INTRODUCTION
Tuberculosis (TB) is an airborne infectious disease that attacks the lungs. The disease is caused by the bacillus Mycobacterium tuberculosis and has become one of the top 10 leading causes of death in the world (Adigun and Singh, 2022; Bhargava and Bhargava, 2020). According to the World Health Organization, in 2019, an estimated 10 million people suffered from TB, of which 206,030 people suffered from multidrug-resistant TB (MDR-TB) and about 1.2 million of these people died (Calnan et al., 2022). In addition, the emergence of drug-resistant TB can hinder TB treatment. Resistance to TB drugs can be MDR-TB involving resistance to first-line drugs such as isoniazid, rifampicin, and pyrazinamide, and extensive drug-resistant TB involving resistance to fluoroquinolones and second-line injectable drugs. Resistance to TB drugs is caused by inappropriate and nonadherent patients in treatment (Alexander and De, 2007; Seung et al., 2015; Teixeira et al., 2020). COVID-19 is an infectious disease similar to TB, which attacks the lungs with symptoms such as cough, fever, and shortness of breath, which is currently a worldwide pandemic (Selvina, 2020). TB patients with COVID-19 suffer from severe pulmonary fibrosis (Tamuzi et al., 2020). TB drug resistance in combination with COVID-19 infections has prompted the development of new antituberculosis drugs that are less resistant, more active, and more effective.
Standard TB treatment is carried out by taking first-line drugs, namely, pyrazinamide, isoniazid, rifampicin, and ethambutol for 2 months, followed by taking isoniazid and rifampicin for 4 months (Abdel-Aziz and Abdel-Rahman, 2010; Sotgiu et al., 2015). Pyrazinamide is a drug with a strong inhibitory effect on M. tuberculosis and is an important factor in shortening TB treatment from 9–12 to 6 months. Pyrazinamide can kill the semidormant M. tuberculosis population in an acidic pH environment, which is not killed by other TB drugs (Zhang, 2004; Zhang et al., 2003). Based on these advantages, pyrazinamide analogs need to be developed. Pyrazinamide analogs substituted with 6-chloro on the pyrazine ring exhibit good activity (Dolezal et al., 2009, 2008; Servusová et al., 2012; Zitko et al., 2016). Substitution on the N atom of the pyrazinamide with ethylphenyl, cycloheptyl, octyl, adamantyl, and 4-chlorobenzyl has been accomplished by a general method involving thionyl chloride, and the analogs showed better activity against M. tuberculosis (Onajole et al., 2013; Pieroni et al., 2011; Semelkova et al., 2015; Zitko et al., 2013). Thionyl chloride is included in the list of the Chemical Weapons Convention, so it is very difficult to import in various countries, especially in Indonesia (Kuitunen et al., 2020; Wirth, 2001). Synthesis of the pyrazinamide analogs was also carried out using 1,1’-carbonyldiimidazole (CDI) reagent as a substitute for thionyl chloride. However, CDI is less reactive than thionyl chloride and is easily hydrolyzed in the presence of water (Bouz et al., 2019).
This study reports on the synthesis of pyrazinamide analogs using the Yamaguchi esterification reaction method and reports on their antituberculosis activity (Kamadatu and Santoso, 2016). It also reports on the docking studies of these compounds in the active site of M. tuberculosis protein.
MATERIALS AND METHODS
General
All chemicals used were purchased from Sigma-Aldrich, Tokyo Chemical Industry, and Merck and were used without further purification. The solvents used for elutions in column chromatography and chromatotron are distilled before use. Melting point analysis was carried out on a Fisher–Johns melting point apparatus and is uncorrected. The infrared spectrum was obtained using FT-IR Shimadzu 8400s measured in KBr solids. Jeol Resonance nuclear magnetic resonance (NMR) Spectrometer (400 MHz for 1H NMR and 100 MHz for 13C NMR) was used to obtain NMR spectra with tetrametilsilan as standard analyzed in deuterated chloroform solvent. High-resolution mass spectra (HRMS) were obtained from the Waters Q-TOF Xevo instrument.
General procedure for the synthesis of pyrazinamide analogs (5a–h)
A solution of pyrazine-2-carboxylic acid or 6-chloropyrazine-2-carboxylic acid (1 eq) in THF (200 ml) was added to a mixture of 2,4,6-trichlorobenzoyl chloride (1 eq) and triethylamine (TEA) (1 eq). The reaction mixture was stirred at room temperature for 20 minutes, followed by the addition of dimethylaminopyridine (DMAP) (1 eq) and aniline or amine (0.25 eq). The mixture was then heated at reflux at 66°C/55°C for 1 hour. After the reaction (TLC) was completed, the mixture was cooled, filtered, diluted with distilled water, and then extracted with dichloromethane (2 × 25 ml). The organic phase was washed successively with 5% hydrochloric acid (25 ml), 5% NaOH (25 ml), 5% NaHCO3 (25 ml), and distilled water (25 ml) and then dried over anhydrous magnesium sulfate. The solvent was removed under reduced pressure to give off-white solids. Compounds 5a, c, d, e were purified using chromatotron or column chromatography (ethyl acetate: n-hexane), while compound 5b was obtained in a pure state without purification.
N-(4-ethylphenyl)pyrazine-2-carboxamide (5a)
White solid; yield = 50%; mp = 133°C–134°C; IR (KBr) υmax 3,345 (N-H), 3,088 (C-H aromatic), 2,960 and 2,928 (C-H aliphatic), 1,678 (C=O), 1,522 (C=N) cm-−1. 1H NMR (400 MHz, CDCl3) δ 9.62 (bs, 1H, NH), 9.49 (d, J = 1.4 Hz, 1H, ArH), 8.77 (d, J = 2.4 Hz, 1H, ArH), 8.56 (dd, J = 2.4, 1.5 Hz, 1H, ArH), 7.65 (d, J = 8.4 Hz, 2H, ArH), 7.22 (d, J = 8.4 Hz, 2H, ArH), 2.63 (q, J = 7.6 Hz, 2H, CH2), 1.22 (t, J = 7.6 Hz, 3H, CH3).−13C NMR (100 MHz, CDCl3) δ 160.6, 147.5, 144.7, 144.6, 142.5, 141.1, 134.9, 128.6, 120.0, 28.5, 15.8. HRESIMS m/z (pos): 228.1135 C13H13N3O (calcd. 228.1137).
N-cycloheptylpyrazine-2-carboxamide (5b)
Yellow solid; yield = 86%; mp = 105°C–106°C; IR (KBr) υmax 3,358 (N-H), 3,090 (C-H aromatic), 2,926 and 2,857 (C-H aliphatic), 1,659 (C=O), 1,528 (C=N) cm−1. 1H NMR (400 MHz, CDCl3) δ 9.37 (d, J = 1.3 Hz, 1H, ArH), 8.70 (d, J = 2.5 Hz, 1H, ArH), 8.48 (dd, J = 2.4, 0.9 Hz, 1H, ArH), 7.75 (bs, 1H, NH), 4.10–4.15 (m, 1H, CH), 2.01–1.98 and 1.67–1.53 (m, 12H, CH2). 13C NMR (100 MHz, CDCl3) δ 161.7, 147.1, 144.9, 144.5, 142.5, 50.5, 35.1, 28.1, 24.1. HRESIMS m/z (pos): 220.1461 C12H17N3O (calcd. 220.1450).
N-octylpyrazine-2-carboxamide (5c)
White solid; yield = 6%; mp = 57°C–58°C; IR (KBr) υmax 3,310 (N-H), 3,055 (C-H aromatic), 2,955 and 2,918 (C-H aliphatic), 1,661 (C=O), 1,535 (C=N) cm−1. 1H NMR (400 MHz, CDCl3) δ 9.39 (d, J = 1.2 Hz, 1H, ArH), 8.72 (d, J = 2.4 Hz, 1H, ArH), 8.50 (dd, J = 2.2, 1.4 Hz, 1H, ArH), 7.80 (bs, 1H, NH), 3.46 (q, J = 6.8 Hz, 2H, CH2), 1.40–1.24 and 1.65–1.58 (m, 12H, CH2), 0.85 (t, J = 6.8 Hz, 3H, CH3). 13C NMR (100 MHz, CDCl3) δ 162.9, 147.2, 144.7, 144.5, 142.6, 39.6, 31.9, 29.8, 29.6, 29.3, 29.3, 27.1, 22.7, 14.2. HRESIMS m/z (pos): 236.1762 C13H21N3O (calcd. 236.1763).
N-(4-chlorobenzyl)pyrazine-2-carboxamide (5d)
White solid; yield = 14%; mp = 134°C–135°C; IR (KBr) υmax 3,364 (N-H), 3,102 (C-H aromatic), 2,957 and 2,922 (C-H aliphatic), 1,661 (C=O), 1,530 (C=N) cm−1. 1H NMR (400 MHz, CDCl3) δ 9.43 (d, J = 1.3 Hz, 1H, ArH), 8.75 (d, J = 2.4 Hz, 1H, ArH), 8.51 (t, J = 1.8 Hz, 1H, ArH), 8.12 (bs, 1H, NH), 7.33–7.27 (m, 4H, ArH), 4.64 (d, J = 6.2 Hz, 2H, CH2).−13C NMR (100 MHz, CDCl3) δ 163.1, 147.5, 144.6, 144.3, 142.7, 136.4, 133.6, 129.3, 129.0, 42.9. HRESIMS m/z (pos): 250.0650 C12H11N3O37Cl (calcd. 250.0556) and 248.0592 C12H11N3O35Cl (calcd. 248.0591).
N-(adamantan-1-yl)pyrazine-2-carboxamide (5e)
Yellow solid; yield = 14 %; mp = 133°C–134°C; IR (KBr) υmax 3,308 (N-H), 3,100 (C-H aromatic), 2,909 and 2,895 (C-H aliphatic), 1,662 (C=O), 1,524 (C=N) cm−1. 1H NMR (400 MHz, CDCl3) δ 9.35 (d, J = 1.1 Hz, 1H, ArH), 8.67 (d, J = 2.5 Hz, 1H, ArH), 8.46 (dd, J = 2.4, 1.5 Hz, 1H, ArH), 7.58 (bs, 1H, NH), 2.12 (s, 9H, CH and CH2), 1.70 (s, 6H, CH2). 13C NMR (100 MHz, CDCl3) δ 161.8, 146.8, 145.5, 144.1, 142.3, 52.1, 41.5, 36.4, 29.5. HRESIMS m/z (pos): 258.1604 C12H17N3O (calcd. 258.1606).
6-Chloro-N-(4-ethylphenyl)pyrazine-2-carboxamide (5f)
Brownish orange solid; yield = 75%; mp = 103°C–104°C; IR (KBr) υmax 3,370 (N-H), 3,044 (C-H aromatic), 2,959 and 2,916 (C-H aliphatic), 1,694 (C=O), 1,535 (C=N) cm-1. 1H NMR (400 MHz, CDCl3) δ 9.39 (s, 1H, ArH), 9.35 (bs, 1H, NH), 8.79 (s, 1H, ArH), 7.65 (d, J = 8.8 Hz, 2H, ArH), 7.23 (d, J = 8.8 Hz, 2H, ArH), 2.65 (q, J = 8.0 Hz, 2H, CH2), 1.24 (t, J = 8.0 Hz, 3H, CH3). 13C NMR (100 MHz, CDCl3) δ 159.3, 147.5, 144.2, 142.3, 141.5, 138.8, 134.6, 128.6, 120.2, 28.5, 15.7. HRESIMS m/z (pos): 264.0656 C13H13N3O37Cl (calcd. 264.0718) and 262.0606 C13H13N3O35Cl (calcd. 262.0747).
6-Chloro-N-cycloheptylpyrazine-2-carboxamide (5g)
White solid; yield = 71%; mp = 47°C–48°C; IR (KBr) υmax 3,273 (N-H), 3,065 (C-H aromatic), 2,920 and 2,857 (C-H aliphatic), 1,653 (C=O), 1,530 (C=N) cm−1. 1H NMR (400 MHz, CDCl3) δ 9.28 (s, 1H, ArH), 8.73 (s, 1H, ArH), 7.56 (bs, 1H, NH), 4.17–4.11 (m, 1H, CH), 2.05–2.00 (m, 2H, CH2), 1.71–1.55 (m, 10H, CH2). 13C NMR (100 MHz, CDCl3) δ 160.5, 147.5, 147.1, 144.5, 142.1, 50.9, 35.1, 28.1, 24.2. HRESIMS m/z (pos): 256.0997 C12H17N3O37Cl (calcd. 256.1031) and 254.0967 C12H17N3O35Cl (calcd. 254.1060).
6-Chloro-N-octylpyrazine-2-carboxamide (5h)
White solid; yield = 73%; mp = 48°C–49°C; IR (KBr) υmax 3,329 (N-H), 3,090 (C-H aromatic), 2,945 and 2,916 (C-H aliphatic), 1,670 (C=O), 1,530 (C=N) cm−1. 1H NMR (400 MHz, CDCl3) δ 9.28 (s, 1H, ArH), 8.73 (s, 1H, ArH), 7.62 (bs, 1H, NH), 3.46 (q, J = 6.8 Hz, 2H, CH2), 1.63 (p, J = 7.5 Hz, 2H, CH2), 1.37–1.23 (m, 10H, CH2), 0.86 (t, J = 8.0 Hz, 3H, CH3). 13C NMR (100 MHz, CDCl3) δ 161.7, 147.6, 147.2, 144.2, 142.1, 39.8, 31.9, 29.6, 29.3, 29.3, 27.0, 22.7, 14.2. HRESIMS m/z (pos): 272.1357 C13H21N3O37Cl (calcd. 272.1344) and 270.1252 C13H21N3O35Cl (calcd. 270.1373).
Antitubercular activity
The resazurin microtiter method (REMA) was used to determine the minimum concentration of the pyrazinamide analogs (5a–h) that could inhibit the growth of M. tuberculosis. Each compound (5a–h) was dissolved in 20% dimethyl sulfoxide solution in water to obtain a stock solution with a final concentration of 1,000 ppm, which was then diluted to obtain a test solution with a concentration of 25, 12.5, 6.25, 3.13, 1.56, and 0.78 g/ml. The test solutions were then placed on a 96-well microplate. As much as 100 μl of M. tuberculosis H37Rv suspension was added to each well, with several wells serving as the positive control, media control, solvent control, compound control, and comparison compound (rifampicin). The microplate was then closed and sealed with a plastic bag and incubated at 37°C for 7 days. After that, 30 μl of resazurin 0.01% solution was added to each well and incubated at 37°C for 1 day. A color change is then observed (a color change from blue to pink indicates bacterial growth). The test was carried out in triplicate for each compound and each concentration.
Molecular docking study
The crystal structure of M. tuberculosis PanD protein (MtbPanD) complexed with 6-chloro-pyrazinoic acid (NMJ) was downloaded from the Protein Data Bank (PDB ID: 6P02) (Sun et al., 2020). The receptor was prepared by removing all water molecules, adding polar hydrogens, and adding Kollman charge using the AutoDockTools package. The 2D structure of the ligands (5a–h) was drawn using MarvinSketch version 20.18. The MMFF94 (Merck Molecular Force Field) from MarvinSketch was then used to minimize the energy of each ligand (Gentile et al., 2020). The docking simulation was carried out using AutoDock 4.2.6 software (The Scripps Research Institute, La Jolla, CA, USA) (Santoso et al., 2022). The active site of the MtbPanD was determined using a grid box centered on × : 47.435, y: −44.96, and z: 0.783 with an xyz dimension size of 22 × 28 × 22 and spacing of 0.375 Å. The Lamarckian genetic algorithm was used to search for the best ligand binding pose with a GA run of 100 (Harini et al., 2021). The docking results were analyzed using BIOVIA Discovery Studio Visualizer copyright © 2019 (Dassault Systèmes Biovia Corp, VélizyVillacoublay, France) (Yadav et al., 2017).
RESULTS AND DISCUSSION
Chemistry
Eight pyrazinamide analogs (5a–h) have been successfully synthesized by the Yamaguchi esterification reaction, as shown in Table 1 (Kamadatu and Santoso, 2016; Maharani and Santoso, 2015). The reaction is initiated by the deprotonation of the pyrazine-2-carboxylic acid (1a) or 6-chloro-pyrazine-2-carboxylic acid (1b) using to form triethylammonium carboxylate salt. The carboxylate anion then reacts with the carbonyl group of 2,4,6-trichlorobenzoyl chloride (2) via a nucleophilic substitution mechanism to form an asymmetric acid anhydride (3), which then reacts with 4- DMAP via a substitution mechanism to form a DMAP-substituted acyl. The amine or aniline then reacts with the DMAP-substituted acyl via a nucleophilic substitution mechanism to give the pyrazinamide analogs (5a–h) (Dhimitruka and SantaLucia, 2006). The yield of the pyrazinamide analogs (5a–h) ranged from 6% to 86%, and the structure of the target compounds was confirmed by IR, HRMS, and NMR.
Antitubercular activity
Pyrazinamide analogs (5a–h) were tested for bacterial growth inhibition activity against M. tuberculosis strain H37Rv using the resazurin microtiter REMA test (Annuur et al., 2018). The results of the bioactivity test were expressed as minimum inhibition concentration (MIC), which was the minimum concentration of the test compound capable of inhibiting bacterial growth, as seen in Table 2. Based on the Global Program for discovering new TB drugs, active compounds must have a MIC value below 6.25 μg/ml (Boechat et al., 2011). Pyrazinamides 5d, g showed the best inhibition activity among the compounds tested with a MIC value of <6.25 μg/ml and had better activity than pyrazinamide itself (MIC = 100 μg/ml). Pyrazinamide 5f had moderate activity with a MIC value of <12.5 μg/ml, which is still better than pyrazinamide. Meanwhile, pyrazinamides 5a–c, e, h did not inhibit the growth of M. tuberculosis at each test concentration, resulting in a MIC value of >25 μg/ml.
The substitution of 4-chlorobenzyl (5d) on the amide group of pyrazinamide resulted in the strongest antituberculosis activity among analog compounds without Cl substitution on the pyrazine ring. The presence of the Cl atom at position-4 of the benzene ring and the extension of the amide bridge with –CH2- could increase the inhibition of M. tuberculosis growth, while the presence of para-ethyl without –CH2- moiety on 5a gave a lower inhibition activity. Its activity is the same as that of the alkyl-substituted compounds (5b, c, e). The presence of Cl in position-6 of the pyrazine ring increased the M. tuberculosis inhibition activity. The inhibition activity of 5f and 5g was 2-fold and 4-fold, respectively, more potent compared to that of 5a and 5b. However, the activity of 5h was still the same as that of 5c.
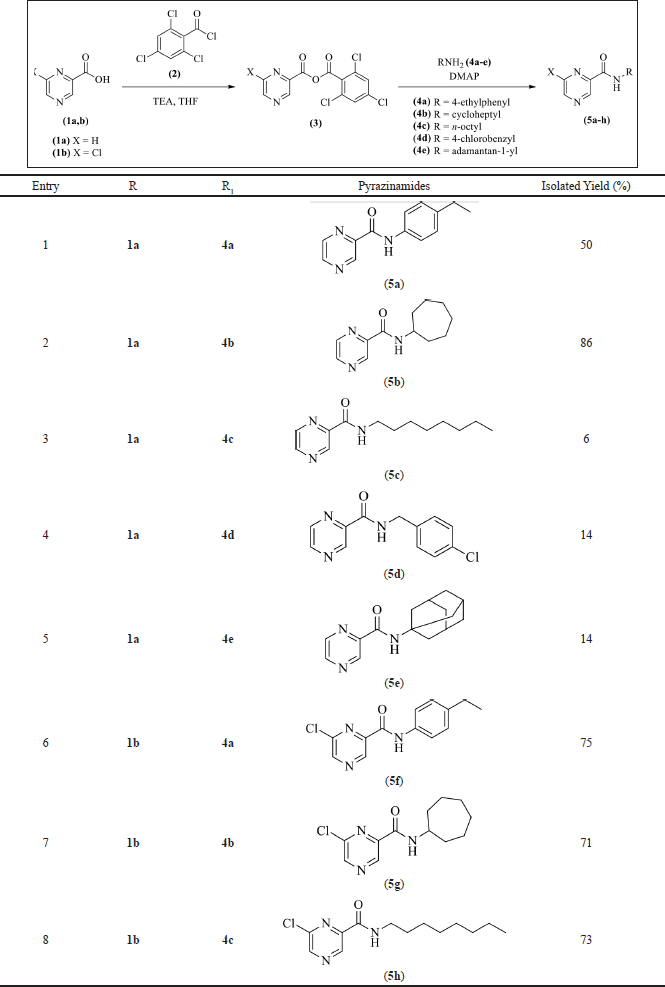 | Table 1. Synthesis of pyrazinamides 5a–h. [Click here to view] |
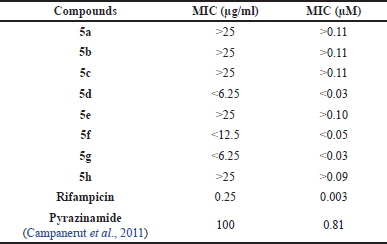 | Table 2. Test results of pyrazinamides 5a–h activity against M. tuberculosis H37Rv. [Click here to view] |
Molecular docking study
PanD is an aspartate decarboxylase that converts L-aspartate to β-alanine in the pantothenate biosynthetic pathway. β-Alanine is necessary for the formation of vitamin B5 and coenzyme A in M. tuberculosis (Sun et al., 2020). CoA plays an important role in the biosynthesis and catabolism of lipids, which supplies the bacillus with its main source of energy during infection (Evans et al., 2016). In the pyrazinamide mode of action, PanD has been suggested as one of its target proteins. Pyrazinamide is a prodrug converted to its active form, pyrazinoic acid (POA), by the enzyme PncA. POA is thought to influence PanD activity in several postulated actions (Butman et al., 2020; Gopal et al., 2020). The molecular docking study was used to investigate the atomic behavior of the pyrazinamides 5a–h at the active site of the MtbPanD located at the junction of two neighboring subunits (PDB ID: 6P02), as can be seen in Figure 1A (Hassan et al., 2020). Before docking the target compounds (5a–h) to the MtbPanD active site, the docking protocol was validated by redocking the native ligands (NMJ) at the MtbPanD active site. The redocking process was considered successful if the root-mean-square deviation (RMSD) was less than 2 Å, indicating that the protocol used can reproduce the original ligand-target interaction (Morris et al., 2009). An RMSD value of 0.43 Å was obtained from the redocking of NMJ at the MtbPanD active site at the junction of subunits 1 and 2, where the difference between crystallographic and redocked NMJ conformations is shown in Figure 1B.
Based on the results, the redocked NMJ produced a binding energy value of −5.83 kcal/mol and had almost the same interaction as the crystallographic NMJ, as shown in Figure 2. The redocked NMJ formed four hydrogen bonds with three MtbPanD key residues, namely, Ala74, Ala 75, and Arg54* (Sun et al., 2020). The N1 atom of the pyrazine ring is bound to Arg45*, the deprotonated carboxylate is bound to Ala74, and the oxygen atom of the carbonyl group (CO) group is bound to both Ala75 and Arg45*. Besides, the positive charge of the protonated amine at Arg54* attracted the negative charge of the deprotonated carboxylate at the redocked NMJ, resulting in a noncovalent interaction called an attractive charge. In addition, the 6-chlorine pyrazine ring formed several hydrophobic interactions, namely, π-σ interaction with Pyr25, alkyl interaction with Ile88*, and π-alkyl interactions with Val47*, Ile86*, and Ile88* residues. The sole difference between the crystallographic and redocked NMJ conformations was the type of contact made with the Tyr58 residue. The crystallographic NMJ interacted with Tyr58 via π-alkyl interaction, whereas redocked NMJ interacted with this residue via van der Waals interaction.
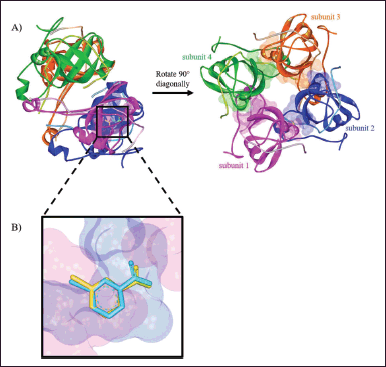 | Figure 1. (A) The active sites of MtbPanD tetramer are located at the junction of two neighboring subunits (PDB ID: 6P02), which are displayed by residue surfaces in various colors depending on the subunits. (B) The crystallographic NMJ conformation (cyan) is superimposed with the redocked NMJ conformation (yellow) in the active site of MtbPanD at the junction of subunits 1 (magenta) and 2 (blue). [Click here to view] |
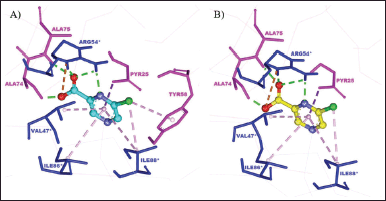 | Figure 2. The binding mode of (A) crystallographic NMJ and (B) redocked NMJ at the MtbPanD active site between subunits 1 and 2 (PDB ID: 6P02). The residues of subunits 1 and 2 are displayed in magenta and blue, respectively. [Click here to view] |
Based on the redocking results, the docking protocol can be applied to simulate the docking of pyrazinamides 5a–h to the MtbPanD active site. Table 3 shows the binding energies and interacting residues of the pyrazinamides 5a–h obtained from their docking simulation. According to the docking results, almost all the pyrazinamides 5a–h have lower binding energies than the pyrazinamide itself (−5.21 kcal/mol). except 5e with a value of -2.83 kcal/mol. The large binding energy value is possible because 5e did not interact with the MtbPanD key residue except for van der Waals interaction, which is a relatively weaker electric attraction than covalent bond and electrostatic interaction (Bronowska, 2011; Tallei et al., 2021). The two most active pyrazinamides 5d, g have lower binding energy than both pyrazinamide (-5.21 kcal/mol) and NMJ (−5.83 kcal/mol). As seen in Figure 3, due to the presence of the amide bridge in the structure of 5d, g, the aromatic cores of 5d, g entered different spaces from the pyrazine core of NMJ at the MtbPanD active site. The amide bridge of 5d, g was in the same space as the pyrazine core of NMJ, whereas the pyrazine core of these two compounds was in the space following the tunnel where the Cl atom of NMJ is located. The 4-chlorobenzyl and cycloheptyl moieties of 5d and 5g, respectively, entered the cavity adjacent to where the carboxylate group of NMJ is located.
The oxygen atom of the CO in 5d formed two hydrogen bonds with key residue Arg54* (Fig. 4A). Both pyrazine core and 4-chlorobenzyl moiety of 5d are bound to the hydrophobic cavity residues. The pyrazine core formed π-σ and π-alkyl interactions with Ile88* and Val47*, respectively. The benzene core of 4-chlorobenzyl moiety formed π-π T-shaped interaction with His11* and π-σ interactions with Ile86* and Pyr25 residues, whereas its Cl atom is bound to Ile60 via alkyl interaction along with Tyr22 and Tyr58 via π-alkyl interactions. In addition, both pyrazine and benzene cores formed π-cation interaction with Arg54* and Lys9*, respectively. The remaining interactions were with Ala74, Ala75, Gly73, Asn72, Val56, Tyr90*, and Thr45* residues.
In contrast to 5d (Fig. 4B), 5g formed hydrogen bonds with Tyr90* and Thr57 residues rather than key residue Arg54*. It only interacted with the key residue Arg54* through π-cation interaction. The Cl atom and the pyrazine core of 5g interacted with Val47* and Ile88* residues via both π-σ and alkyl interactions. In addition, the Cl atom interacted with Thr45* via a halogen interaction, which is a C-X interaction with structural properties akin to a weak hydrogen bond (Balasubramaniyan et al., 2018). Meanwhile, the cycloheptyl moiety of 5g only formed an alkyl interaction with Ile86. The van der Waals interactions were observed between 5g Tyr58, Pyr25, Gly73, Asn72, Val56, Ile60, Tyr22, Lys9*, His11*, and Ile46* residues.
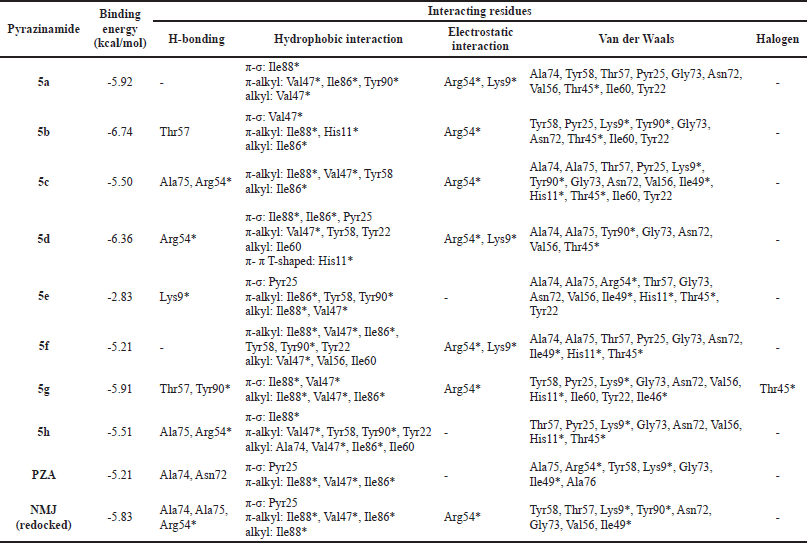 | Table 3. The binding energies and interacting residues of the pyrazinamides 5a–h. [Click here to view] |
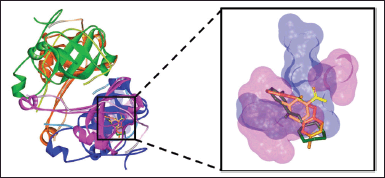 | Figure 3. The binding conformations of 5d (orange), 5g (dark green), and NMJ (yellow) in the active site of MtbPanD at the interface of subunits 1 (magenta) and 2 (blue) (PDB ID: 6P02). [Click here to view] |
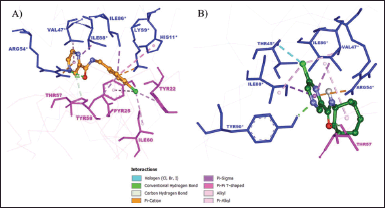 | Figure 4. The binding mode of (A) 5d and (B) 5g at the active site of MtbPanD between subunits 1 and 2 (PDB ID: 6P02). Subunit 1 residues are displayed in magenta, while subunit 2 residues are shown in blue. [Click here to view] |
CONCLUSION
Eight pyrazinamide analogs 5a–h have been successfully synthesized by the Yamaguchi esterification method in up to 86% yield. Pyrazinamides 5d, g exhibited the best antitubercular activity with a MIC value of <6.25 μg/ml. The molecular docking experiments using pyrazinamides 5a–h at the active site of the MtbPanD protein (PDB ID: 6P02), POA (PZA-activated form) targeted protein, revealed the probable binding interactions at the MtbPanD active site. The docking results showed that 5d and 5g had lower binding energy than the 6P02 native ligand and pyrazinamide. These compounds are regarded as promising candidates for further optimization as antituberculosis drugs.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
ACKNOWLEDGEMENTS
The authors would like to thank the Institut Teknologi Sepuluh Nopember for the WCP-like fund.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Abdel-Aziz M, Abdel-Rahman HM. Synthesis and anti-mycobacterial evaluation of some pyrazine-2-carboxylic acid hydrazide derivatives. Eur J Med Chem, 2010; 45(8):3384–8.
Adigun R, Singh R. Tuberculosis. StatPearls, StatPearls Publishing, Treasure Island, FL, 2022..
Alexander PE, De P. The emergence of extensively drug-resistant tuberculosis (TB): TB/HIV coinfection, multidrug-resistant TB and the resulting public health threat from extensively drug-resistant TB, globally and in Canada. Can J Infect Dis Med Microbiol, 2007; 18(5):289–91.
Annuur RM, Titisari DA, Dinarlita RR, Fadlan A, Ersam T, Nuryastuti T, Santoso M. Synthesis and anti-tuberculosis activity of trisindolines. AIP Conf Proc, 2018; 2049(1):020088.
Balasubramaniyan S, Irfan N, Umamaheswari A, Puratchikody A. Design and virtual screening of novel fluoroquinolone analogs as effective mutant DNA GyrA inhibitors against urinary tract infection-causing fluoroquinolone resistant Escherichia coli. RSC Advances 2018; 8(42):23629–47.
Bhargava A, Bhargava M. Tuberculosis deaths are predictable and preventable: comprehensive assessment and clinical care is the key. J Clin Tuberc Other Mycobact Dis, 2020; 19:100155.
Boechat N, Ferreira VF, Ferreira SB, de Lourdes G Ferreira M, de C da Silva F, Bastos MM, Dos S Costa M, Lourenço MCS, Pinto AC, Krettli AU, Aguiar AC, Teixeira BM, da Silva NV, Martins PRC, Bezerra FAFM, Camilo ALS, da Silva GP, Costa CCP. Novel 1,2,3-triazole derivatives for use against Mycobacterium tuberculosis H37Rv (ATCC 27294) strain. J Med Chem, 2011; 54(17):5988–99
Bouz G, Semelková L, Jan?ourek O, Kone?ná K, Paterová P, Navrátilová L, Kubí?ek V, Kuneš J, Doležal M, Zitko J. Derivatives of 3-aminopyrazine-2-carboxamides: synthesis, antimicrobial evaluation, and in vitro cytotoxicity. Molecules, 2019; 24(7):1212.
Butman HS, Kotzé TJ, Dowd CS, Strauss E. Vitamin in the crosshairs: targeting pantothenate and coenzyme a biosynthesis for new antituberculosis agents. Front Cell Infect Microbiol, 2020; 10:605662.
Calnan M, Moran A, Jassim AlMossawi H. Maintaining essential tuberculosis services during the COVID-19 pandemic, Philippines. Bull World Health Organ, 2022; 100(2):127–34.
Dhimitruka I, SantaLucia J. Investigation of the yamaguchi esterification mechanism. Synthesis of a Lux-S enzyme inhibitor using an improved esterification method. Org Lett, 2006; 8(1):47–50.
Campanerut PAZ, Ghiraldi LD, Spositto FLE, Sato DN, Leite CQF, Hirata MH, Hirata RDC, Cardoso RF. Rapid detection of resistance to pyrazinamide in Mycobacterium tuberculosis using the resazurin microtitre assay. J Antimicrob Chemother, 2011; 66(5):1044–6.
Doležal M, Cmedlova P, Palek L, Vinsova J, Kuneš J, Buchta V, Jampilek J, Krá?ová K. Synthesis and antimycobacterial evaluation of substituted pyrazinecarboxamides. Eur J Med Chem, 2008; 43(5):1105–13.
Doležal M, Zitko J, Kesetovicová D, Kuneš J, Svobodová M. Substituted N-phenylpyrazine-2-carboxamides: Synthesis and antimycobacterial evaluation. Molecules, 2009; 14(10):4180–9.
Evans JC, Trujillo C, Wang Z, Eoh H, Ehrt S, Schnappinger D, Boshoff HIM, Rhee KY, Barry CE, Mizrahi V. Validation of CoaBC as a bactericidal target in the coenzyme a pathway of Mycobacterium tuberculosis. ACS Infect Dis, 2016; 2(12):958–968.
Gentile D, Patamia V, Scala A, Sciortino MT, Piperno A, Rescifina A. Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar Drugs, 2020; 18(4):225.
Gopal P, Sarathy JP, Yee M, Ragunathan P, Shin J, Bhushan S, Zhu J, Akopian T, Kandror O, Lim TK, Gengenbacher M, Lin Q, Rubin EJ, Grüber G, Dick T. Pyrazinamide triggers degradation of its target aspartate decarboxylase. Nat Commun, 2020; 11(1):1661.
Harini K, Jayashree S, Tiwari V, Vishwanath S, Sowdhamini R. Ligand docking methods to recognize allosteric inhibitors for G-protein-coupled receptors. Bioinform Biol Insights, 2021; 15:117793222110377.
Hassan NW, Saudi MN, Abdel-Ghany YS, Ismail A, Elzahhar PA, Sriram D, Nassra R, Abdel-Aziz MM, El-Hwash SA. Novel pyrazine based anti-tubercular agents: design, synthesis, biological evaluation and in silico studies. Bioorg Chem, 2020; 96:103610.
Bronowska AK. Thermodynamics of ligand-protein interactions: Implications for molecular design. In: Moreno Pirajn JC, (ed.). Thermodynamics - interaction studies - Solids, Liquids and Gases InTech, London, pp 1–48, 2011.
Kamadatu L, Santoso M. Synthesis and cytotoxicity of 4-allyl-2 methoxyphenol derivatives. IPTEK J Proc Ser, 2016; 2(1):193–4.
Kuitunen M-L, Cecilia Altamirano J, Siegenthaler P, Hannele Taure T, Antero Häkkinen V, Sinikka Vanninen P. Derivatization and rapid GC-MS screening of chlorides relevant to the chemical weapons convention in organic liquid samples. Anal Methods, 2020; 12(19):2527–35.
Maharani N, Santoso M. Synthesis of 2-phenylethyl esters by one-pot the yamaguchi esterification. Adv Environ Biol, 2015; 9(23):214–8.
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem, 2009; 30(16):2785–91.
Onajole OK, Pieroni M, Tipparaju SK, Lun S, Stec J, Chen G, Gunosewoyo H, Guo H, Ammerman NC, Bishai WRR, Kozikowski AP. Preliminary structure-activity relationships and biological evaluation of novel antitubercular indolecarboxamide derivatives against drug-susceptible and drug-resistant Mycobacterium tuberculosis strains. J Med Chem, 2013; 56(10):4093–103.
Pieroni M, Tipparaju SK, Lun S, Song Y, Sturm AW, Bishai WR, Kozikowski AP. Pyrido[1,2-a]benzimidazole-based agents active against tuberculosis (TB), multidrug-resistant (MDR) TB and extensively drug-resistant (XDR) TB. ChemMedChem, 2011; 6(2):334–42.
Santoso M, Ong LL, Aijijiyah NP, Wati FA, Azminah A, Annuur RM, Fadlan A, Judeh ZMA. Synthesis, α-glucosidase inhibition, α-amylase inhibition, and molecular docking studies of 3,3-di(indolyl)indolin-2-ones. Heliyon, 2022; 8:e09045.
Selvina. New paradigm of COVID-19 with pulmonary tuberculosis: a brief review. Eur J Med Health Sci, 2020; 2(6).
Semelkova L, Konecna K, Paterova P, Kubí?ek V, Kunes J, Novakova L, Marek J, Naesens L, Peško M, Krá?ová K, Doležal M, Zitko J. Synthesis and biological evaluation of N-alkyl-3-(alkylamino)-pyrazine-2-carboxamides. Molecules, 2015; 20(5):8687–711.
Servusová B, Eibinová D, Doležal M, Kubí?ek V, Paterová P, Peško M, Krá?ová K. Substituted N-benzylpyrazine-2-carboxamides: Synthesis and biological evaluation. Molecules, 2012; 17(11):13183–98.
Seung KJ, Keshavjee S, Rich ML. Multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis. Cold Spring Harb Perspect Med, 2015; 5(9):a017863.
Sotgiu G, Centis R, D’ambrosio L, Migliori GB. Tuberculosis treatment and drug regimens. Cold Spring Harb Perspect Med, 2015; 5(5):a017822.
Sun Q, Li X, Perez LM, Shi W, Zhang Y, Sacchettini JC. The molecular basis of pyrazinamide activity on Mycobacterium tuberculosis PanD. Nat Commun, 2020; 11(1):339.
Tallei TE, Tumilaar SG, Lombogia LT, Adam AA, Sakib SA, Emran TB, Idroes R. Potential of betacyanin as inhibitor of SARS-CoV-2 revealed by molecular docking study. IOP Conf Ser: Earth Environ Sci, 2021; 711(1):012028.
Tamuzi JL, Ayele BT, Shumba CS, Adetokunboh OO, Uwimana-Nicol J, Haile ZT, Inugu J, Nyasulu PS. Implications of COVID-19 in high burden countries for HIV/TB: a systematic review of evidence. BMC Infect Dis, 2020; 20(1):744.
Teixeira C, Ventura C, Gomes JRB, Gomes P, Martins F. Cinnamic derivatives as antitubercular agents: Characterization by quantitative structure–activity relationship studies. Molecules, 2020; 25(3):456.
Wirth DD. Thionyl chloride. In: Encyclopedia of reagents for organic synthesis. John Wiley & Sons Ltd, New York, NY, 2001.
Yadav S, Pandey SK, Singh VK, Goel Y, Kumar A, Singh SM. Molecular docking studies of 3-bromopyruvate and its derivatives to metabolic regulatory enzymes: implication in designing of novel anticancer therapeutic strategies. PLoS One, 2017; 12(5):e0176403.
Zhang Y. Persistent and dormant tubercle bacilli and latent tuberculosis. Front Biosci, 2004; 9:1136–56.
Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother, 2003; 52(5):790–5.
Zitko J, Barbora S-V, Paterová P, Navrátilová L, Trejtnar F, Kuneš J, Doležal M. Design, synthesis and anti-mycobacterial evaluation of some new N-phenylpyrazine-2-carboxamides. Chem Pap, 2016; 70(5):649–57.
Zitko J, Servusová B, Paterová P, Mandíková J, Kubí?ek V, Ku?era R, Hrabcová V, Kuneš J, Soukup O, Doležal M. Synthesis, antimycobacterial activity and in vitro cytotoxicity of 5-chloro-N-phenylpyrazine-2-carboxamides. Molecules, 2013; 18(12):14807–25.