INTRODUCTION
Enterovirus A71 (EV-A71) is a member of the Picornaviridae family and is one of the main human pathogens causing hand, foot, and mouth disease. It is highly infectious, mainly affecting young children, and can be fatal in patients aged 36–60 months (Ooi et al., 2010). The virus spreads between people via direct contact with nose and throat discharges and blister fluid, and it can also be transmitted through an orofecal route (Tang and Holmes, 2017). In Thailand, the EV-A71 B5 subgenotype (EV-A71B5) was identified as the cause of the large-scale hand, foot, and mouth disease epidemic that occurred in 2012. There are no existing vaccines to protect Thai children against EV-A71B5. In mainland China, three inactivated vaccines that prevent the EV-A71 C4 subgenotype were submitted to the China Food and Drug Administration between 2015 and 2016 for approval, which have since been approved for commercial use (Mao et al., 2016; Zhou et al., 2016). However, inactivated vaccines have some disadvantages, such as unfavorable immunogenicity effects due to the inactivation process using chemical or physical methods (DeZure et al., 2016) and potentially unwanted adverse effects (Lin et al., 2015a). Several EV-A71 vaccines without the risk of virulence reversion to their pathogenic forms as well as more effective vaccines, such as virus-like particle (VLP)-based (Kim et al., 2019), recombinant protein (Han et al., 2014), and subunit protein vaccines (Cao et al., 2013), are currently being developed.
EV-A71 is a nonenveloped, single-stranded, positive RNA virus with a particle diameter of approximately 25–40 nm (Wang et al., 2016; Xue et al., 2017). The single open reading frame of EV-A71 produces three polyproteins: P1, P2, and P3. During viral replication, the P1 structural polyprotein is divided into VP0, VP1, and VP3. VP0 is then further autocatalyzed into VP2 and VP4 during virus maturation (Wang et al., 2012). P2 encodes nonstructural proteins 2A, 2B, and 2C, whereas P3 encodes nonstructural proteins 3A, 3B, 3C, and 3D (Solomon et al., 2010). Since the P1 structural polyprotein (VP1–VP4) conserves the region of several immunodominant epitopes (Foo et al., 2008; Kiener et al., 2012, 2014; Zhao et al., 2013), comprises major targets for EV-A71-specific immunoglobulin (Ig) M and IgG antibodies (Aw-Yong et al., 2016), and has the potency to stimulate robust immunogenicity and heterotypic immunity against different the subgenotypes of EV-A71, it can serve as a candidate for EV-A71 vaccine development (Han et al., 2014). 3Dp°l (RNA-dependent RNA polymerase) consists of RdRp-specific T-cells responses to protect against EV-71 infection (Dang et al., 2014) and improves the safety and stability of live-attenuated vaccines (Li et al., 2021). No evidence was available regarding the expression of P1 and 3Dpol to develop an EV-A71 vaccine.
The baculovirus expression system (BES) has been broadly utilized for vaccine production. Previous studies have reported that the structural proteins of EV-A71 can self-assemble into VLPs in BES via the expression of recombinant P1 and 3CD proteins under the regulation of polyhedron and various exogenous promoters (Kim et al., 2019; Lin et al., 2015a). The polyhedron promoter (pPH) is one of the strongest known promoters (Cox, 2012), and GP41 was designed to regulate 3CD protease proteins, which can enhance the processing of P1 proteins into mature VLPs and convert these into self-assembling VLPs that produce higher VLP yield (Kim et al., 2019). Further, the VLPs of the various subgenotypes of EV-A71 have been produced in insect cells, namely, Sf9, Hi5, and Sf21 (Kim et al., 2019; Lin et al., 2015a; Zhao et al., 2017), using different protein purification processes, such as ultracentrifugation and multistep chromatography (Zhao et al., 2017). Similarly, using 6 × His-tag (His6) affinity chromatography and tobacco etch virus (TEV) protease for fusing two proteins of interest can be an alternative yet a simple and common method for purifying recombinant fusion proteins (Goh et al., 2017).
Our purpose was to prepare a novel recombinant protein P1-3Dp°l of EV-A71B5 using BES under a dual promoter system (pPH and GP41) and study its production and expression in Spodoptera frugiperda (Sf9) cells. Finally, we evaluated protein purification using gravity affinity chromatography, and the particle size was determined by a transmission electron microscope (TEM). Using concatenation of Bagg and Albino (BALB/c) mice, we also determined immunogenicity and neutralization activity against EV-A71 of the B5 subgenotype commonly found in Thailand.
MATERIALS AND METHODS
Cell media and reagents
Fetal bovine serum (FBS), pFastBac1 transfer vector, Sf-900 II SFM, Cellfectin® II reagent, and Roswell Park Memorial Institute (RPMI) 1640 Medium were obtained from Invitrogen, Waltham, MA. TEV protease enzyme was obtained from Applied Biological Materials, Richmond, Canada. Phenylmethylsulfonyl fluoride (PMSF) and Ni-NTA agarose resin were obtained from Gold Biotechnology, St. Louis, MO, and western blotting luminal reagent was obtained from Santa Cruz Biotech, Heidelberg, Germany.
Gene design, gene optimization, and recombinant bacmid P1-3Dp°l construction
The His6 and the TEV protease cleavage site were fused to P1 polyprotein under the control of pPH and contained a poly(A) tail. The 3D polymerase gene was regulated by an additional GP41 promoter and contained a poly(A) tail. The coding sequence of the fusion gene was optimized for S. frugiperda MNPV codon usage and synthesized by the Beijing Genomics Institute (Beijing, China). The gene sequences for P1 and 3Dp°l of the EV-A71B5 were submitted to GenBank (accession no. MT944986). We cloned five His6 genes, the TEV protease cleavage site, P1, GP41, and 3Dp°l into a pUC57 vector to construct pUC57-His6-TEV-P1-GP41-3Dp°l, which was then transformed in Escherichia coli DH5α. The pUC57-His6-TEV-P1-GP41-3Dp°l plasmid was extracted and digested using EcoRI and HindIII to release the His6-TEV-P1-GP41-3Dp°l fragment. The His6-TEV-P1-GP41-3Dp°l fragment was cloned into a pFastBac1 vector under the regulation of a pPH and transformed in E. coli DH5α to create the pFBac-His6-TEV-P1-GP41-3Dp°l expression cassette (Fig. 1). To confirm correct gene insertion, we used restriction enzyme for digestion and subsequently transformed the inserted gene into E.coli DH10Bac to generate the recombinant bacmid P1-3Dp°l. The positive clones, which produced large white colonies as opposed to blue colonies (nonrecombinant bacmid DNA) were selected and confirmed by polymerase chain reaction (PCR). We identified the recombinant bacmid P1-3Dp°l via PCR using a TaKaRa Ex TaqTM kit (Takara, Japan) with M13 forward (M13F; 5′-GTTTTCCCAGTCACGAC-3′) and M13 reverse (M13R; 5′-CAGGAAACAGCTATGAC-3′) and specific primers P1_3DF (5′-GAATTCATGAGCTACTATCACCACCATCAC-3′)and P1_3DR (5′-AAGCTTTTAGAAGA GCTCGAGCCAGTTG-3′) to recognize His6-TEV-P1-GP41-3Dp°l. The parameters for PCR with M13F and M13R were as per a previously reported method (Issaro et al., 2019). PCR with other gene-specific primers was performed in the following condition: initial denaturation at 94°C for 10 minutes; 25 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C–62°C for 30 seconds, and extension at 72°C for 1 minute 30 seconds; final extension at 72°C for 20 minutes.
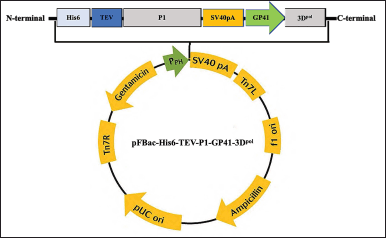 | Figure 1. Schematic representation of pFBac-His6-TEV-P1-GP41-3Dpol construction (modified picture from Invitrogen, USA). [Click here to view] |
Expression of the recombinant bacmid P1-3Dp°l in Sf9 cells and protein purification using Ni-NTA agarose resin
The recombinant bacmid P1-3Dp°l was transfected into Sf9 cells to generate recombinant baculoviruses and create viral stocks. Each viral stock was amplified and examined by an end-point dilution method (O’Reilly et al., 1992). To optimize the conditions for P1-3Dp°l expression, we investigated the multiplicity of infection (MOI), cell density, and harvest time in 125 ml shaker flasks containing 22.5 ml Sf-900 II SFM (Invitrogen, USA) at 27.3°C ± 0.5°C with shaking at 105.0 ± 0.5 rpm. We evaluated harvest time at an MOI of 10 and cell density of 1.5 × 106 cells/ml, after which cells were collected at 24, 48, 72, 96, and 120 hours after infection. The protein samples were harvested from each supernatant and cell lysate layer and were then separated by 12% (v/v) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and characterized by western blotting. The P1-3Dp°l protein was produced in Sf9 cells at the highest level and purified under native conditions using gravity affinity chromatography (Ni-NTA agarose).
We used a slightly modified version of the protein purification procedure reported by Issaro et al. (2019) and Shah et al. (2013). First, we harvested and separated the cell culture supernatant from the Sf9 cell pellet by centrifugation. The cell culture supernatant was then filtered through a 0.22 μm filter followed by buffer exchange using the binding buffer (50 mM Tris-Base, 100 mM NaCl, 10 mM imidazole, 10% glycerol, and 1 mM PMSF, pH 8.0) in ultrafiltration (Merck Millipore, USA). Next, the Sf9 cell pellet was collected and equilibrated in the binding buffer on ice for 30 minutes, following which the cells were freeze-thawed and homogenized on ice three times (1–2 minutes each time). The mixture was then incubated on ice for 20 minutes before centrifugation at 15,000 rpm at 4°C for 30 minutes to harvest the clear supernatant. The clear supernatant from the cell lysate or cell culture supernatant was exchanged in the binding buffer and then mixed with Ni-NTA agarose. The mixture was incubated at 4°C overnight with gentle shaking and loaded onto a column for gravity chromatography. The column was washed using a washing buffer with increasing imidazole concentration (10–100 mM in Tris buffer at pH 8.0). We then treated P1-3Dp°l on resin with TEV protease and incubated it at 4°C overnight to remove His6-TEV. The reaction mixture was applied to the column and the native of P1-3Dp°l protein was collected in the flow-through fraction. The P1-3Dp°l protein was dialyzed and concentrated with a 10 kDa molecular weight cutoff membrane (Merck Millipore, USA). The P1-3Dp°l protein was detected via 12% (v/v) SDS-PAGE, analyzed through western blotting, and quantified using a bicinchoninic acid protein assay kit (Merck Millipore, USA).
Protein transfer and immunoblotting
Proteins were separated via 12% (v/v) SDS-PAGE. P1-3Dp°l expression was determined at 24, 48, 72, 96, and 120 hours after infection at MOI 10. The protein bands were stained using Coomassie Brilliant Blue R-250 (PanReac, Spain). For western blotting, the proteins were transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA, USA) and blocked with 5% skim milk in Tris-buffered saline (TBS) with 1% tween (TBS-T) for 1 hour. The membranes were then probed with goat anti-mouse monoclonal antibody against VP0 (MAB 979, Merck Millipore, USA) or goat anti-mouse monoclonal antibody against VP1 (MA5-16317, Thermo Fisher Scientific, Waltham, MA) for 16 hours at 4°C. The membrane was then washed with TBS-T and subsequently incubated with a goat anti-mouse horse radish peroxidase- (HRP-) conjugated antibody (12-349, Merck Millipore, USA) for 1 hour at room temperature. Proteins bands were developed using western blotting luminal reagent (Santa Cruz Biotech, Germany) and imaged using a gel documentation system (GE, ImageQuant LAS500). The intensity of the P1-3Dp°l protein band was assessed using ImageJ software (National Institutes of Health, Bethesda, MD).
Electron microscopy detection
To observe the formation of purified P1-3Dp°l protein, we deposited 10 μl of P1-3Dp°l sample onto a Formvar-coated, 300-mesh Cu grid. The P1-3Dp°l sample was added onto the Cu grid and incubated at 4°C for 2 minutes and a filter paper was then used to get rid of the overspilled solution. The Cu grid was negatively stained with 2% uranyl acetate solution for 2 minutes, which was then removed using filter paper. The stained grid was dried for 16–18 hours and observed under a TEM at 80 kV (Philips/TECNAI 20, Holland). The diameter of the particle size of P1-3Dp°l proteins was determined using ImageJ software (National Institutes of Health).
Mouse immunization and P1-3Dpol protein-specific IgG and IgM antibody responses
Six- to eight-week-old BALB/c female mice in each group (n = 5 per group) were immunized via intramuscular injection with one of the following: 5 μg or 10 μg of P1-3Dp°l and phosphate-buffered saline (PBS) formulated with Imject™ Alum Adjuvant (Thermo Fisher Scientific). Mice were immunized with the same dose at days 0, 14, and 28, following which their blood was harvested on day 0 and every 14 days thereafter (Fig. 7A). Mice sera were inactivated before storage at −80°C. IgG and IgM antibodies against P1-3Dp°l proteins were detected in all mouse serum samples using indirect enzyme-linked immunosorbent assay (ELISA). Briefly, a 96-well microtiter plate was coated with 50 μl of 5.0 and 0.625 μg/ml P1-3Dp°l protein, followed by 50 μl of 1:200 and 1:800 serum diluted in coating buffer for the detection of IgG and IgM, respectively. Subsequently, 50 μl of a 1:10,000 dilution of goat anti-mouse IgG (Merck Millipore, Burlington, MA) and a 1:5,000 dilution of goat anti-mouse IgM (Boster Bio, USA) were conjugated with HRP and visualized using 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate (Biolegend, San Diego, CA). A 100-μl aliquot of 2 N H2SO4 was added to stop the reaction. The color density was read within 30 minutes at 450 nm using a Fluostar Omega microplate reader (Biomedizinische Geräte Prof. Gurath Labtech, Ortenberg, Germany). For western blotting, the pooled sera from each group were collected on day 42 to confirm the presence of P1-3Dp°l protein-specific IgG and IgM antibodies at a dilution of 1:300 and 1:150, respectively.
Measuring interferon (IFN)-γ and interleukin (IL)-4 levels in mouse serum using ELISA
Serum samples were collected on day 42 in each group after the last immunization to examine the levels of IFN-γ and IL-4 using two-site sandwich ELISA kits (EliKine™, Abbkine, China) according to the manufacturer’s instructions. The corresponding standards for IFN-γ and IL-4 levels were provided in the kit and a standard curve was drawn. This curve was used to calculate the concentrations of IFN-γ and IL-4 levels in mouse serum.
Cell lines, virus strains, and microneutralization assay
Vero E6 cells (ATCC®, CRL-1586™) were cultured in minimum essential media (MEM; Gibco BRL, USA) with 10% heat-inactivated FBS, 1% glutamine, 100 U/ml penicillin, and 100 μg/ml of streptomycin. EV-A71B5 was propagated and titrated in Vero cells using the focus assay.
The virus titer was determined using a focus assay performed in quadruplicate. Briefly, the Vero cells were treated with 50 μl of the 10-fold serially diluted virus. After incubation at 37°C for 2 hours, 100 μl of 2% carboxymethyl cellulose (CMC) overlay medium in MEM supplemented with 2% heat-inactivated FBS and 100 U penicillin and streptomycin was added to each well. The plates were incubated at 37°C for 3 days, and all media were subsequently removed. The infected cells were fixed with 3.7% formaldehyde and 1% Triton X-100 in PBS. Next, 1:5,000 dilution of rabbit polyclonal antibody (GTX124261, GeneTex, USA) to EV71 and 1:1,000 dilution of anti-rabbit IgG HRP-linked antibody (7074, Cell signaling technology, USA) were added. Lastly, HRP substrate (3,3’-diaminobenzidine; Sigma, USA) was added to each well.
A microneutralization assay was performed using Vero cells to quantify the neutralizing antibodies. Briefly, the Vero cells were incubated for 18–24 hours at 37°C with 5% CO2. The serum samples to be assayed were heat-inactivated at 56°C for 30 minutes and then twofold serially diluted with RPMI. The heat-inactivated sera were mixed with the calibrated virus dose (2,000 FFU/ml) at an equal volume and incubated at room temperature for 1 hour. After incubation, the mixture was transferred to Vero cells, followed by incubation at 37°C with 5% CO2 for 90 minutes. Subsequently, a 2% CMC overlay medium in MEM supplemented with 2% heat-inactivated FBS and 100 U penicillin and streptomycin was added to each well. The plates were subsequently incubated at 37°C for 3 days. Finally, the infected cells were fixed and stained as described above. The cells and virus control were analyzed simultaneously. The neutralization titer was defined as the reciprocal of the serum dilution that resulted in 50% inhibition of viral replication compared with the virus control.
Statistical analysis
All data were analyzed using a two-way analysis of variance, followed by a Bonferroni post hoc test. A p-value of <0.05 indicated statistical significance. All visualization and statistical analyses were analyzed using the GraphPad Prism software version 9.0 for Windows (GraphPad Software, San Diego, CA).
RESULTS
Construction of recombinant bacmid P1-3Dp°l and baculovirus generation
We cloned the His6-TEV-P1-GP41-3Dp°l with EcoRI and HindIII restriction sites into the pUC57 vector. The double digestion of the pUC57 vector using EcoRI and HindIII released a 4.5-kb fragment of His6-TEV-P1-GP41-3Dp°l (Fig. 2A lanes 2 and 3 and B lane 1). The gene fragment of His6-TEV-P1-GP41-3Dp°l was then cloned into a pFastBac vector (4.8 kb in size; Fig. 2A lanes 5 and 6 and B lane 3). The correct transfer vector was transformed and manipulated in E. coli DH5α to create pFBac-His6-TEV-P1-GP41-3Dp°l of 9.3 kb after digestion using only one restriction enzyme (Fig. 3A) and was then transformed into E. coli DH10Bac. The largest white colony containing recombinant bacmid DNA was selected as opposed to blue colonies containing nonrecombinant bacmid DNA, following which PCR was performed using M13F and M13R primers as well as specific primers to identify His6-TEV-P1-GP41-3Dp°l. PCR of the recombinant bacmid DNA yielded a 4.5 kb fragment for His6-TEV-P1-GP41-3Dp°l amplified using the P1_3DF and P1_3DR primers and a 300 bp fragment for nonrecombinant bacmid DNA (negative control) amplified using M13F and M13R primers (Fig. 3B). The produced recombinant bacmid P1-3Dp°l production was then used for transfecting Sf9 cells.
Optimization of protein expression in Sf9 cells
To set the optimal conditions for P1-3Dp°l expression, we assessed the expression levels in the supernatant and cells using western blotting at various time points after infection at MOI 10 with a titer of 5.1 × 107 pfu/ml and a cell density of 1.5 × 106 cells/ml. The P1-3Dp°l bands were detected using goat anti-VP1 mouse monoclonal antibody (Fig. 4A). The highest production levels of the recombinant P1-3Dp°l protein were obtained at 96 hours, as determined by the ratio of band intensities on the western blot of P1-3Dp°l in the supernatant to that in the cell lysate (Fig. 4B).
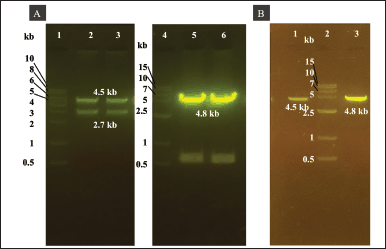 | Figure 2. Agarose gel electrophoresis analysis of His6-TEV-P1-GP41-3Dp°l and pFastBac vectors double-digested using EcoRI and HindIII. (A) Lane 1: 1.0 kb DNA ladder. Lanes 2–3: approximately 4.5 kb pUC57-His6-TEV-P1-GP41-3Dp°l vector double-digested with EcoRI and HindIII. Lane 4: 1.5 kb DNA ladder. Lanes 5–6: approximately 4.8 kb pFastBac vector double-digested with EcoRI and HindIII. (B) Lane 1: purified PCR product of His6-TEV-P1-GP41-3Dp°l. Lane 2: 1.5 kb DNA ladder. Lane 3: purified PCR product of pFastBac vector. [Click here to view] |
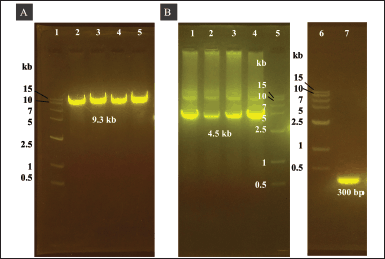 | Figure 3. Agarose gel electrophoresis of pFBac-His6-TEV-P1-GP41-3Dp°l expressed in E. coli DH5α and PCR amplification of the recombinant bacmid DNA in DH10Bac cells. (A) Agarose gel electrophoresis of pFBac-His6-TEV-P1-GP41-3Dp°l after digestion with EcoRI. Lane 1: 1.5 kb DNA ladder. Lanes 2–5: approximately 9.3 kb pFBac-His6-TEV-P1-GP41-3Dp°l digested with EcoRI. (B) Agarose gel electrophoresis of PCR products. Lanes 1–4: pFBac-His6-TEV-P1-GP41-3Dp°l amplified using the P1_3DF and P1_3DR primers; the expected size of the PCR product was approximately 4.5 kb. Lane 5: 1.5 kb DNA ladder. Lanes 6: 1.5 kb DNA ladder. Lane 7: nonrecombinant bacmid control amplified using the M13 forward and reverse primers; the expected size of the PCR product was 300 bp. [Click here to view] |
Protein purification and western blotting
The supernatants and cell lysates of Sf9 cells at 96 hours postinfection were harvested (35 ml) and purified via gravity chromatography using Ni-NTA agarose resin. (Supplementary Figure S1). The purified protein was indicated by the band pattern on 12% (v/v) SDS-PAGE after cleavage using TEV protease (Fig. 5A). Goat anti-mouse EV71 antibody detected VP0 and VP1 at sizes of 36 kDa (Fig. 5B and C).
Additionally, bands of intermediate proteins (VP0 + VP3) were observed at approximately 60 kDa (Fig. 5B and Supplementary Figure S2), and the protein profile in Figure 5C shows faint bands of VP1 protein, resulting from the low concentration of antigen along with low expression levels of VP1 in the sample. The total yield of purified P1-3Dp°l protein was 0.17 mg/ml, and it was stored at −80°C without any loss of activity.
Characterization of P1-3Dp°l
Using TEM, we examined the purified P1-3Dp°l protein after infecting Sf9 cells with recombinant bacmid P1-3Dpol to generate VLPs. Spherical-shaped structures were observed, with the average diameter of P1-3Dp°l protein being 33 nm (Fig. 6A and B). Particles are indicated using a black arrow and the bar size was 50 and 200 nm (Fig. 6A). The results demonstrated that the recombinant protein P1-3Dp°l, i.e., P1-3Dp°l protein successfully assembled under the expression of P1 polyprotein with 3D polymerase protein using a strong GP41 promoter upstream of the 3D polymerase.
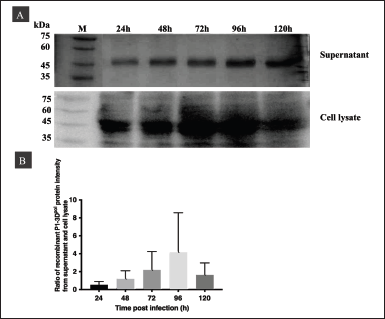 | Figure 4. Optimization of P1-3Dp°l expressed in insect cells at MOI 10 pfu/ml and different postinfection time points (24–120 hours) in cell culture supernatants and cell lysates. (A) Western blotting of P1-3Dp°l expressed at MOI 10 pfu/ml in cell culture supernatants and cell lysates. (B) Western blot analysis shows the ratio of protein intensity of P1-3Dp°l in the supernatant to that in the cell lysate at 24, 48, 72, 96, and 120 hours postinfection. [Click here to view] |
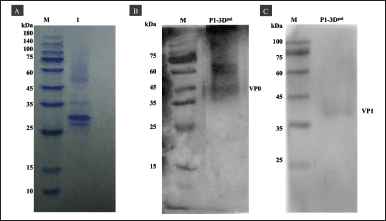 | Figure 5. SDS-PAGE and western blotting of P1-3Dp°l purification under native conditions. (A) SDS-PAGE of purified P1-3Dp°l using Ni-NTA resin after cleavage with TEV protease. (B and C) Western blotting of P1-3Dp°l with VP0 and VP1 anti-mouse monoclonal EV71 antibody, respectively. [Click here to view] |
Immunogenicity of P1-3Dp°l
Indirect ELISA was used to measure the levels of IgG and IgM antibodies in mice immunized with either 5 or 10 μg/mouse P1-3Dp°l formulated with ImjectTM Alum adjuvant or PBS formulated with ImjectTM Alum adjuvant. IgG levels reached a maximum at day 28 after the primary booster, but IgG was not induced in control mice injected with PBS formulated with ImjectTM Alum adjuvant. Both dose regimes formulated with ImjectTM Alum adjuvant induced significantly higher levels of IgG at day 28 than PBS formulated with ImjectTM Alum adjuvant (p < 0.0001) (Fig. 7B). In addition, IgG levels induced by a dose of 10 μg/mouse P1-3Dp°l with ImjectTM Alum adjuvant were higher than those induced by 5 μg/mouse P1-3Dp°l with ImjectTM Alum adjuvant (p < 0.001). While the IgM levels in mice immunized with 10 μg/mouse P1-3Dp°l formulated with ImjectTM Alum adjuvant reached a maximum on day 28 and declined by day 42. In contrast, IgM levels in mice immunized with 5 μg/mouse P1-3Dp°l formulated with ImjectTM Alum adjuvant increased continuously and reached the maximum on day 42. Apart from this, no significant differences were noted between the two dosages in terms of IgM levels (p < 0.05) (Fig. 7C).
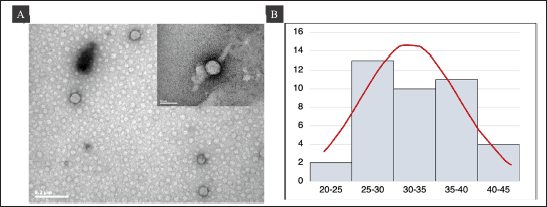 | Figure 6. TEM analysis of VLP at 80 kV. Bar = 200 and 50 nm. (A) TEM image of purified P1-3Dp°l protein. (B) Histogram of the size distribution of P1-3Dp°l. [Click here to view] |
To verify the presence of P1-3Dp°l-specific antibodies, the western blot results of anti-P1-3Dp°l mouse IgG and IgM were analyzed, and P1-3Dp°l at dosages of 5 and 10 μg/mouse was identified after 42 days in pooled sera samples from immunized mice, as assessed by the presence of a band at approximately 36 kDa, which reacted with P1-3Dp°l protein (Fig. 8A, B, E, and F). Further, anti-P1-3Dp°l mouse IgG and IgM could not be identified in the pooled sera of mice injected with PBS formulated with ImjectTM Alum adjuvant (Fig. 8C and D).
IFN-γ and IL4 levels in mouse serum
To detect cellular immune responses in immunized mice, two-site sandwich ELISA was used to detect changes in IFN-γ [T helper type 1 (Th1) cytokine] and IL-4 [T helper type 2 (Th2)] cytokine in the sera of mice injected with 5 or 10 μg/mouse P1-3Dp°l and PBS formulated with ImjectTM Alum adjuvant. The concentrations of IFN-γ and IL-4 in mouse sera were examined on day 42 (Fig. 9). The results showed that IFN-γ levels in the sera of mice injected with 10 μg/mouse P1-3Dp°l with ImjectTM Alum adjuvant were significantly increased compared with those in the sera of mice injected with PBS with ImjectTM Alum adjuvant as the negative control (p < 0.01, Fig. 9A). Furthermore, although IL-4 levels in the sera of mice injected with 10 μg/mouse P1-3Dp°l with ImjectTM Alum adjuvant were increased compared with those in the sera of mice injected with PBS ImjectTM Alum adjuvant, this increase was not significant (Fig. 9B). Hence, the administration of P1-3Dp°l at dosages of 10 μg/mouse could effectively induce IFN-γ in mouse sera.
Neutralizing antibody titer in mouse serum
Mouse serum was collected on days 0, 14, 28, and 42 after immunization with PBS (negative control), 5 μg/mouse P1-3Dp°l protein, or 10 μg/mouse P1-3Dp°l protein. Both 5 and 10 μg/mouse P1-3Dp°l formulated with ImjectTM Alum adjuvant induced a significantly greater number of neutralizing antibodies than PBS (p < 0.0001) (Fig. 10). Conversely, PBS formulated with ImjectTM Alum adjuvant did not induce the neutralizing antibodies. These results indicate that the antibody of P1-3Dp°l protein at a dosage of 5 and 10 μg/mouse was effective in neutralizing the EV-A71B5 virus. The neutralization titer against EV-A71B5 was not significantly different between dosages of 5 μg/mouse and 10 μg/mouse (p > 0.05) (Fig. 10).
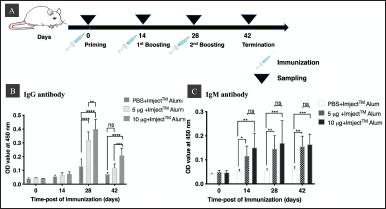 | Figure 7. Immunogenicity profile of immunized and control mice. (A) Schematic diagram of the immunization and mice bleeding schedule (n = 5). (B and C) IgG and IgM induced by PBS and 5 or 10 P1-3Dp°l μg/mouse. The sera were harvested at days 0, 14, 28, and 42 after immunization. Data are expressed as mean (± SD) of optical density values at 450 nm (n = 5), *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and ns, not significant. Asterisks (*) above the bar indicate statistically significant differences compared with PBS. [Click here to view] |
DISCUSSION
We used a simplified strategy for producing P1-3Dp°l of EV-A71B5 in Sf9 cells under the BES system. The gene construct comprised His6 with a TEV protease cleavage site, P1 under pPH control with a poly(A) tail, and 3D polymerase under GP41 promoter control with a poly(A) tail in the same recombinant DNA molecule. Two poly(A) tails are important for simultaneous gene expression (Wu et al., 2012) in two different promoter systems. Our results corroborate those of previous studies using a dual promoter system to successfully express VLPs employing baculovirus expressing the structural proteins P1 and 3CD in various insect cells (Kim et al., 2019; Lin et al., 2015a). In summary, using a dual promoter system containing pPH and GP41 led to high levels of protein expression (Kim et al., 2019).
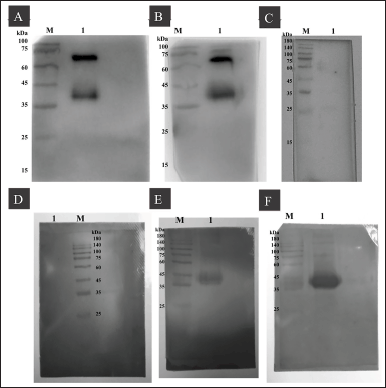 | Figure 8. Western blotting for analyzing IgG and IgM reactivity against P1-3Dp°l protein. (A–C) Western blotting for analyzing the IgG reactivity of each group against P1-3Dp°l protein in the pooling sera and then diluting at 1:300. (A) Lane M: protein ladder; lane 1: mice immunized with 5 μg/mouse P1-3Dp°l. (B) Lane M: protein ladder; Lane 1: mice immunized with 10 μg/mouse P1-3Dp°l. (C) Lane M: protein ladder; mice immunized with PBS. (D–F) Western blotting for analyzing the IgM reactivity of each group against P1-3Dp°l protein in the pooling sera and then diluting at 1:150. (D) Lane M: protein ladder; Lane 1: mice immunized with PBS. (E) Lane M: protein ladder; Lane 1: mice immunized with 5 μg/mouse P1-3Dp°l (F) Lane M: protein ladder; Lane 1: mice immunized with 10 μg/mouse P1-3Dp°l. [Click here to view] |
To further upgrade this protein purification method, we used His6 and TEV protease enzymes in the dual promoter system. This method made the protein easier to purify at a laboratory scale and could be used for developing a vaccine against EV-A71B5. The recombinant bacmid P1-3Dp°l expressed in Sf9 cells was evaluated at different postinfection time points at MOI 10. The highest and most efficient protein production was observed at 96 hours (Fig. 4). Hence, this time point can be used for P1-3Dp°l protein expression. Further, along with P1-3Dp°l of EV-A71B5, P1, and 3Dp°l were efficiently expressed under pPH and GP41 promoter control, respectively, and the P1-3Dp°l protein could be purified using gravity affinity chromatography (Ni-NTA agarose). We used SDS-PAGE and western blotting to characterize the banding pattern of purified P1-3Dp°l after the cleavage of His6 using TEV protease. More interestingly, we found that P1-3Dp°l could convert to capsid proteins VP0 and VP1 at the same size of 36 kDa (Fig. 5B and C) without EV-A71 3C protease mediating the cleavage of P1; however, the mechanism underlying this phenomenon is unclear. In the protein profile shown in Figure 5B, we can observe a protein band with a size of approximately 60 kDa that could correspond to EV71 proteins of VP0 + VP3 intermediates derived from the partially processed structure of EV-A71 VLP produced in Sf9 cells (Luo et al., 2021; Somasundaram et al., 2016). In this study, faint bands of VP1 protein, caused by the low concentration of antigen and low expression levels of VP1 in the sample, were observed (Fig. 5C) after cleavage using TEV protease and purification steps. The low protein expression level may be attributed to the gene construction method, insect cell lines, and the culture conditions used (Kim et al., 2019; Yang et al., 2020). Furthermore, some differences in the particle size were found in this study (Fig. 6B), which may result in the different assembly pathways that affect the natural stability of protein–protein interactions (Lin et al., 2015b) and may be related to our gene construction. In addition, the process design includes the cell culture system and protein storage conditions, such as pH, temperature, and ionic strength, which affect the properties of protein conformation (Effio and Hubbuch, 2015; Fernandes et al., 2013; Kim and Kim, 2017). We believed that the protein–protein interactions of gene construction, cell culture type, and purification process affected the production levels of P1-3Dp°l and its particle size, consequently affecting its antigenicity and immunogenicity (Zhao et al., 2017). Hence, this is the first study to develop a novel recombinant protein, P1-3Dp°l, specific to the B5 subgenotype of EV-A71 (Thailand strain) through gene expression encoding for genes encoding for P1 and 3Dp°l with exogenous promoters in Sf9 cells.
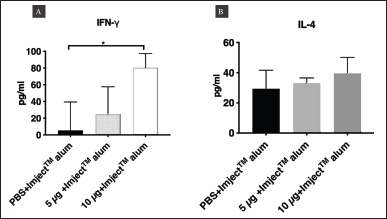 | Figure 9. Two-site sandwich ELISA for assessing the levels of IFN-γ and IL-4 in the three groups of BALB/c mice. (A) The levels of IFN-γ and (B) IL-4 in the sera of mice injected with PBS, 5 μg/mouse P1-3Dp°l, or 10 μg/mouse P1-3Dp°l on day 42. The level of IFN-γ and IL-4 are represented as mean (±SD) values (n = 5), *p < 0.05. Asterisk (*) above the bar represents statistically significant differences compared with PBS. [Click here to view] |
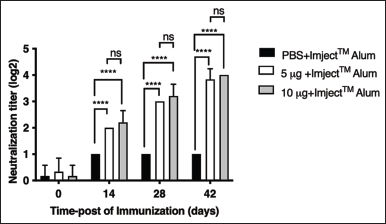 | Figure 10. Titers of neutralizing antibodies in the three groups of BALB/c mice injected with PBS, 5 μg/mouse P1-3Dp°l, or 10 μg/mouse P1-3Dp°l. The serum antibody samples were collected on days 0, 14, 28, and 42 after immunization. The data represent the mean (±SD) values (n = 5), *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and ns, not significant. Asterisks (*) above the bar represent statistically significant differences compared with PBS. [Click here to view] |
To date, there are no reports about P1 and 3Dp°l of EV-A71B5 that can process the polyprotein into the structure proteins VP1 and VP0 to candidate EV-A71 vaccine. 3Dp°l is involved in the RdRp-specific T-cell responses against EV-71 infection (Dang et al., 2014), improves the safety and stability of live-attenuated vaccines (Li et al., 2021), and stimulates both cellular and humoral immune responses (Collen et al., 1998). To study mice immunogenicity of P1-3Dp°l using the injecting BALB/c mice with doses of 5 and 10 μg/mouse P1-3Dp°l formulated with ImjectTM Alum adjuvant induced a strong humoral immune response. Mice injected with 10 μg/mouse EV-A71B5 P1-3Dp°l formulated with ImjectTM Alum adjuvant produced higher levels of IgG and IgM than those injected with 5 μg/mouse P1-3Dp°l. Therefore, the dosage of P1-3Dp°l is crucial for stimulating an effective immune response in mice. In addition, western blotting of IgG and IgM against P1-3Dp°l in mouse serum samples revealed that P1-3Dp°l had good antigenicity. Anasir et al. (2019) summarized the data of IgG- and IgM-binding epitopes of EV-A71 found in the VP1 region. Our results showed that VP1 protein promotes humoral immune response.
Both Th1 and Th2 responses are essential immune responses against antigens, and the production of major cytokines, such as IFN-γ, IL-2, IL-4, and IL-10, plays a vital role in the immune response against EV-A71 (Chung et al., 2008; Zhou et al., 2015). IFN-γ, a Th1 cytokine, is critical for a cellular immune response involving the intracellular killing of EV-A71 (Chang et al., 2006), whereas IL-4, a Th2-type cytokine, is related to the humoral immune responses (Ch’ng et al., 2011). In our study, 10 μg/mouse P1-3Dp°l formulated with ImjectTM Alum adjuvant induced the highest IFN-γ levels and neutralizing antibody titer, but IL-4 levels were low, which was similar to the results reported previously (Wang et al., 2013). Notably, In et al. (2017) reported that an inactivated vaccine dosage of 10 μg formulated with adjuvants resulted in excellent induction of neutralization antibody titer and a mixed Th1/Th2 immune response. Cytokines play an important role in activating Th cells (Paintlia et al., 2002) and are commonly known as inflammatory metabolites as IFN-γ plays a crucial role in preventing pathogenic infection and is responsible for activating Th1 cells (Xu et al., 2016). In our study, we found that P1-3Dp°l induces the expression of IFN-γ, indicating that the P1-3Dp°l protein is responsible for the Th1 immune responses and is associated with the antigen-induced immune response against viral antigens that is effective in inducing CD8+ IFN-γ-specific immune responses against EV-A71 and neutralizing titer (Chung et al., 2008; Lin et al., 2009a, 2009b). In contrast, IL4 is a marker of Th2 cells and results in the stimulation of B cells, including cell proliferation, differentiation, and maturation. Th2 cell responses to antibody production influenced the proliferation of CD4+ T-cells (Debock et al., 2013). In this study, a dosage of 10 μg/mouse P1-3Dp°l protein was able to induce IFN-γ and was superior to a dosage of 5 μg/mouse P1-3Dp°l protein. This finding indicates that 10 μg/mouse P1-3Dp°l protein could induce a Th1-type cytokine (IFN-γ). Moreover, P1-3Dp°l induced the production of IgG and IgM after mice immunization, as confirmed by ELISA and western blotting. Finally, they successfully induced both IgG and IgM antibodies, the Th1 cytokine IFN-γ, and a neutralization titer against EV-A71B5 at a 10 μg/mouse dose. Our results support that 10 μg/mouse P1-3Dp°l formulated with ImjectTM Alum adjuvant warrants further study.
CONCLUSION
We successfully constructed a novel recombinant bacmid P1-3Dp°l, expressed it in Sf9 cells, and described a simplified strategy for P1-3Dp°l protein purification using gravity affinity chromatography at the laboratory scale. The results demonstrated the effectiveness of P1-3Dp°l in eliciting the production of neutralizing antibodies, IgG, and IgM, as well as the Th1-cytokine IFN-γ, which provides a basis for the development of vaccines against EV-A71B5 commonly found in Thailand.
ACKNOWLEDGMENT
The authors would like to thank the Faculty of Pharmaceutical Sciences at Burapha University for its facilities and support.
AUTHORS’ CONTRIBUTIONS
N.I. designed the research studies, performed experiments, and wrote the manuscript. A.K. coordinated and performed animal experiments. A.J. and P.L. were involved in virus culture, performed microneutralization experiments, and drafted the manuscript. W.N. and M.N.T. were involved in coordination, performed data analysis, and participated in revising the manuscript. All authors have read and approved the final manuscript.
FINANCIAL SUPPORT
This research was supported by the vaccine grant research from the National Vaccine Institute, Thailand 2019–2021 (grant number 2562.1/2) to N.I.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVAL
Animal studies were conducted at the Animal Health Service Center, Faculty of Veterinary Medicine, Chiang Mai University, and approval was acquired from the respective institutional animal care and use committees (Approval No. MC002/2562[01/2561-11-06]).
DATA AVAILABILITY
The data presented in this study are available in this article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Anasir MI, Poh CL. Advances in antigenic peptide-based vaccine and neutralizing antibodies against viruses causing hand, foot, and mouth disease. Int J Mol Sci, 2019; 20(6):1256. CrossRef
Aw-Yong KL, Sam IC, Koh MT, Chan YF. Immunodominant IgM and IgG epitopes recognized by antibodies induced in enterovirus A71-associated hand, foot, and mouth disease patients. PLoS One, 2016; 11(11):e0165659. CrossRef
Debock I, Jaworski K, Chadlaoui H, Delbauve S, Passon N, Twyffels L, Leo O, Flamand V. Neonatal follicular Th cell responses are impaired and modulated by IL-4. J Immunol, 2013; 191(3):1231–9. CrossRef
Cao YG, Li ZH, Yue YY, Song NN, Peng L, Wang LX, Lu X. Construction and evaluation of a novel Bacillus subtilis spores-based enterovirus 71 vaccine. J Appl Biomed, 2013; 11(2):105– 13. CrossRef
Ch’ng WC, Saw WT, Yusoff K, Shafee N. Immunogenicity of a truncated enterovirus 71 VP1 protein fused to a Newcastle disease virus nucleocapsid protein fragment in mice. Acta Virol, 2011; 55(3):227–33. CrossRef
Chang LY, Hsiung CA, Lu CY, Lin TY, Huang FY, Lai YH, Chiang YP, Chiang BL, Lee CY, Huang LM. Status of cellular rather than humoral immunity is correlated with clinical outcome of enterovirus 71. Pediatr Res, 2006; 60(4):466–71. CrossRef
Chung YC, Ho MS, Wu JC, Chen WJ, Huang JH, Chou ST, Hu YC. Immunization with virus-like particles of enterovirus 71 elicits potent immune responses and protects mice against lethal challenge. Vaccine, 2008; 26(15):1855–62. CrossRef
Collen T, Baron J, Childerstone A, Corteyn A, Doel TR, Flint M, Garcia-Valcarcel M, Parkhouse RM, Ryan MD. Heterotypic recognition of recombinant FMDV proteins by bovine T-cells: the polymerase (P3Dp°l) as an immunodominant T-cell immunogen. Virus Res, 1998; 56(2):125–33. CrossRef
Cox MM. Recombinant protein vaccines produced in insect cells. Vaccine, 2012; 30(10):1759–66. CrossRef
Dang S, Gao N, Li Y, Li M, Wang X, Jia X, Zhai S, Zhang X, Liu J, Deng H, Dong T. Dominant CD4-dependent RNA-dependent RNA polymerase-specific T-cell responses in children acutely infected with human enterovirus 71 and healthy adult controls.Immunol, 2014; 142(1):89–100. CrossRef
DeZure AD, Berkowitz NM, Graham BS, Ledgerwood JE. Whole-inactivated and virus-like particle vaccine strategies for chikungunya virus. J Infect Dis, 2016; 214:S497–9. CrossRef
Effio CL, Hubbuch J. Next generation vaccines and vectors: designing downstream processes for recombinant protein-based virus-like particles. Biotechnol J, 2015; 10(5):715–27. CrossRef
Fernandes F, Teixeira AP, Carinhas N, Carrondo MJ, Alves PM. Insect cells as a production platform of complex virus-like particles. Expert Rev Vaccines, 2013; 12(2):225–36. CrossRef
Foo DG, Macary PA, Alonso S, Poh CL. Identification of human CD4 T-cell epitopes on the VP1 capsid protein of enterovirus 71. Viral Immunol, 2008; 21(2):215–24. CrossRef
Goh HC, Sobota RM, Ghadessy FJ, Nirantar S. Going native: complete removal of protein purification affinity tags by simple modification of existing tags and proteases. Protein Expr Purif, 2017; 129:18–24. CrossRef
Han X, Ying XL, Zhou SL, Han T, Huang H, Jin Q, Yang F, Sun QY, Sun XX. Characterization of the enterovirus 71 P1 polyprotein expressed in Pichia pastor as a candidate vaccine. Hum Vaccin Immunother, 2014; 10(8):2220–6. CrossRef
In HJ, Lim H, Lee JA, Kim HJ, Kim JW, Hyeon JY, Yeo SG, Lee JW, Yoo JS, Choi YK, Lee SW. An inactivated hand-foot-and-mouth disease vaccine using the enterovirus 71 (C4a) strain isolated from a Korean patient induces a strong immunogenic response in mice. PLoS One, 2017; 12(5):e0178259. CrossRef
Issaro N, Wu F, Weng L, Zhou M, Fang Z, Huang S, Rajamanickm V, Liu M, Tian H, Li X, Jiang C. Induction of immune responses by a novel recombinant fusion protein of enterovirus A71 in BALB/c mice. Mol Immunol, 2019; 105:1–8. CrossRef
Kiener TK, Jia Q, Lim XF, He F, Meng T, Chow VT, Kwang J. Characterization and specificity of the linear epitope of the enterovirus 71 VP2 protein. Virol J, 2012; 9(1):55. CrossRef
Kiener TK, Jia Q, Meng T, Chow VTK, Kwang J. A novel universal neutralizing monoclonal antibody against enterovirus 71 that targets the highly conserved “knob” region of VP3 protein. PLoS Negl Trop Dis, 2014; 8(5):e2895. CrossRef
Kim H, Kim HJ. Yeast as an expression system for producing virus-like particles: what factors do we need to consider? Lett Appl Microbiol, 2017; 64(2):111–23. CrossRef
Kim HJ, Son HS, Lee SW, Yoon Y, Hyeon JY, Chung GT, Lee JW, Yoo JS. Efficient expression of enterovirus 71 based on virus-like particles vaccine. PloS one, 2019; 14(3):e0210477. CrossRef
Li ML, Shih SR, Tolbert BS, Brewer G. Enterovirus A71 Vaccines. Vaccines, 2021; 9(3):199. CrossRef
Lin SY, Yeh CT, Li WH, Yu CP, Lin WC, Yang JY, Wu HL, Hu YC. Enhanced enterovirus 71 virus-like particle yield from a new baculovirus design.Biotechnol Bioeng, 2015a; 112(10):2005–15. CrossRef
Lin YJ, Liu WT, Stark H, Huang CT. Expression of enterovirus 71 virus-like particles in transgenic enoki (Flammulina velutipes). Appl Microbiol Biotechnol, 2015b; 99(16):6765–74. CrossRef
Lin YW, Chang KC, Kao CM, Chang SP, Tung YY, Chen SH. Lymphocyte and antibody responses reduce enterovirus 71 lethality in mice by decreasing tissue viral loads. J Virol, 2009a; 83(13):6477–83. CrossRef
Lin YW, Wang SW, Tung YY, Chen SH. Enterovirus 71 infection of human dendritic cells. Exp Biol Med, 2009b; 234(10):1166–73. CrossRef
Luo J, Huo C, Qin H, Hu J, Lei L, Pan Z. Chimeric enterovirus 71 virus-like particle displaying conserved coxsackievirus A16 epitopes elicits potent immune responses and protects mice against lethal EV71 and CA16 infection. Vaccine, 2021; 39(30):4135– 43. CrossRef
Mao QY, Wang Y, Bian L, Xu M, Liang Z. EV71 vaccine, a new tool to control outbreaks of hand, foot and mouth disease (HFMD). Expert Rev Vaccines, 2016; 15(5):599–606. CrossRef
O’Reilly D, Miller LK, Luck VA. Baculovirus expression vector. A laboratory manual. Oxford University Press, New York, NY, pp 41–6, 1992.
Ooi MH, Wong SC, Lewthwaite P, Cardosa MJ, Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol, 2010; 9(11):1097– 105. CrossRef
Paintlia MK, Kaur S, Gupta I, Ganguly NK, Mahajan RC, Malla N. Specific IgA response,T-cell subtype and cytokine profile in experimental intravaginal trichomoniasis. Parasitol Res, 2002; 88(4):338–43. CrossRef
Shah NB, Hutcheon ML, Haarer BK, Duncan TM. F1-ATPase of Escherichia coli: the ε- inhibited state forms after ATP hydrolysis, is distinct from the ADP-inhibited state, and responds dynamically to catalytic site ligands.J Biol Chem, 2013; 288(13):9383–95. CrossRef
Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis, 2010; 10(11):778–90. CrossRef
Somasundaram B, Chang C, Fan YY, Lim PY, Cardosa J, Lua L. Characterizing Enterovirus 71 and Coxsackievirus A16 virus-like particles production in insect cells. Methods, 2016; 95:38–45. CrossRef
Tang JW, Holmes CW. Acute and chronic disease caused by enteroviruses. Virulence, 2017; 8(7):1062–5. CrossRef
Wang M, Jiang S, Wang Y. Recombinant VP1 protein expressed in Pichia pastoris induces protective immune responses against EV71 in mice. Biochem Biophys Res Commun, 2013; 430(1):387–93. CrossRef
Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat Struct Mol Biol, 2012; 19(4):424–9. CrossRef
Wang X, Xiao X, Zhao M, Liu W, Pang L, Sun X, Cen S, Yang BB, Huang Y, Sheng W, Zeng Y. EV71 virus-like particles produced by co-expression of capsid proteins in yeast cells elicit humoral protective response against EV71 lethal challenge. BMC Res Notes, 2016; 9(1):42. CrossRef
Wu TY, Chen YJ, Teng CY, Chen WS, Villaflores O. A bi-cistronic baculovirus expression vector for improved recombinant protein production. Bioeng Bugs, 2012; 3(2):129–32. CrossRef
Xu X, Huang H, Wang Q, Cai M, Qian Y, Han Y, Wang X, Gao Y, Yuan M, Xu L, Yao C, Xiao L, Shi B. IFN-γ-producing Th1-like regulatory T cells may limit acute cellular renal allograft rejection: paradoxical post-transplantation effects of IFN-γ. Immunobiol, 2017; 222(2):280–90. CrossRef
Xue L, Liu JN, Wang Q, Zhang C, Xu L, Luo J, Wang J, Qin C, Liu Y, Su Z. Purification and assembling a fused capsid protein as an enterovirus 71 vaccine candidate from inclusion bodies to pentamer-based nanoparticles. Biochem Eng J, 2017; 117:139–46. CrossRef
Yang Z, Gao F, Wang X, Shi L, Zhou Z, Jiang Y, Ma X, Zhang C, Zhou C, Zeng X, Liu G, Fan J, Mao Q, Shi L. Development and characterization of an enterovirus 71 (EV71) virus-like particles (VLPs) vaccine produced in Pichia pastoris. Hum Vaccin Immunother, 2020; 16(7):1602–10. CrossRef
Zhao D, Sun B, Sun S, Fu B, Liu C, Liu D, Chu Y, Ma Y, Bai L, Wu Y, Zhou Y, Su W, Hou A, Cai L, Xu F, Kong W, Jiang C. Characterization of human enterovirus71 virus-like particles used for vaccine antigens. PLoS One, 2017; 12(7):e0181182. CrossRef
Zhao M, Bai Y, Liu W, Xiao X, Huang Y, Cen S, Chan PK, Sun X, Sheng W, Zeng Y. Immunization of N terminus of enterovirus 71 VP4 elicits cross-protective antibody responses. BMC Microbiol, 2013; 13(1):287. CrossRef
Zhou SL, Ying XL, Han X, Sun XX, Jin Q, Yang F. Characterization of the enterovirus 71 VP1 protein as a vaccine candidate. J Med Virol, 2015; 87(2):256–62. CrossRef
Zhou Y, Li JX, Jin PF, Wang YX, Zhu FC. Enterovirus 71: a whole virion inactivated enterovirus 71 vaccines. Expert Rev Vaccines, 2016; 15(7):803–13. CrossRef
DATA AVAILABILITY
Supplementary data can be downloaded from the journal’s website