, antibiotic resistance-related diseases are annually responsible for 700,000 deaths worldwide [3]. This number is predicted to increase in the coming years if more effective antibiotics are not discovered. Therefore, further research into new sources of antibiotics is required to overcome this problem.
Soil provides a favorable habitat for numerous microorganisms, including bacteria, fungi, viruses, and protozoa. Generally, 1 g of soil contains bacteria at up to 106–108 colony-forming units (CFU)/g [4]. Therefore, this environment is a potential source for isolating bacteria, particularly those capable of producing antibiotics. Notably, some new antibiotics, such as surfactin [5], bicyclomycin [6], bacitracin [7], and lysocin E [8] are reportedly produced by soil bacteria. Bacteria antibiotics have received significant attention from the scientific communities because of their structural and functional variations. Most bacterial antibiotics are synthesized through the polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) pathways. NRPS and PKS are multimodular enzymes that are commonly involved in the biosynthesis of polyketides and nonribosomal peptides. The NRPS and PKS modules comprise multiple domains that regulate the condensation and elongation of peptides or polyketide polymer chains [9]. For example, bioactive compounds, such as fengycin, iturin, and surfactin, are synthesized through the NRPS pathway, whereas macrolactin, difficidin, and bacillaene are synthesized through the PKS pathway [10]. The detection of genes involved in the production of active microbial compounds may provide valuable insights into the ability of bacteria to produce specific types of metabolites and their potential medicinal applications [11].
Muna Island is located in Central Buton Regency, Southeast Sulawesi, Indonesia. The island has a natural karst hills landscape, which is further distinguished by its dry soils and calcareous rock [12]. Such environmental characteristics allow microbial colonization that may lead to the synthesis of unique antibacterial compounds. Arid environments are the habitats for numerous novel metabolite-producing bacteria [13]. To the best of our knowledge, no research has yet been reported on the diversity and bioprospecting of bacteria in the Soil of Muna Island. Therefore, the present study aimed to analyze the antibacterial and antibiofilm activities of the soil bacteria originating from Muna Island against multidrug-resistant (MDR) strains, along with the molecular identification and detection of microbial active compound-related genes of the most promising isolate.
MATERIALS AND METHODS
Soil collection and MDR strains
A soil sample was obtained from Muna Island, Southeast Sulawesi, Indonesia (GPS location: latitude 4°54’48.8”S and longitude 122°39’56.7”E). The tested isolates of MDR strains, namely Klebsiella pneumoniae strain M19, Pseudomonas aeruginosa strain M19, Escherichia coli strain M4, methicillin-resistant Staphylococcus aureus (MRSA), and Bacillus subtilis strain M18 were acquired from Dr. Kariadi Central General Hospital (provided by Dr. Rhesi Kristiana, MERO Foundation, Indonesia).
Isolation of soil bacteria
Soil bacteria were isolated using the serial dilution method [14]. In brief, 1 g of the soil sample was placed in 9 ml of sodium chloride solution (0.85%) at 10−1 dilution and serially diluting to 10−5. Subsequently, approximately 100 μl of each suspension diluted from 10−3 to 10−5 was platted on the nutrient agar (NA) medium and incubated at ±28°C for 48 hours. Purified bacterial colonies were further used for preliminary antibacterial screening.
Antagonism assay
Primary screening for the antibacterial activity of soil bacterial isolates toward five MDR strains was performed using a dual culture assay [15]. Each target bacteria was previously cultured on nutrient broth (NB) media for 24 hours at 37°C (OD600 = 0.6) and then 1% was inoculated into molten NA and poured into sterile Petri dishes. After the medium solidified, colonies of soil bacteria were streaked around on it, and the Petri dishes were incubated at 37°C overnight. Antibacterial activity was indicated by the formation of an inhibition zone around the colonies. The diameter of the inhibition zone was measured and presented in millimeters. The test was performed in triplicate.
Metabolite extraction
Metabolites from two of the most potent soil bacteria with the broadest antibacterial spectra were extracted to obtain the crude extracts. The isolates were precultured overnight on NB media, and 1% (v/v) of the preculture was inoculated into fresh NB media. Following this, the culture was incubated and shaken at 120 rpm at 28°C overnight. Ethyl acetate was added equally to 2 l of the culture and then shaken for 1 hour at 180 rpm. Subsequently, the upper layer was then separated and evaporated at 50°C [16]. The percentage yield of the crude extract obtained was determined using the following formula:
Antibacterial test of bacterial crude extract (disc-diffusion assay)
The antibacterial activity of metabolite extracts from two potential isolates was evaluated by dripping 20 μl (80 mg/ml) of the extract dissolved in dimethyl sulfoxide (DMSO) on a sterile paper disc, then placing it on Mueller Hinton Agar (MHA) medium that had been inoculated with 1% (v/v) target strains. DMSO and tetracycline (200 μg/ml) were used as negative and positive controls, respectively. The diameters of the inhibitory zones formed after 24 hours of incubation are expressed in millimeters. The test was performed in triplicate [17].
Determination of the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values
MIC and MBC of each extract were determined using a microbroth-dilution assay [18]. The turbidity of the target suspension in the NaCl solution was adjusted to a McFarland standard of 0.5, which is equivalent to 1 × 108 CFU/ml. Subsequently, two-fold dilutions of extracts were prepared at concentrations of 0.07–10 mg/ml, then added to a microtitre plate containing Mueller Hinton broth (MHB) medium to a final volume of 200 μl/well. The plate was incubated in a shaker at 150 rpm at 37°C for 24 hours. MIC is defined as the lowest extract concentration required to inhibit bacterial growth (clear visible medium). The MBCs were determined by plating 100 μl of the treated culture on the MHA plates. The spread suspension was transferred from the MIC well to the well containing the highest concentration of extract. MBC was defined as the concentration of an extract that killed all the bacteria, as indicated by the absence of bacterial growth on the MHA plates.
Antibiofilm assessment
Briefly, extracts at concentrations of ¼ × MIC, ½ × MIC, 1 × MIC, and 2 × MIC were placed in sterile microtitre 96-well plates with brain heart infusion (BHI) medium. Furthermore, bacterial targets at 0.5 McFarland standard, which is equivalent to 1 × 108 CFU/ml, were inoculated in each well following the incubation period at 37°C with shaking at 120 rpm overnight. Thereafter, the BHI medium was discarded, and the cells were washed two times with NaCl solution (0.85%). Crystal violet (0.1%) was then added to each well followed by 30 minutes of incubation at 37°C. The crystal violet was then removed, and the stained biofilm was washed with 99% DMSO before calculating its absorbance using an enzyme-linked immunosorbent assay reader (ThermoFischer) at a wavelength of 595 nm [18].
Molecular identification of the potential isolates
The potential isolates were identified based on their 16S rRNA sequences. Bacterial genomic DNA was extracted using a DNA extraction kit (Geneproof), following the kit instructions. Amplification of the 16S rRNA sequences was performed using the 1387R (5’-GGGCGGWGTGTACAAGGC-3’) and 63F (5’-CAGGCCTAACACATGCAAGTC-3’) primers with amplified products of approximately 1,300 bp in size [19]. A total of 50 μl of polymerase chain reaction (PCR) mixture was prepared by mixing 13 μl nuclease-free water, 5 μl 1387R primer (10 μM), 5 μl 63F primer (10 μM), 2 μl genomic DNA (100 ng/μl), and 25 μl 2× GoTaq Green® Master Mix (Promega). The PCR cycling conditions were as follows: predenaturation at 94°C for 5 minutes, followed by 30 cycles of denaturation (94°C for 30 seconds), annealing (55°C for 45 seconds), extension (72°C for 1 minute 45 seconds), and final extension (72°C for 10 minutes) [20]. Electrophoresis of the PCR products was performed on a 1.5% agarose gel at a voltage of 50 V, with 1 × TAE buffer for 50 minutes. DNA was visualized using fluorosafe dyes and observed using a UV transilluminator. The PCR products were sequenced, and the chromatogram quality of the 16S rRNA sequences was analyzed using seqtrace 0.9.0 software. The sequences were aligned using the Nucleotide BLAST, available at https://blast.ncbi.nlm.nih.gov/Blast.cgi. The sequences were also deposited in the NCBI database. A phylogenetic tree was constructed using the Molecular Evolutionary Genetics Analysis software version 11.0, using the neighbor-joining tree method and 1,000 bootstrap replications.
Detection of antibiotic-biosynthetic genes
Ten genes involved in different antibiotic-biosynthetic pathways, namely NRPS, polyketide synthase type 1 (PKS 1) mlnA, dhBE, bacD, dfnD, srfA, ituA, fenA, and baeR were amplified using specific primers for PCR screening (Table 1). A total of 50 μl PCR mixture was prepared by mixing 2 μl bacterial genomic DNA (100 ng/μl), 5 μl reverse primer (10 μM), 5 μl forward primer (10 μM), 25 μl 2× GoTaq Green® Master Mix (Promega), and adjusted with 13 μl nuclease-free water. The amplified DNA fragments were visualized on a 1.5% agarose gel and observed using a UV transilluminator.
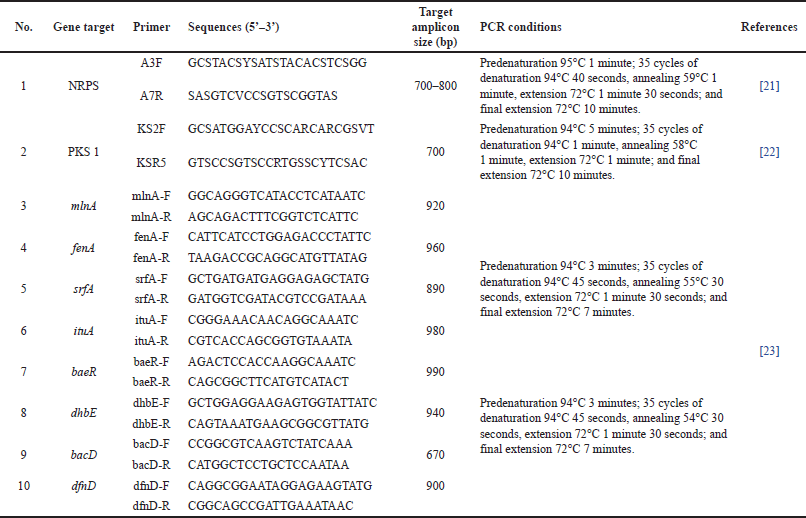 | Table 1. Specific primers, PCR conditions, and amplicon sizes used in this study. [Click here to view] |
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
The most potent extract (M7) was used for analyzing its compound profiling using LC-MS/MS analysis (Xevo G2- XS quadripole time-of-flight mass spectrometer, Waters, USA) with an electron spray interface (ESI). LC-MS/MS was performed as previously described [20]. Each peak was identified using ESI in positive ion mode. The identified mass was analyzed using the library available in the UNIFI software and the website of Natural Products Atlas (https://www.npatlas.org/).
Statistical analysis
Data obtained through the antagonism test, disc diffusion assay, MIC and MBC determination, and antibiofilm assessment, were analyzed and reflected as the average ± standard deviation from three replicates. Statistical significance was determined using one-way analysis of variance continued by Tukey’s tests. Statistical significance was set at p < 0.05.
RESULTS
Bacterial isolates from Muna Island soil
Soil collected from the Muna Islands contained bacteria at approximately 1.5 × 106 CFU/g. Growing bacterial colonies were then characterized based on their colony morphology (form, color, texture, elevation, size, and margin). Based on their distinctive colony morphologies, 15 isolates were successfully been purified. These isolates were then used for further analyses.
Antibacterial activity of soil bacteria
Fifteen bacterial isolates displayed diverse antibacterial capabilities against five MDR strains (Table 2). Five of the 15 isolates, namely M1, M7, P1, P5, and P6, exhibited antibacterial activity against at least one MDR strain, as indicated by the formation of an inhibitory zone around the endophytic colony (Fig. 1) with inhibitory zone diameters of 4.1 ± 3.5 to 9.3 ± 0.3 mm. The M7 and P1 isolates had the broadest spectra of antibacterial activity because of their ability to inhibit the growth of all MDR strains growth. Therefore, these two isolates were then selected for further analysis.
Extraction yield
The crude extracts of the M7 and P1 isolates had yields of 0.011% and 0.013%, respectively (Table 3). The crude extracts have comparable percentage yields.
Antibacterial activity of crude extract derived from the most potent isolates
At a concentration of 80 mg/ml, the crude extracts of the M7 and P1 isolates inhibited the growth of the five MDR strains. Both extracts exhibited diverse antibacterial activities against the tested strains, as indicated by the range of inhibitory zone sizes (7–8.7 mm). Tetracycline, used as a positive control, also exhibited an inhibitory zone with a diameter of 8–18 mm in all MDR strains. In contrast, DMSO, which was used as a negative control, exhibited no inhibition zones (Table 4).
MIC and MBC of M7 and P1 crude extracts against MDR strains
MIC for M7 and P1 crude extracts against all MDR strains ranged from 312.5 to >10,000 μg/ml (Table 5). Crude extracts of M7 isolate had the lowest MIC of 312.5 μg/ml against the E. coli strain M4 and MRSA, whereas crude extracts of P1 were most effective against the B. subtilis strain M18 and E. coli strain M4. Lower MIC indicates stronger antibacterial activity. In addition, the MBC of both crude extracts was higher than the MIC for all MDR strains. However, both crude extracts were less active against K. pneumoniae strain M19, as shown by the higher MICs and MBCs of all samples compared to those associated with the other strains. This may be because this strain is extremely resistant to the tested concentrations of the crude extracts tested.
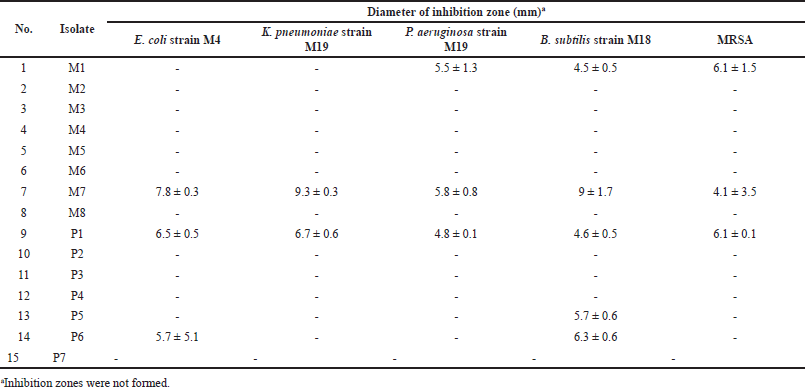 | Table 2. Antibacterial activity of soil bacteria against MDR strains tested using the dual culture assay. [Click here to view] |
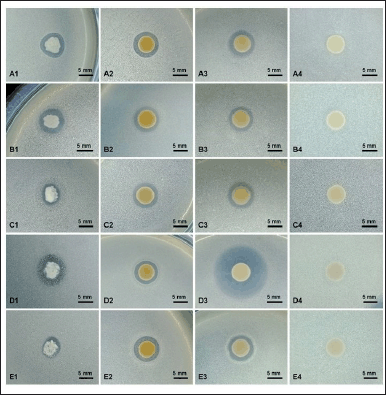 | Figure 1. Antibacterial activity of the M7 isolate against E. coli strain M4 (A), K. pneumoniae strain M19 (B), P. aeruginosa strain M19 (C), B. subtilis strain M19 (D), and MRSA (E); tested with colony (1), crude extract (80 mg/ml) (2) tetracycline (200 μg/ml) as positive control (3), and DMSO 99% as negative control (4). Scale bar, 5 mm. [Click here to view] |
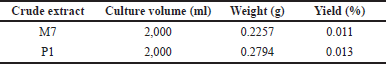 | Table 3. Yield percentages of crude extracts derived from the most potent isolates. [Click here to view] |
Antibiofilm potential
The results of the antibiofilm ability from two selected extracts against all MDR bacterial strains demonstrated that both extracts significantly (p < 0.05) reduced MDR biofilm formation in a concentration-dependent manner. The highest inhibition values were recorded for the M7 and P1 extracts at a concentration of 2 × MIC against MRSA and B. subtilis strain M18, with inhibition values of 65.1% and 67.25%, respectively. In contrast, the lowest reduction in biofilm formation was recorded for the P1 extract at a concentration of ¼ × MIC, with inhibition values of 5.18% against MRSA (Fig. 2A and B).
Molecular identity of selected isolates
The 16S rRNA analysis showed that the most potent isolates, namely P1 and M7 isolates, shared similarities (>97.5%) with Bacillus aerius strain 24K and Priestia (Bacillus) aryabhattai strain B8W22, respectively (Table 6). The 16S rRNA sequences of both isolates could be accessed on the NCBI GenBank database through accession numbers OR066161.1 and OR066162.1. The phylogenetic tree consistently shows that these isolates were closely related to each of their closest related species (Fig. 3).
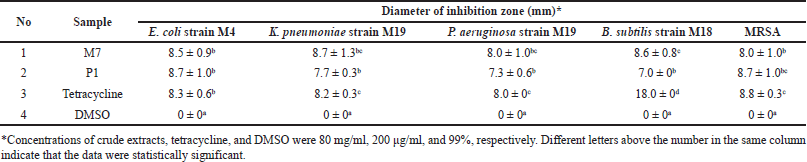 | Table 4. Antibacterial activity of crude extracts of the most potent isolates against MDR strains using the disc-diffusion method. [Click here to view] |
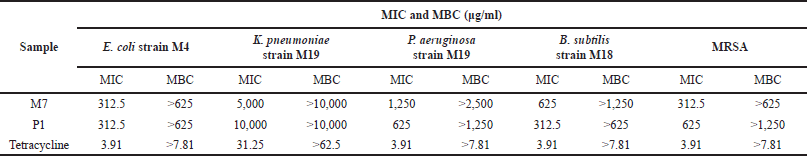 | Table 5. MICs and MBCs of M7 and P1 extracts against MDR strains. [Click here to view] |
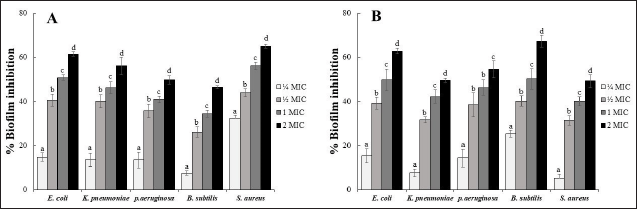 | Figure 2. Effect of selected bacterial crude extracts on the biofilm inhibition against MDR clinical isolates cells biofilm. Inhibition activity of crude extract from M7 (A), and P1 (B) isolates. [Click here to view] |
 | Table 6. Molecular identity of the selected isolates based on 16S rRNA sequences. [Click here to view] |
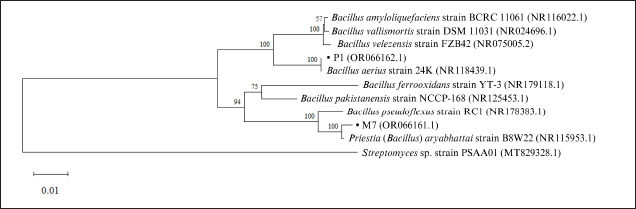 | Figure 3. Genetic relationship between M7 and P1 isolates and other Bacillus-related strains based on 16S rRNA sequences. [Click here to view] |
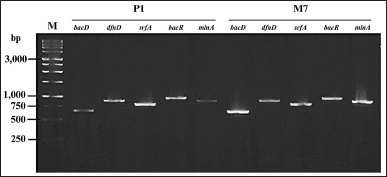 | Figure 4. Bands of secondary metabolite biosynthetic genes of P1, and M7 isolates. M represents 1 kb ladder markers on the 1.5% agarose gel. [Click here to view] |
The presence of metabolite biosynthetic genes
The M7 and P1 isolates were determined to have the mlnA (macrolactin biosynthesis), baeR (bacillaene biosynthesis), dfnD (difficidin biosynthesis), srfA (surfactin biosynthesis), and bacD (bacilysin biosynthesis) genes. Amplification of these genes revealed the formation of an electrophoretic band of approximately 920 bp for mlnA, 900 bp for dfnD, 890 bp for srfA, 990 bp for baeR, and 670 bp for bacD (Fig. 4). However, NRPS, PKS type 1, dhbE, ituA, and fenA were absent in both isolates (Table 7).
Metabolite profile of M7 extract
Because the M7 extract exhibited the most potent antibacterial and antibiofilm activities based on the initial screening, such as disc diffusion, determination of MIC and MBC, and antibiofilm assay, the M7 extract was selected for analyzing its metabolites profile using LC-MS/MS (Supplementary material 1–13). The results revealed that 13 recognized putative compounds were dominant in the extract (Fig. 5). Interestingly, six antibacterial compounds were detected in the extract, including cyclo(D-Pro-L-Tyr), marinoquinoline G, N-carbamoyl-2-hydroxy-3-methoxybenzamide, rancinamycin Ib, dipyrimicin A, and methyl 6-carbamoylphenazine-1-carboxylate (Table 8).
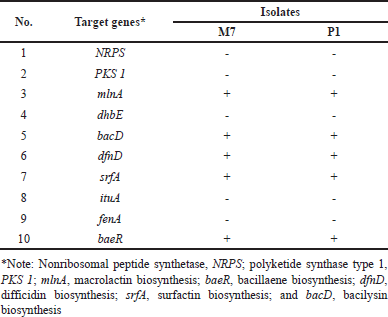 | Table 7. Presence of antibiotic-biosynthetic genes in two selected isolates. [Click here to view] |
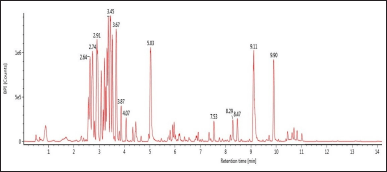 | Figure 5. LC-MS/MS chromatogram profile of M7 extract. [Click here to view] |
DISCUSSION
Soil bacteria could serve as sources of diverse antibacterial compounds that play a significant role in the development of antibiotics. In the present study, 15 soil bacterial isolates from Muna Island, Southeast Sulawesi, were screened for their antibacterial activity against five MDR strains, including MRSA. Bacillus subtilis strain M18, P. aeruginosa strain M19, E. coli strain M4, and K. pneumoniae strain M19. Five out of the 15 isolates (33%) displayed antagonistic activity to the tested MDR strains. This antagonistic potential is supported by the ability of soil bacteria to produce metabolites that can inhibit the MDR strain growth through numerous mechanisms, including the inhibition of cell wall synthesis, disruption of cell membrane integrity, inhibition of nucleic acid and protein synthesis, and inhibition of bacterial metabolic processes [35]. M7 and P1 were the two most potent isolates because they inhibited the growth of all tested MDR strains. The isolates were selected for metabolite extraction to confirm their antibacterial activity. Notably, the two isolates exhibited similar yields after extraction. Similar conditions in terms of culture time, ethyl acetate solvent-to-bacterial culture ratio, and extraction time were likely responsible for the relatively comparable yields.
Furthermore, the M7 and P1 crude extracts at a concentration of 80 mg/ml showed varied antibacterial activities against the MDR strains, as assessed using the disc-diffusion assay. The effectiveness of antibacterial agents is commonly categorized based on their inhibitory zone diameters, such as weak (<5 mm), moderate (5–10 mm), strong (10–20 mm), and very strong (>20 mm) [36]. Based on this categorization, the M7 and P1 extracts possessed moderate antibacterial activity against all MDR strains. Crude extracts of M7 isolates exhibited larger inhibition zones in P. aeruginosa strain M19, K. pneumoniae strain M19, and B. subtilis strain M18 than the inhibition zone associated with the P1 extract. The different effects of these soil bacterial extracts on the inhibition of the target bacteria may be influenced by their resistance to antimicrobial agents. MDR bacteria have developed resistance mechanisms against antimicrobial agents, such as through efflux pumps, antibiotic degradation and inactivation, drug target alteration, antibiotic target protection, and reduction in membrane permeability [37]. However, the M7 and P1 crude extracts displayed a broad antibacterial spectrum, as they inhibited the growth of Gram-negative (K. pneumoniae strain M19, P. aeruginosa strain M19, and E. coli strain M4) and Gram-positive (B. subtilis strain M18 and MRSA) bacteria.
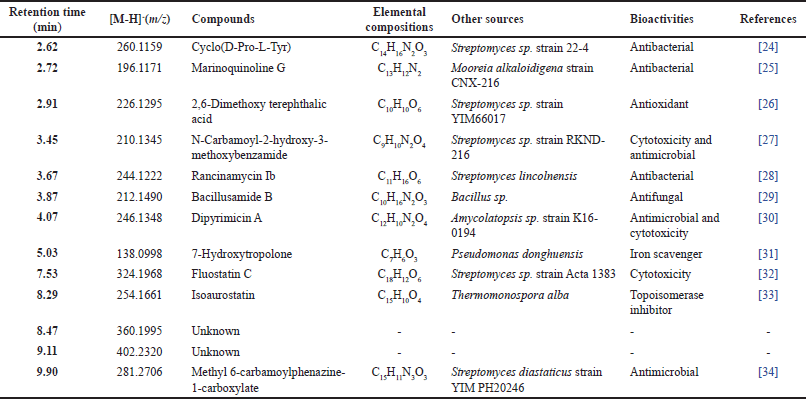 | Table 8. Metabolite profile of M7 extract. [Click here to view] |
The investigation of the antibacterial activity of the two selected isolates was then continued using determining the lowest extract concentration required to inhibit (MIC) and kill (MBC) the target bacteria as one of the pharmacological parameters of antibacterial compounds to be used for medicinal purposes. Lower concentrations of this compound are expected to reduce its toxicity in the human body. The MICs of M7 and P1 crude extracts ranged from 312.5 μg/ml to >10,000 μg/ml. The MIC obtained in this study are also higher than those reported in earlier studies. The MICs of extract from Bacillus safensis MK-12.1 were 3.12–6.25 mg/ml against antibiotic-resistant strains of Acinobacter baumanii, E. coli, P. aeruginosa, and S. aureus [38]. The MICs of the bacterial extracts used in this study are also stronger than that of Bacillus cereus extract, which ranges from 8.34 to 33.34 mg/ml against Shigella dysenteriae, Salmonella typhi, S. aureus, E. coli, and Corynebacterium diphtheriae 8.34–33.34 mg/ml [39]. However, further investigation is necessary to purify the antibacterial compounds present in the M7 and P1 extract. Furthermore, in this study the MBCs of both crude extracts were higher than the MICs, suggesting that higher concentrations were required to completely kill all the target bacteria in the medium. Another study reported that MBC may be equal to or 2–4 fold higher than the MIC [40].
We further investigated the antibiofilm activities of the two potent bacterial extracts. The biofilm structure plays a crucial role in antibiotic resistance mechanisms because it inhibits the penetration of antibiotics and protects microbial cells from the host immunity [41]. In the present study, the M7 and P1 extracts (at a concentration of 2× MIC) showed the best antibiofilm activity against biofilms formed by MRSA and B. subtilis strain M18, respectively. Notably, several antibiofilm agents have been previously reported from numerous soil Bacillus sp. strains, including Bacillus spp. against MRSA and A. baumanii biofilms [42], B. cereus against MRSA biofilm [43], and B. cereus ILBB55 against S. aureus, and P. aeruginosa biofilms clinical strains with diverse antibiofilm capacities [44]. This is likely because the sources of Bacillus spp. isolates were different, thus the active compounds produced by these bacteria exhibit various characteristics and bioactivities.
Isolates M7 and P1 isolates were identified based on the 16S rRNA gene. This gene is used for bacterial identification because it is found in all prokaryotic organisms, contains both variable and conserved regions, and evolves slowly [45]. Sequence alignment of the 16S rRNA gene showed that isolates P1 and M7 were closely related to B. aerius strain 24K and P. (Bacillus) aryabhattai strain B8W22, respectively. According to Stackebrandt and Goebel [46], two organisms with a DNA similarity of >97.5% are closely related at the species level. Interestingly, Bacillus produces various metabolites that have antibacterial effects, such as bacitracin [47], macrolactin [48], and bacilysin [49]. Thus, the metabolites produced by soil bacteria from Muna Island may be an alternative source of novel antibiotics.
The presence of antibiotic-related genes in both isolates suggests that they may have the potential to produce active molecules. Using a molecular approach, M7 and P1 isolates were shown to contain the mlnA, dfnD, baeR, srfA, and bacD genes, which are involved in the biosynthesis of macrolactin, difficidin, bacillaene, surfactin, and bacilysin, respectively. These metabolites are primarily produced by Bacillus spp., have remarkable biological activity, and act as antibacterial agents [50]. Macrolactin, bacillaene, and difficidin are types of polyketides that inhibit bacterial growth through the inhibition of protein synthesis. These compounds have been reported to show antibacterial activity against pathogens, such as S. aureus, E. coli, and P. aeruginosa [51]. Surfactin is an amphiphatic lipopeptide that exhibits an antibacterial mechanism by destabilizing and interrupting membrane integrity [52]. Surfactin from B. cerulans is active against MDR strains, including MRSA, K. pneumoniae, and E. coli [53]. Bacilysin is a peptide antibiotic that exhibits broad antibacterial properties by interfering with glucosamine synthesis [54]. However, the NRPS, PKS type 1, dhbE, ituA, and fenA genes were not detected in the M7 and P1 genomes. This suggests that these genes are not involved in the biosynthesis of antibacterial compounds by the two isolates.
To investigate the potency of the most potent extract (M7), the secondary metabolite constituents present in the corresponding extract were identified using LC-MS/MS analysis. The results indicated that the potential extract contained 13 putative compounds as identified in the library database [Fig. 5]. Interestingly, three compounds namely cyclo(D-Pro-L-Tyr), marinoquinoline G, and rancinamycin Ib have been reported to exhibit high antibacterial activity against the phytopathogenic bacteria, Pontibacillus sp. and S. aureus, respectively [24–25,28]. Furthermore, three putative compounds including N-carbamoyl-2-hydroxy-3-methoxybenzamide, dipyrimicin A, and methyl 6-carbamoylphenazine-1-carboxylate have antimicrobial and cytotoxic properties [27,30,34]. Therefore, our results confirm that the antibacterial and antibiofilm properties of the potential M7 extract are likely due to the involvement of these compounds. In addition, the M7 extract contained several compounds that exhibited some potential bioactivities, including 2,6-dimethoxy terephthalic acid (antioxidant), bacillusamide B (antifungal), 7-hydroxytropolone (iron scavenger), fluostatin C (cytotoxicity), and isoaurostatin (topoisomerase inhibitor) [26,29,31–33].
CONCLUS?ON
Five of the 15 soil bacterial isolates (33%) exhibited various antagonistic activity against the tested MDR strains. Among the 15 isolates, M7 and P1 were the most potent isolates, as indicated by their broadest antibacterial spectra against all MDR strains. The crude extracts from these two isolates inhibited all MDR strains. However, the extracts were more active against E. coli strain M4, B. subtilis strain M18, and MRSA, as indicated by their MIC values. The two selected isolates were identified as Bacilli. The presence of metabolite biosynthesis genes, including mlnA, baeR, dfnD, srfA, and bacD, further suggests the ability of the bacteria to produce antibacterial compounds. These findings demonstrate that naturally occurring Bacillus spp. has a significant potential to produce active constituents against bacteria and disrupt their biofilm structure, enabling the discovery of novel antibiotics. Further characterization and structural elucidation of the active compounds are required to expand our knowledge and enable the development of new antibiotic candidates.
ACKNOWLEDGMENTS
The current study was fully funded by the Directorate of Research and Innovation, IPB University, through the “Riset Kolaborasi Nasional (Ri-Na) 2023” that was awarded to Jepri Agung Priyanto. The authors also thank Dr. Rhesi Kristiana for providing bacterial-resistant strains and Apriza Yuswan for their assistance during this study.
AUTHOR CONTRIBUTIONS
JAP, MEP, and RIA conceived the study. ENWH, JAP, and MEP performed the laboratory experiments. ENWH, JAP, MEP, LOAFH, and JML acquired and analyzed the data. An early version of this manuscript was written by ENWH, and revised by JAP, MEP, RIA, LOAFH, and JML.
CONFLICT OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVAL
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
The 16S rRNA sequences of the potential bacteria can be downloaded from the NCBI GenBank database with the accession IDs OR066161.1, and OR066162.1. All other experimental data are included in this article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Wang N, Luo J, Deng F, Huang Y, Zhou H. Antibiotic combination therapy: a strategy to overcome bacterial resistance to aminoglycoside antibiotics. Front Pharmacol. 2022;13:1–15. CrossRef
2. Mancuso G, Midiri A, Gerace E, Biondo C. Bacterial antibiotic resistance: the most critical pathogens. Pathogens. 2021;10:1–14. CrossRef
3. World Health Organization. No time to wait: securing the future from drug-resistant infections. Geneva, Switzerland: WHO; 2019 [cited June 2023]. Available from: https://www.who.int/publications/i/item/no-time-to-wait-securing-the-future-from-drug-resistant-infections.
4. Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol. 2002;68:2391–2396. CrossRef
5. Barale SS, Ghane SG, Sonawane KD. Purification and characterization of antibacterial surfactin isoforms produced by Bacillus velezensis SK. AMB Express. 2022;12:1–20. CrossRef
6. Chekan JR, Moore BS. Biosynthesis of the antibiotic bicyclomycin in soil and pathogenic bacteria. Biochemistry. 2018;57:897–8. CrossRef
7. Amin A, Khan MA, Ehsanullah M, Haroon U, Azam SM, Hameed A. Production of peptide antibiotics by Bacillus sp. GU 057 indigenously isolated from saline soil. Braz J Microbiol 2012;43(4):1340–6. CrossRef
8. Hamamoto H, Urai M, Ishii K, Yasukawa J, Paudel A, Murai M, et al. Lysocin E is a new antibiotic that targets menaquinone in the bacterial membrane. Nat Chem Biol. 2015;11(2):127–33. CrossRef
9. Ansari MZ, Yadav G, Gokhale RS, Mohanty D. NRPS-PKS: a knowledge-based resource for analysis of NRPS-PKS megasynthases. Nucleic Acids Res. 2004;32:405–13. CrossRef
10. Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol. 2005;56:845–57. CrossRef
11. Zhang W, Zhang F, Li Z, Miao X, Meng Q, Zhang X. Investigation of bacteria with polyketide synthase genes and antimicrobial activity isolated from South China Sea sponges. J Appl Microbiol. 2009;107:567–75. CrossRef
12. Purwaningsih S, Sutisna E, Nugroho AA. Characterization, diversity, and effectiveness phosphate solubilizing bacteria from the soil and rhizosphere on the growth of Glycine max L. in greenhouse. IOP Conf Ser Earth Environ Sci. 2022;976:1–8. CrossRef
13. Mohammadipanah F, Wink J. Actinobacteria from arid and desert habitats: diversity and biological activity. Front Microbiol. 2016;6:1–10. CrossRef
14. Gislin D, Sudarsanam D, Raj GA, Baskar K. Antibacterial activity of soil bacteria isolated from Kochi, India and their molecular identification. J Genet Eng Biotechnol. 2018;16:287–94. CrossRef
15. Zhou L, Song C, Li Z, Kuipers OP. Antimicrobial activity screening of rhizosphere soil bacteria from tomato and genome-based analysis of their antimicrobial biosynthetic potential. BMC Genomics. 2021;22:1–14. CrossRef
16. Priyanto JA, Astuti RI, Nomura J, Wahyudi AT. Bioactive compounds from sponge associated bacteria: anticancer activity and NRPS-PKS gene expression in different carbon sources. American J Biochem Biotechnol. 2017;13(4):148–56. CrossRef
17. Priyanto JA, Prastya ME, Sinarawadi GS, Datu’salamah W, Avelina TY, Yanuar AIA, et al. The antibacterial and antibiofilm potential of Paederia foetida Linn. leaves extract. J Appl Pharm Sci. 2022;12:117–24. CrossRef JAPS.2022.121012
18. Clinical and Laboratory Standars Institute. Performance standards for antimicrobial susceptibility testing. 30th edition. Vol. 40, no. 1. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
19. Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64(2):795–9. CrossRef
20. Priyanto JA, Prastya ME, Astuti RI, Kristiana R. The antibacterial and antibiofilm activities of the endophytic bacteria associated with Archidendron pauciflorum against multidrug?resistant strains. Appl Biochem Biotechnol. 2023;195(11):6653–74. CrossRef
21. Ayuso-Sacido A, Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol. 2004;49:10–24. CrossRef
22. Schirmer A, Gadkari R, Reeves CD, Ibrahim F, DeLong EF, Hutchinson CR. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl Environ Microbiol. 2005;71:4840–9. CrossRef
23. Hazarika DJ, Goswami G, Gautom T, Parveen A, Das P, Barooah M, et al. Lipopeptide mediated biocontrol activity of endophytic Bacillus subtilis against fungal phytopathogens. BMC Microbiol. 2019;19:1–13. CrossRef
24. Watana-Amorn P, Charoenwongsa W, Williams C, Crump MP, Apichaisataienchote B. Antibacterial activity of cyclo(L-Pro-L-Tyr) and cyclo(D-Pro-L-Tyr) from Streptomyces sp. strain 22-4 against phytopathogenic bacteria. Nat Prod Res. 2016;30:1980–3. CrossRef
25. Choi EJ, Nam SJ, Paul L, Beatty D, Kauffman CA, Jensen PR, et al. Previously uncultured marine bacteria linked to novel alkaloid production. Chem Biol. 2015;22:1270–9. CrossRef
26. Zhou H, Yang Y, Peng T, Li W, Zhao L, Xu L, et al. Metabolites of Streptomyces sp., an endophytic actinomycete from Alpinia oxyphylla. Nat Prod Res. 2014;28:265–7. CrossRef
27. Liang L, Wang G, Haltli B, Marchbank DH, Stryhn H, Correa H, et al. Metabolomic comparison and assessment of co-cultivation and a heat-killed inducer strategy in activation of cryptic biosynthetic pathways. J Nat Prod. 2020;83:2696–705. CrossRef
28. Argoudelis AD, Pyke TR, Sprague RW. Rancinamycins, metabolites produced by Streptomyces lincolnensis in sulfur-depleted media. J Antibiot. 1976;29:777–86. CrossRef
29. Yonezawa K, Yamada K, Kouno I. New diketopiperazine derivatives isolated from sea urchin-derived Bacillus sp. Chem Pharm Bull. 2011;59:106–8. CrossRef
30. Izuta S, Kosaka S, Kawai M, Miyano R, Matsuo H, Matsumoto A, et al. Dipyrimicin A and B, microbial compounds isolated from Amycolatopsis sp. K16-0194. J Antibiot. 2018;71:535–7. CrossRef
31. Jiang Z, Chen M, Yu X, Xie Z. 7-Hydroxytropolone produced and utilized as an ironscavenger by Pseudomonas donghuensis. Biometals. 2016;29:817–26. CrossRef
32. Baur S, Niehaus J, Karagouni AD, Katsifas EA, Chalkou K, Meintanis C, et al. Fluostatins C-E, novel members of the fluostatin family produced by Streptomyces strain Acta 1383. J Antibiot. 2006;59:293–7. CrossRef
33. Suzuki K, Yahara S, Maehata K, Uyeda M. Isoaurostatin, a novel topoisomerase inhibitor produced by Thermomonospora alba. J Nat Prod. 2001;64:204–7. CrossRef
34. Hu L, Chen X, Han L, Zhao L, Miao C, Huang X, et al. Two new phenazine metabolites with antimicrobial activities from soil-derived Streptomyces species. J Antibiot. 2019;72:574–7. CrossRef
35. Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4:482–501. CrossRef
36. Ouchari L, Boukeskasse A, Bouizgarne B, Ouhdouch Y. Antimicrobial potential of actinomycetes isolated from the unexplored hot Merzouga desert and their taxonomic diversity. Biol Open. 2019;8:1–7. CrossRef
37. Varela MF, Stephen J, Lekshmi M, Ojha M, Wenzel N, Sanford LM, et al. Bacterial resistance to antimicrobial agents. Antibiotics (Basel). 2021;10(5):1–22. CrossRef
38. Iqbal S, Qasim M, Begum F, Rahman H. Isolation and molecular identification of Bacillus safensis (MK-12.1) exhibiting broad-spectrum antibacterial activity against multi-drug resistant isolates. J Bacteriol Mycol. 2021;9:87–92. CrossRef
39. Amin M, Rakhisi Z, Ahmady AZ. Isolation and identification of Bacillus species from soil and evaluation of their antibacterial properties. Avicenna J Clin Microbiol Infect. 2015;2:1–4. CrossRef
40. Maiti PK, Das S, Sahoo P, Mandal S. Streptomyces sp SM01 isolated from Indian soil produces a novel antibiotic picolinamycin effective against multi drug resistant bacterial strains. Sci Rep. 2020;10:1–12. CrossRef
41. Patel R. Biofilms, and antimicrobial resistance. Clin Orthopaedics Related Res. 2005;437:41–7. CrossRef
42. Elamary R, Salem WM. Optimizing and purifying extracellular amylase from soil bacteria to inhibit clinical biofilm-forming bacteria. PeerJ. 2020;8:1–22. CrossRef
43. Dhanam S, Arumugam T, Rajasekar S. Bioflm effects of the soil Bacillus cereus metabolites: isolation, characterization and antimicrobial activity against methicillin?resistant Staphylococcus aureus. Int J Peptide Res Ther. 2021;27:2371–9. CrossRef
44. Raffaelli S, Abreo E, Altier N, Vazquez A, Albores S. Bioprospecting the antibiofilm and antimicrobial activity of soil and insect gut bacteria. Molecules. 2022;27:1–13. CrossRef
45. Srinivasan R, Karaoz U, Volegova M, Mackichan J, Kato-Maeda M, Miller S, et al. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS One. 2015;10:1–22. CrossRef
46. Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Lancet. 1994;168:882–3. CrossRef
47. Luo C, Chen M, Luo K, Yin X, Onchari MM, Wang X, et al. Genome sequencing and genetic engineering reveal the contribution of bacitracin produced by Bacillus paralicheniformis CPL618 to anti-Staphylococcus aureus activity. Curr Microbiol. 2023;80:1–12. CrossRef
48. Romero-Tabarez M, Jansen R, Sylla M, Lünsdorf H, Häussler S, Santosa DA, et al. 7-O-malonyl macrolactin A, a new macrolactin antibiotic from Bacillus subtilis active against methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and a small-colony variant of Burkholderia cepacia. Antimicrob Agents Chemother. 2006;50:1701–9. CrossRef
49. Islam T, Rabbee MF, Choi J, Baek KH. Biosynthesis, molecular regulation, and application of bacilysin produced by Bacillus species. Metabolites. 2022;12:1–13. CrossRef
50. Stoica RM, Moscovici M, Tomolescu C, C???ric? A, B?beanu N, Popa O, et al. Antimicrobial compounds of the genus Bacillus: a review. Rom Biotechnol Lett. 2019;24:1111–9. CrossRef
51. Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol. 2019;10:1–19. CrossRef
52. Seydlová G, Svobodová J. Review of surfactin chemical properties and the potential biomedical applications. Cent Eur J Med. 2008;3:123–33. CrossRef
53. Das P, Mukherjee S, Sen R. Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. J Appl Microbiol. 2008;104:1675–84. CrossRef
54. Özcengiz G, Ö?ülür I. Biochemistry, genetics and regulation of bacilysin biosynthesis and its significance more than an antibiotic. N Biotechnol. 2015;32:612–9. CrossRef