INTRODUCTION
Human African trypanosomiasis (HAT) is one of the neglected tropical diseases that represent a serious threat to humans, especially in low-income countries of Africa, Asia, and South America (Molyneux et al., 2017). The geographical distribution of the sleeping sickness or HAT is restricted by a distribution of tsetse flies, which is found in the rural areas of more than 36 African countries (Brun and Blum, 2012; De Koning, 2020; Gayer et al., 2007). This vector-borne parasitic disease, caused by two subspecies of Trypanosoma brucei parasite: Trypanosoma brucei gambiense (accounts for 97% of cases) and Trypanosoma brucei rhodesiense, is considered fatal if left untreated (Pays, 1999; Steverding, 2010). Drug toxicities, resistance, and route of administration (either intramuscular and/or intravenous) raise the urgent need for new drugs orally effective against HAT (Álvarez-Bardón et al., 2020). Although extracts of natural products present many challenges to drug discovery programs worldwide (because of the chemical diversity found in a single extract), it is still known that bioactive compounds from natural products offer an incomparable and safer source for various disease therapies.
Moreover, the genus Nephthea has been reported to be rich in secondary metabolites such as sterols, sesquiterpenes, and diterpenes. Many of these metabolites recently showed antiprotozoal activities against various parasitic species such as trypanosome, malaria, and leishmania (Allam et al., 2021; Álvarez-Bardón et al., 2020).
Metabolomics is a valuable analytical tool used to comprehensively and highly accurately determine metabolites in a given metabolome (Bakr et al., 2021; Jacob et al., 2019; Said et al., 2021). It allows rapid identification (dereplication) and quantification of secondary metabolites in extracts. The combination of biological activity testing of crude extracts with metabolomics accelerates drug discovery, partly because crude extracts show higher biological activity and partly due to the ability to discriminate between complex mixtures of metabolites (Abdelmohsen et al., 2014; Kjer et al., 2010; Lotfy et al., 2021; Tawfike et al., 2013; Yuliana et al., 2011).
Ornithine decarboxylase (ODC), a proven drug target for the treatment of sleeping sickness, is the enzyme responsible for the metabolism of polyamines (such as putrescine, spermidine, and spermine) which are essential for cellular replication and hence parasite survival (Ibezim et al., 2017a). Trypanosome-specific metabolic and cellular pathways represent excellent molecular targets. The ability to synthesize polyamines, putrescine, and spermidine is vital for the proliferation of bloodstream forms in trypanosomes. In this process, ODC has a crucial function. This enzyme is considered the best-validated drug target in T. brucei, which is the target of eflornithine, a drug used clinically for treating HAT.
TTherefore, we aim to evaluate the in vitro antitrypanosomal activity of the crude extract and its derived fractions; petroleum ether and ethyl acetate of Nephthea mollis assisted by Liquid Chromatography-High Resolution-Electrospray Ionization - Mass Spectrometry (LC-HR-ESI-MS) metabolomics profiling to identify the secondary metabolites responsible for the antitrypanosomal potential of the soft coral. Additionally, we performed in silico molecular docking simulations within ODC active site to explore the binding mode of dereplicated compounds.
MATERIAL AND METHODS
Soft coral material
The soft coral N. mollis (Kingdom: Animalia, Phylum: Cnidaria, Class: Anthozoa, Subclass: Octocorallia, Order: Alcyonacea, Family: Nephtheidae) was collected by Prof. Mohamed A. Abu El-Regal, Professor of Biological Oceanography, Marine Biology Department, Faculty of Marine Science, King Abdulaziz University, Jeddah, Saudi Arabia, by scuba diving from the coasts of Hurghada, Egypt, at a depth of 12 m in March, 2018. It was then stored at −20°C ± 1°C until investigation. Part of the specimen used in the study was kept in the herbarium of the Pharmacognosy Department, Faculty of Pharmacy, Minia University, Minia, Egypt, under registration number Mn-Ph-Cog-47.
Chemicals and reagents
Solvents used in this study (purchased from El-Nasr Company for Pharmaceuticals and Chemicals, Egypt), including ethanol, ethyl acetate, and petroleum ether (b.p. 60°C–80°C), were of analytical grade and distilled before use. Solvents of HPLC grade, such as acetonitrile and methanol, were obtained from SDFCL sd fine-Chem Limited, India. Suramin (Germanin®) was obtained from Bayer AG, Leverkusen, Germany.
Extract preparation
The freeze-dried soft coral material (45 g) was extracted with ethanol until exhaustion. The crude ethanol extract (15 g, 33%) was suspended in distilled water and fractionated successively with petroleum ether and ethyl acetate. The organic phase in each step was separately concentrated under a vacuum, yielding the petroleum ether fraction (6 g), the ethyl acetate fraction (1 g), and the aqueous fraction (5 g). The crude extract and its derived fractions, petroleum ether and ethyl acetate, were kept at 4°C ± 1°C for antitrypanosomal assay and other analysis.
In vitro antitrypanosomal assay
The antitrypanosomal activity was tested against T. brucei following the protocol of Huber and Koella (1993). A number of 104 trypanosomes per milliliter of T. brucei strain TC 221 was cultivated in a complete Baltz medium. Trypanosomes were tested in 96-well plate chambers against different concentrations of test extracts (20 µL of stock solutions 10-200 µg/ml) diluted with 1 % Dimethyl Sulfoxide (DMSO) to a final volume of 200 µL. For control, 1% DMSO, as well as parasites without any test extracts, were used simultaneously in each plate to show an absence of any activity due to 1% DMSO. The plates were then incubated at 37°C ± 1 °C in an atmosphere of 5% CO2 for 24 hours. After adding 20 µl of AlamarBlue, the activity was measured after 48 and 72 hours by light absorption using an MR 700 Microplate Reader at a wavelength of 550 nm with a reference wavelength of 650 nm. Linear interpolating three independent measurements quantified IC50 values of the tested extracts. Suramin was used as a positive control (IC50 0.23 µg/ml) (Abdelmohsen et al., 2010; Cos et al., 2006).
LC-MS metabolomics profiling
Metabolomics profiling of the crude extract of N. mollis was carried out as described by Abdelmohsen et al. (2010) (Elmaidomy et al., 2019) using an ACQUITY ultra-performance liquid chromatography system connected to a SYNAPT G2 HDMS quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, USA). Chromatographic separation was carried out using a BEH C18 column (2.1 × 100 mm, 1.7 μm particle size; Waters, Milford, USA) accompanied by a guard column (2.1 × 5 mm, 1.7 μm particle size). The mobile phase used during the separation consisted of purified water (A) and acetonitrile (B) with 0.1% formic acid added to each solvent. A gradient elution started at a flow rate of 300 μl/minute with 10% B linearly increasing to 100% B within 30 minutes and remaining isocratic for the next 5 minutes before linearly decreasing back to 10% B for the following 1 minute. The volume injected was 2 μl, and the column temperature was adjusted to 40°C ± 1°C. The obtained raw data were separated into positive and negative ionization modes using MSConvert software. Accordingly, the files were imported to the data mining software MZmine version 2.10 for peak picking, followed by deconvolution, deisotoping, alignment, and formula prediction. In MZmine, a framework for differential analysis of MS data, the RAW data are imported by selecting the ProteoWizard-converted positive or negative files in mzML format followed by peaks detection in the sample using the chromatogram builder with a minimum time span set to 0.2 minutes, the minimum height and m/z tolerance to 1.0 × 104 and 0.001 m/z or 5.0 ppm, respectively. Mass ion peaks were isolated with a centroid detector threshold, greater than the noise level set to 1.0 × 104 and an MS level of one. For all remaining steps, select all files under peak lists before executing each step. Chromatogram deconvolution was then performed to detect the individual peaks. The local minimum search algorithm (chromatographic threshold: 95%, search minimum in RT range: 0.4 minutes, minimum relative height: 5%, minimum absolute height: 3.0 × 104, minimum ratio of peak top/edge: 3 and peak duration range: 0.2–5 minute) was applied. The isotopic peaks grouper was used (m/z tolerance: 0.001 m/z or 5.0 ppm, retention time tolerance: 0.1 absolute (minute), maximum charge: 2, and representative isotope: most intense) to identify the isotopes. This step will only deisotope peaks detected in the original search, i.e., those assigned a peak ID.
Filtering is helpful to set certain parameters when only considering a certain RT window, e.g., 5–40 minutes or m/z range window, or discard IDs that are only present in the sample. For chromatographic alignment and gap-filling, the retention time normalizer (m/z tolerance: 0.001 m/z or 5.0 ppm, retention time tolerance: 0.5 absolute (minute), and minimum standard intensity: 5.0 × 103) was used to reduce interbatch variation. The peak lists were all aligned using the join aligner parameters set to m/z tolerance: 0.001 m/z or 5.0 ppm, weight for m/z: 20, retention time tolerance: 5.0 relative (%), and weight for RT: 20. The values for the weight of m/z and RT should be kept the same; this means that both RT and m/z are given equal importance. Missing peaks were detected using the gap-filling peak finder with an intensity tolerance of 25%, m/z tolerance of 0.001 m/z or 5.0 ppm and retention time tolerance of 0.5 absolute (minute), followed by creating a file called “Neg-gap filled” if negative mode and “Pos-gap filled” if positive mode. An adduct search was performed for Na-H, K-H, NH4, formate, and ACN+H (RT tolerance: 0.2 absolute (minute), m/z tolerance: 0.001 m/z or 5.0 ppm, max relative adduct peak height: 30%).
Additionally, a complex search was performed (ionization method: [M+H]+ for ESI positive mode and [M-H]− for ESI negative mode, retention time tolerance: 0.2 absolute (minute), m/z tolerance: 0.001 m/z or 5.0 ppm and with a maximum complex peak height of 50%). The processed data set was then subjected to molecular formula prediction and peak identification to search for unidentified features and select atoms C, H, N, O, and other elements. Adjust parameters with heuristics element count with all three suboptions to get the isotope pattern filter working with all features with isotope peaks (Mahmoud et al., 2021).
Excel macros were written to enable the subtraction of background peaks. The Excel macro was used to dereplicate each m/z ion peak with compounds in the customized database (using RT and m/z threshold of ±3 ppm), which provided details on the putative identities of all metabolites. The macro was then utilized to identify the top 20 features (ranked by peak intensity) and to correspond putative identities in the sample by creating a list for the sample.
The Dictionary of Natural Products (DNP), METLIN, and MarinLit databases were used to identify compounds (Ancheeva et al., 2018).
In silico molecular docking
In silico molecular docking simulations were performed for dereplicated compounds of N. mollis within the ODC (PDB ID: 1NJJ) active site. RCSB PDB deposited crystal structure resolution was 2.45 Å and it was used as a PDB entry for ODC since it was cocrystallized with a small molecule natural substrate capable of stimulating the activity of ODC in a biochemical and cell-based assay. Structures of dereplicated compounds were prepared in ChemDraw® Ultra (v. 8, 2013) and were transferred as smiles to Builder software embedded in Molecular Operating Environment (MOE 2014.0901; Chemical Computing Group, Montreal, QC, Canada) software. Protein-ligand complex was retrieved from the protein data bank, and the cocrystallized water molecules and nonessential small molecules were deleted. After carefully examining the topologies, the retained protein-ligand complex was subjected to energy minimization using GROMACS 4.5.5 and the GROMOS force field 53a6 (Berman et al., 2000; Scholz et al., 2015). The cocrystallized ligands were removed from the protein and redocked using the London dG scoring function implemented in MOE. Their structures were protonated at a cut-off value of 15 Å. Validation of the docking process of cocrystallized ligand (ornithine ORX) and MD simulations of all test compounds from N. mollis were done by the Triangle Matcher method, which was preferred because it has been reported to be more systematic in its ligand atoms alignment than other methods like Alpha Triangle. The poses generated from the placement stage were refined using the Forcefield refinement scheme according to its default settings. The scheme uses the molecular mechanics’ setup and the reaction field functional form to perform energy minimization and treat the solvent effect (Ibezim 2017a, 2017b).
The free energy of binding of a given pose is estimated by the London dG scoring function from the sum of each of these terms: average gain/loss of rotational/translational entropy, energy due to loss of flexibility of the ligand, H-bond energies, energy due to metal-ligation and desolvation energy of atoms. London dG rescores 1 for only 10 retained docking poses of each compound, taking the lowest free energy of binding.
The resultant docking poses for each compound were examined and arranged according to their free binding energy value (S; Kcal/mol) and the root-mean-square deviation (RMSD; Å), using the lowest RMSD values relative to the crystal structure binding poses (Jackson et al., 2003; Shady et al., 2020).
RESULTS AND DISCUSSION
Antitrypanosomal activity
Using the protocol of Huber and Koella (1993), the crude extract, and its derived fractions, petroleum ether, and ethyl acetate of N. mollis were tested for their in vitro antitrypanosomal effects against T. brucei. The results revealed that the crude extract and ethyl acetate fraction of the soft coral N. mollis displayed remarkable in vitro antitrypanosomal effect against T. brucei with IC50 values of 6.4 and 3.7 µg/ml, respectively, while the petroleum ether fraction shows a weak antitrypanosomal activity with IC50 value of (>50 µg/ml). This encouraged us to undergo further metabolomics analysis of the active crude extract to reveal its constituents.
The proposed mechanism of action of sesquiterpenes in the parasite cells may be due to the formation of thiol adducts with components found in the intracellular medium (namely trypanothione, glutathione, and thiol groups in proteins). The parasite’s cells become more vulnerable to oxidative stress with a reduction in trypanothione and, hence, it was suggested that compounds induced programmed cell death in the tested parasite (Zeouk et al., 2020). In another way, the proposed mechanism of action of diterpenes in the parasite cells may be due to their considerable permeabilization in the membrane of the parasite (Ueno et al., 2018).
Metabolomics analysis
The crude extract of N. mollis was subjected to metabolomics profiling for the first time. The chemical profiling of dereplicated compounds was consistent with literature data on secondary metabolites of N. mollis (Figs. 1 and 2). The dereplicated compounds were identified using databases (DNP, METLIN, and MarinLit databases) (Macintyre et al., 2014). Thirty-three compounds were preliminarily identified based on their mass and summarized in Table 1 and Figure 3. Most of the identified compounds (27 compounds) were identified as sesquiterpenes type of compounds, in addition to a few diterpenes and terpenoid-related carboxylic acids compounds.
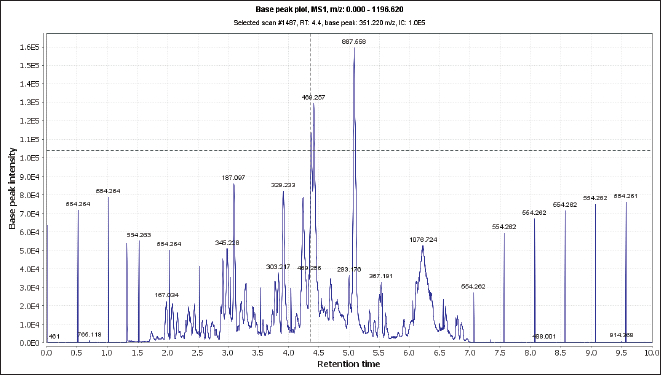 | Figure 1. Total ion chromatogram of the crude extract of N. mollis (negative mood). [Click here to view] |
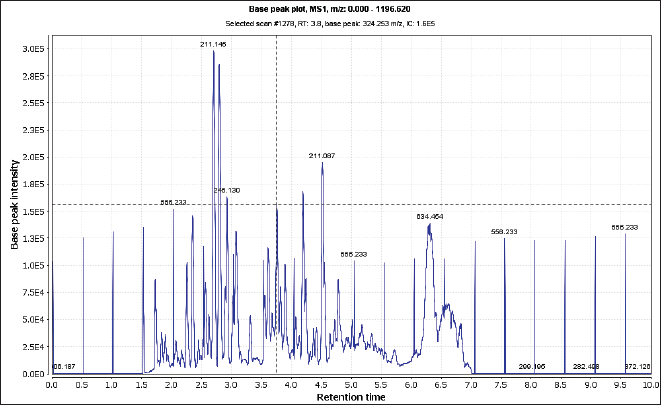 | Figure 2. Total ion chromatogram of the crude extract of N. mollis (positive mood). [Click here to view] |
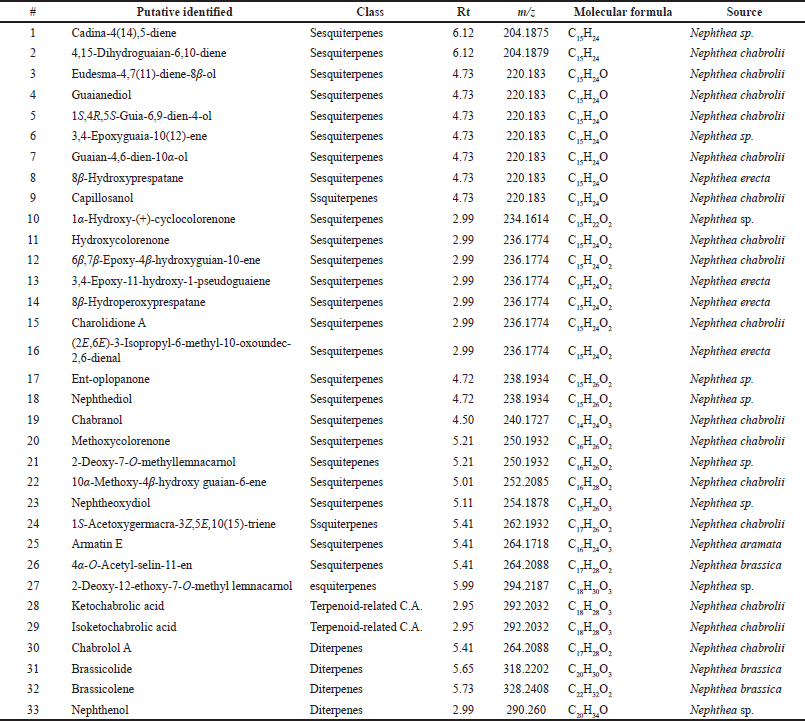 | Table 1. A list of identified metabolites in soft coral N. mollis. [Click here to view] |
The mass ion peak at m/z 204.187 for the suggested formula C15H24 was identified as cadina-4(14),5-diene (1), which was formerly reported from the soft coral Nephthea sp. (Kitagawa et al., 1987). Likewise, the mass ion peak at m/z 204.187 for the predicted molecular formula C15H24 was also dereplicated as the sesquiterpene: 4,15-dihydro guaian-6,10-diene (2), which was previously obtained from Nephthea chabrolii (Amir et al., 2012). Additionally, the mass ion peak at m/z 220.183, in accordance with the molecular formula C15H24O, was dereplicated as eudesma-4,7(11)-diene-8β-ol (3) and/or guaianediol (4) and/or 1S,4R,5S-guia-6,9-dien-4-ol (5) and/or 3,4-epoxyguaia-10 (12)-ene (6) and/or guaian-4,6-dien-10α-ol (7) and/or 8β-hydroxyprespatane (8) and/or capillosanol (9). All of these sesquiterpenes were previously obtained from N. chabrolii (Amir et al., 2012) except sesquiterpenes (6) and (8) which were obtained from Nephthea sp. and Nephthea erecta, respectively (Amir et al., 2012) Similarly, the mass ion peak at m/z 234.161 for the predicted molecular formula C15H22O2 was dereplicated as the sesquiterpenes: 1α-hydroxy-(+)-cyclocolorenone (10), which was formerly characterized from Nephthea sp. (Hu et al., 2011). Furthermore, the mass ion peak at m/z 236.177, in accordance with the molecular formula C15H24O2, was dereplicated as hydroxycolorenone (11) and/or 6β,7β-epoxy-4β-hydroxyguaian-10-ene (12) and/or 3,4-epoxy-11-hydroxy-1-pseudoguaiene (13) and/or 8β–hydroperoxyprespatane (14) and/or chabrolidione A (15) and/or (2E,6E)-3-isopropyl-6-methyl-10-oxoundec-2,6-dienal (16). Sesquiterpenes (11), (12), and (15) were previously obtained from N. chabrolii (Hu et al., 2011; Su et al., 2007), while sesquiterpenes (13), (14), and (16) were obtained from N. erecta (Abdelhafez et al., 2020). Moreover, the mass ion peak at m/z 238.193, in agreement with the molecular formula C15H26O2 was dereplicated as sesquiterpenes; ent-oplopanone (17) and/or nephthediol (18) both were previously reported from Nephthea sp. (Kitagawa et al., 1987). Similarly, the mass ion peak at m/z 240.172 for the predicted molecular formula C14H24O3, was dereplicated as the sesquiterpenes; chabranol (19) was formerly characterized from N. chabrolii (Amir et al., 2012). Moreover, the mass ion peak at m/z 250.193, in agreement with the molecular formula C16H26O2, was dereplicated as the sesquiterpenes. As for methoxycolorenone (20) and/or 2-deoxy-7-O-methyllemnacarnol (21), the former was previously reported from N. chabrolii (Handayani et al., 1997), while the latter was previously reported from Nephthea sp. (Kapojos et al., 2008). Another compound was dereplicated as 10α-methoxy-4β-hydroxy guaian-6-ene (22), on account of the observed mass ion peak at m/z 252.208 and in accordance with the molecular formula C16H28O2. This sesquiterpene was formerly characterized from N. chabrolii (Amir et al., 2012). Additionally, the mass ion peak at m/z 254.187 for the suggested formula C15H26O3 was identified as nephtheoxydiol (23). These sesquiterpenes were formerly reported from the soft coral Nephthea sp. (Kitagawa et al., 1987). Likewise, the mass ion peak at m/z 262.193 for the predicted molecular formula C17H26O2 was dereplicated as the sesquiterpenes: 1S-acetoxygermacra-3Z,5E,10(15)-triene (24), which was previously characterized from N. chabrolii (Zhang et al., 2001). Similarly, the mass ion peak at m/z 264.171, in consonance with the predicted formula C16H24O3, was dereplicated as armatin E (25), a sesquiterpene earlier reported in Nephthea armata (El-Gamal et al., 2004), while that at m/z 264.208 was characterized as 4α-O-acetyl-selin-11-en (26), a sesquiterpene having the molecular formula C17H28O2. This compound was identified from Nephthea brassica (Duh et al., 1999). The mass ion peak at m/z 294.218 for the predicted molecular formula C18H30O3 was dereplicated as the sesquiterpenes: 2-deoxy-12-ethoxy-7-O-methyl lemnacarnol (27), which was previously characterized from Nephthea sp. (Kapojos et al., 2008). Besides the sesquiterpenes constituents, an assortment of other structurally different metabolites has also been described encompassing terpenoid-related carboxylic acids: ketochabrolic acid (28) and/or isoketochabrolic acid (29), which were identified from the mass ion peak at m/z 292.203, in agreement with the molecular formula C18H28O3. Both compounds were previously reported from N. chabrolii (Su et al., 2007). Moreover, as a common biosynthetic feature of Nephthea soft corals, LC-HR-ESI-MS analysis indicated the presence of a variety of diterpenes. In that respect, the mass ion peak at m/z 264.208, consistent with the formula C17H28O2, was dereplicated as chabrolol A (30) that was previously identified from N. chabrolii (Zhang et al., 2001). Likewise, the mass ion peak at m/z 318.220 for the suggested molecular formula C20H30O3 was characterized as brassicolide (31), while that at m/z 328.240 for the suggested molecular formula C22H32O2 was characterized as brassicolene (32), and both diterpenes (31) and (32) were formerly obtained from N. brassica (Duh et al., 1999). Another diterpene with the molecular formula C20H34O, in good agreement with the mass ion peak at m/z 290.260, was characterized as nephthenol (33), which was previously reported from Nephthea sp. (Kitagawa et al., 1987).
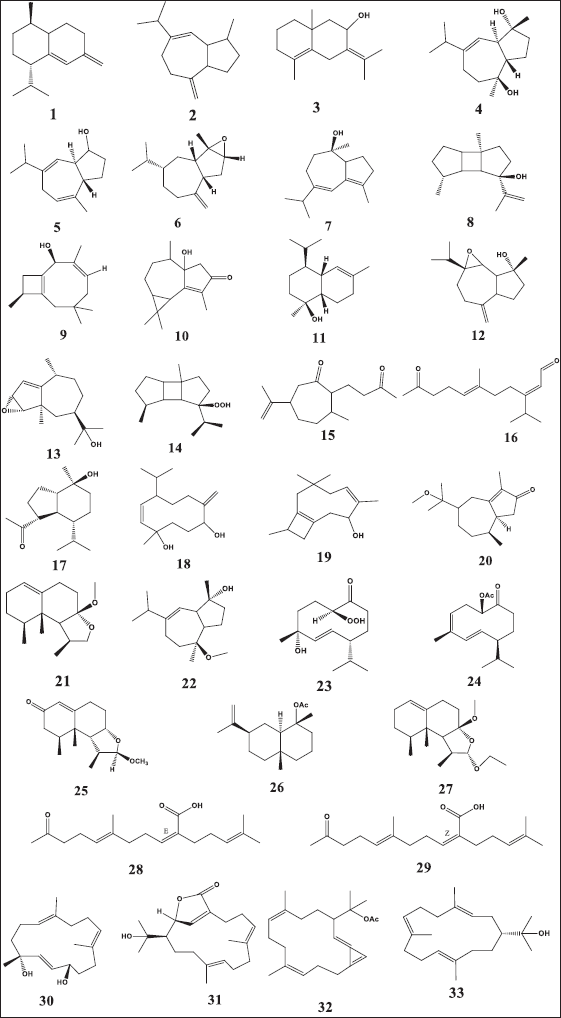 | Figure 3. Chemical structure of dereplicated compounds 1–33. [Click here to view] |
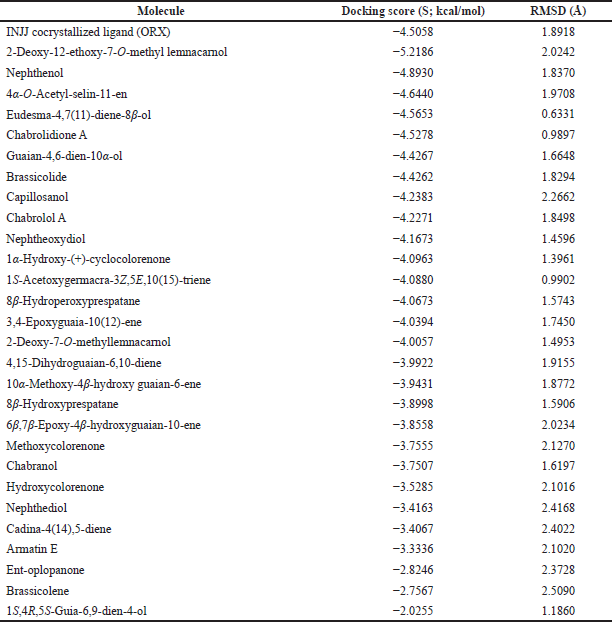 | Table 2. Binding free energy (S; kcal/mol) and binding accuracy (RMSD; Å) of N. mollis and cocrystallized ligand within ODC active site (PDB ID: 1NJJ). [Click here to view] |
Molecular docking
For further exploration and to get a better idea about the possible target affected by dereplicated compounds of N. mollis to afford their antitrypanosomal activity, we performed in silico molecular docking simulations within ODC. The X-ray structure of ODC showed ligands; D-ornithine, ORX, a substrate analog, was placed within the ODC active site, and Geneticin (G418) as a weak non-competitive inhibitor occupying an allosteric site of ODC. Molecular docking simulations performed within the active site of ODC showed a number of binding interactions (H-bonding and H-pi interactions) between ORX and a number of amino acid residues (GLU 274, ASP 332, GLY 276, ARG 277, TYR 389, GLY 237, and HIS 197) with binding free energy (expressed as docking score, S) of −4.5058 kcal/mol, as listed in Table 2.
Docking of dereplicated compounds within ODC active site revealed a number of interesting observations. First of all, binding free energy for five of the identified compounds (2-deoxy-12-ethoxy-7-o-methyl lemnacarnol, nephthenol, 4α-o-acetyl-selin-11-en, eudesma-4,7(11)-diene-8β-ol, and chabrolidione A) was better than ORX, as listed in Table 2.
Additionally, as found with ORX, most of the test compounds showed either H-bonding and/or H-pi binding interactions with key amino acid residues: GLU 274, ARG 277, and HIS 197 (Table 3 and Fig. 4). Finally, a visual inspection of the best docking poses of 6,7-epoxy-4-hydroxyguaian-10-ene, chabrolidione A, and nephtheoxydiol showed a good overlay with ORX within ODC active site, as shown in Figure 5. Overall, the docking results showed how good the overlay of the dereplicated compounds of N. mollis with ORX within ODC active site was, which could be used for explaining their antitrypanosomal activity.
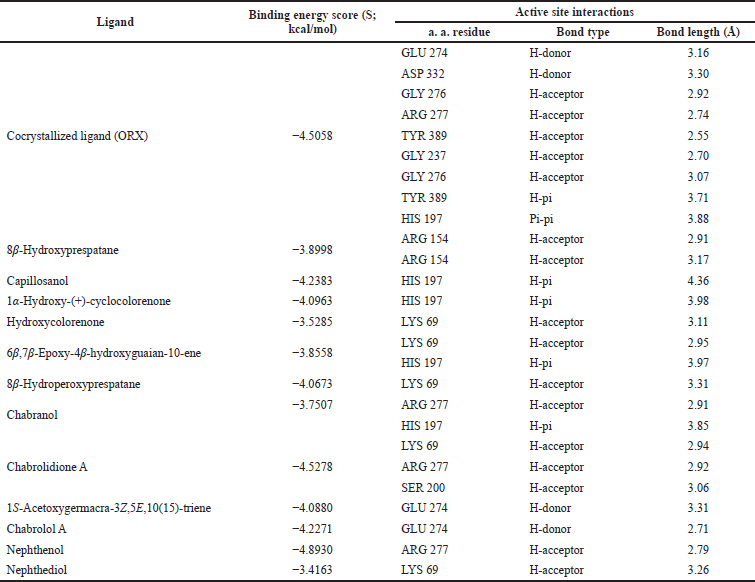 | Table 3. Binding free energy (S; kcal/mol) and binding interactions for co-crystallized ligand (ORX) and Nephthea mollis within ornithine decarboxylase (ODC) active site (PDB ID: 1NJJ; 2.45 Å). [Click here to view] |
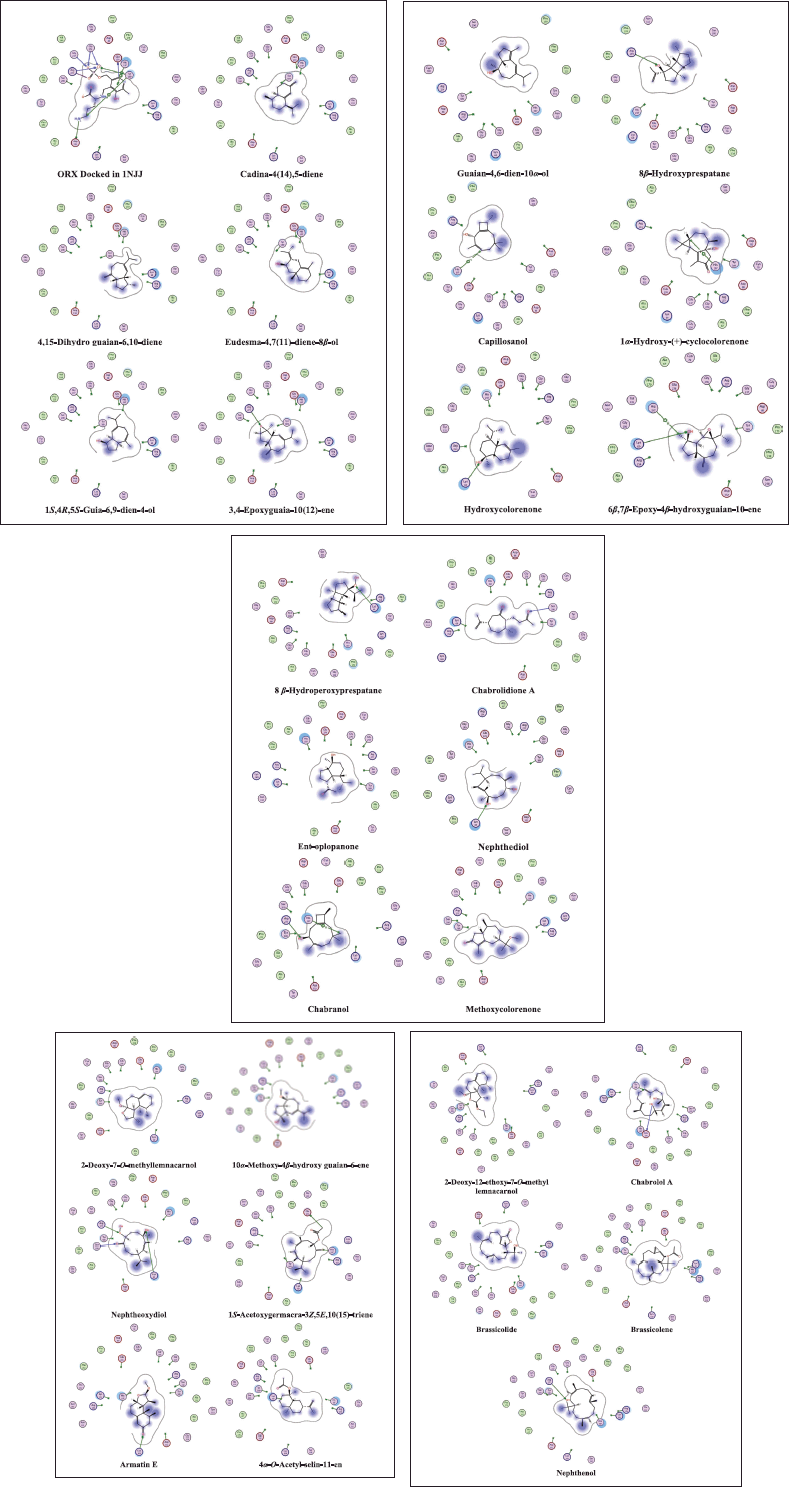 | Figure 4. 2D interaction diagrams in the active site 1NJJ made by the cocrystallized ligand (ORX) and the dereplicated compounds. The MOE software generates the diagrams. [Click here to view] |
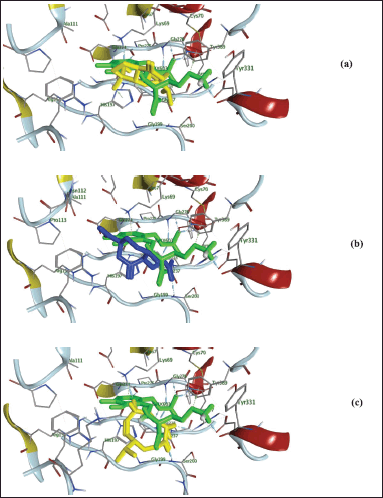 | Figure 5. 3D presentation of 6,7-epoxy-4-hydroxyguaian-10-ene (yellow-color; (a), chabrolidione A blue-color; (b), nephtheoxydiol yellow-color; (c) overlay within 1NJJ active site and its ligand (ORX; green-color). [Click here to view] |
CONCLUSION AND FUTURE PERSPECTIVES
This study highlighted the promising antitrypanosomal activity of the soft coral N. mollis extracts as a possible natural therapy for HAT. The crude extract and its derived ethyl acetate fraction displayed remarkable in vitro antitrypanosomal activity against T. brucei. Thirty-three compounds of sesquiterpenes and diterpenes class of compounds were mined with the help of LC–MS-based metabolomics. Docking studies within ODC crystal structure provided insights about how the dereplicated compounds bind and interact with various amino acid residues lining ODC active site, which was further evidenced by in vitro assays against T. brucei. Thus, further work on the extract of N. mollis will be included in our plans to purify the compounds responsible for their antitrypanosomal activity and perform in vivo experiments and safety studies for the purified compounds. Moreover, it should be further investigated for a supportive role in the pharmaceutical field toward developing new antitrypanosomal drugs.
ACKNOWLEDGMENTS
We thank Prof. RuAngelie Edrada-Ebel (Head of the Natural Products Metabolomics Group, University of Strathclyde) for the Macro software used in the dereplication study. The authors uploaded the article on the preprint server link here: https://assets.researchsquare.com/files/rs-1090567/v1/779390fd-82d0-4c1b-b8e1 49f8261e3005.pdf?c=1637766316; DOI: https://doi.org/10.21203/rs.3.rs-1090567/v1. The authors will inform the preprint server after the publication of this article in JAPS.
CONFLICTS OF INTEREST
The authors reported no potential conflicts of interest.
FUNDING
There is no funding to report.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral concerning jurisdictional claims in published institutional affiliation.
AUTHORS’ CONTRIBUTIONS
K.M.A. did conceptualization, data curation, and writing-original draft. Y.A.M. did formal analysis and methodology. U.R.A. carried out writing review and editing. A.I.K. did investigation, A.E.A. validation, M.A.F. project administration, and E.S.E. supervision. All authors discussed the results and contributed to the final manuscript.
REFERENCES
Abdelhafez OH, Ali TFS, Fahim JR, Desoukey SY, Ahmed S, Behery FA, Kamel MS, Gulder TA, Abdelmohsen UR. Anti-inflammatory potential of green synthesized silver nanoparticles of the soft coral Nephthea sp. supported by metabolomics analysis and docking studies. Int J Nanomed, 2020; 15(1):5345–60. CrossRef
Abdelmohsen UR, Cheng C, Viegelmann C, Zhang T, Grkovic T, Ahmed S, Quinn RJ, Hentschel U, Edrada-Ebel RJMd. Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora sp. EG49. Mar Drugs, 2014; 12(3):1220–44. CrossRef
Abdelmohsen UR, Pimentel-Elardo SM, Hanora A, Radwan M, Abou-El-Ela SH, Ahmed S, Hentschel UJ. Isolation, phylogenetic analysis and anti-infective activity screening of marine sponge-associated Actinomycetes. Mar Drugs, 2010; 8(3):399–412. CrossRef
Allam KM, Khedr AI, Allam AE, Abdelkader MSA, Elkhayat ES, Fouad MA. Chemical, and biological diversity in Nephthea soft corals in the current decade: a review. JABPS, 2021; 4(3):124–33. CrossRef
Álvarez-Bardón M, Pérez-Pertejo Y, Ordóñez C, Sepúlveda-Crespo D, Carballeira NM, Tekwani BL, Murugesan S, Martinez-Valladares M, García-Estrada C, Reguera RMJMD. Screening marine natural products for new drug leads against trypanosomatids and malaria. Mar Drugs, 2020; 18(4):187. CrossRef
Amir F, Koay YC, Yam WS. Chemical constituents and biological properties of the marine soft coral Nephthea: a review (Part 1). Trop J Pharm Res, 2012; 11(3):485–98. CrossRef
Ancheeva E, Daletos G, Proksch P. Lead compounds from mangrove-associated microorganisms. Mar Drugs, 2018; 16(9):319. CrossRef
Bakr RO, Tawfike A, El-Gizawy HA, Tawfik N, Abdelmohsen UR, Abdelwahab MF, Alshareef WA, Fayez SM, El-Mancy SM, El-Fishawy AM, Abdelkawy MA, Fayed MA. The metabolomic analysis of five Mentha species: cytotoxicity, anti-helicobacter assessment, and the development of polymeric micelles for enhancing the anti-helicobacter activity. RSC Adv, 2021; 11(13):7318–30. CrossRef
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucl Acids Res, 2000; 28(1):235–42. CrossRef
Brun R, Blum J. Human African trypanosomiasis. Infect Dis Clin, 2012; 26(2):261–73. CrossRef
Cos P, Vlietinck AJ, Berghe DV, Maes L. Anti-infective potential of natural products: how to develop a stronger in vitro proof-of-concept’. J Ethnopharmacol, 2006; 106(3):290–302. CrossRef
De Koning PH. The drugs of sleeping sickness: their mechanisms of action and resistance, and a brief history. Trop Med Infect Dis, 2020; 5(1):14. CrossRef
Duh CY, Wang SK, Weng YL, Chiang MY, Dai FJ. Cytotoxic terpenoids from the Formosan soft coral Nephthea brassica. J Nat Prod, 1999; 62(11):1518–21. CrossRef
El-Gamal AA, Wang SK, Dai CF, Duh CY. New nardosinanes and 19-oxygenated ergosterols from the soft coral Nephthea armata collected in Taiwan. J Nat Prod, 2004; 67(9):1455–8. CrossRef
Elmaidomy AH, Mohammed R, Hassan HM, Owis AI, Rateb ME, Khanfar MA, Krischke M, Mueller MJ, Abdelmohsen UR. Metabolomic profiling and cytotoxic tetrahydrofurofuran lignans investigations from Premna odorata Blanco. Metabolites, 2019; 9(10):223. CrossRef
Gayer M, Legros D, Formenty P, Connolly MA. Conflict and emerging infectious diseases. Emerg Infect Dis, 2007; 13(11): 1625–31. CrossRef
Handayani D, Edrada RA, Proksch P, Wray V, Witte L, van Ofwegen L, Kunzmann A. New oxygenated sesquiterpenes from the Indonesian soft coral Nephthea chabrolii. J Nat Prod, 1997; 60(7):716–8. CrossRef
Hu J, Yang B, Lin X, Zhou X, Yang X, Long L, Liu Y. Chemical and biological studies of soft corals of the Nephtheidae family. Chem Biodivers, 2011; 8(6):1011–32. CrossRef
Huber W, Koella JC. A comparison of three methods of estimating EC50 in studies of drug resistance of malaria parasites. Acta Trop, 1993; 55(4):257–61. CrossRef
Ibezim A, Debnath B, Ntie-Kang F, Mbah CJ, Nwodo NJ. Binding of antitrypanosomal natural products from African flora against selected drug targets: a docking study. Med Chem Res, 2017a; 26(3):562–79. CrossRef
Ibezim A, Nwodo NJ, Nnaji NJ, Ujam OT, Olubiyi OO, Mba CJ. In silico investigation of morpholines as a novel class of trypanosomal triosephosphate isomerase inhibitors. Med Chem Res, 2017b; 26(1):180–9. CrossRef
Jackson LK, Goldsmith EJ, Phillips MA. X-ray structure determination of Trypanosoma brucei ornithine decarboxylase bound to D-ornithine and to G418: insights into substrate binding and ODC conformational flexibility. J Biol Chem, 2003; 278(24):22037–43. CrossRef
Jacob M, Lopata AL, Dasouki M, Abdel Rahman AM. Metabolomics toward personalized medicine. Mass Spectrom Rev, 2019; 38(3):221–38. CrossRef
Kapojos MM, Mangindaan REP, Nakazawa T, Oda T, Ukai K, Namikoshi MJC. Three new nardosinane type sesquiterpenes from an Indonesian soft coral Nephthea sp. Chem Pharm Bull, 2008; 56(3):332–4. CrossRef
Kitagawa I, Cui Z, Son BW, Kobayashi M, Kyogoku YJC. Marine natural products. XVII. Nephtheoxydiol, a new cytotoxic hydroperoxy-germacrane sesquiterpene, and related sesquiterpenoids from an Okinawan soft coral of Nephthea sp. (Nephtheidae). Chem Pharm Bull, 1987; 35(1):124–35. CrossRef
Kjer J, Debbab A, Aly AH, Proksch P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat Protoc, 2010; 5(3):479–90. CrossRef
Lotfy MM, Sayed AM, AboulMagd AM, Hassan HM, El Amir D, Abouzid SF, El-Gendy AO, Rateb ME, Abdelmohsen UR, Alhadrami H, Mohammed R. Metabolomic profiling, biological evaluation of Aspergillus awamori, the river Nile-derived fungus using epigenetic and OSMAC approaches. RSC Adv, 2021; 11(12):6709–19. CrossRef
Macintyre L, Zhang T, Viegelmann C, Martinez IJ, Cheng C, Dowdells C, Abdelmohsen UR, Gernert C, Hentschel U, Edrada-Ebel R. Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar Drugs, 2014; 12(6):3416–48. CrossRef
Mahmoud BK, Hamed ANE, Samy MN, Abdelmohsen UR, Attia EZ, Fawzy MA, Refaey RH, Salem MA, Pimentel-Elardo SM, Nodwell JR, Desoukey SY, Kamel MS. Metabolomic profiling and biological investigation of Tabebuia Aurea (Silva Manso) leaves, family Bignoniaceae. Nat Prod Res, 2021; 35(22):4632–7. CrossRef
Molyneux DH, Savioli L, Engels D. Neglected tropical diseases: progress towards addressing the chronic pandemic. Lancet, 2017; 21(1):312–25. CrossRef
Pays E. Expression and function of surface proteins in Trypanosoma brucei. Mol Biochem Parasitol, 1999; 91(1):3–36. CrossRef
Said AAE, Afifi AH, Ali TF, Samy MN, Abdelmohsen UR, Fouad MA, Attia EZ. NS3/4A helicase inhibitory alkaloids from Aptenia cordifolia as HCV target. RSC Adv, 2021; 11(52):32740–9. CrossRef
Scholz C, Knorr S, Hamacher K, Schmidt B. DOCKTITE-a highly versatile step-by-step workflow for covalent docking and virtual screening in the molecular operating environment. J Chem Inf Model, 2015; 55(2):398–406. CrossRef
Shady NH, Elfakharany Z, Salem MA, Ahmed S, Fouad MA, Kamel MS, Krischke M, Mueller MJ, Abdelmohsen UR. Dereplication analysis and antitrypanosomal potential of the red sea sponge Amphimedon sp. supported by molecular modeling. Rev Bras Farmacogn, 2020; 30:290–4. CrossRef
Steverding D. The development of drugs for treatment of sleeping sickness: a historical review. Parasit Vectors, 2010; 3(1):1–9. CrossRef
Su JH, Dai CF, Huang HH, Wu YC, Sung PJ, Hsu CH, Sheu JH. Terpenoid-related metabolites from a formosan soft coral Nephthea chabrolii. Chem Pharm Bull, 2007; 55(4):594–7. CrossRef
Tawfike AF, Viegelmann C, Edrada-Ebel R. Metabolomics and dereplication strategies in natural products. Methods Mol Biol, 2013; 1055:227–44. CrossRef
Ueno AK, Barcellos AF, Costa-Silva TA, Mesquita JT, Ferreira DD, Tempone AG, Romoff P, Antar GM, Lago JHG. Antitrypanosomal activity and evaluation of the mechanism of action of diterpenes from aerial parts of Baccharis retusa (Asteraceae). Fitoterapia, 2018; 125:55–8. CrossRef
Yuliana ND, Khatib A, Choi YH, Verpoorte R. Metabolomics for bioactivity assessment of natural products. Phytother Res, 2011; 25(2):157–69. CrossRef
Zeouk I, Sifaoui I, López-Arencibia A, Reyes-Batlle M, Bethencourt-Estrella CJ, Bazzocchi IL, Bekhti K, Lorenzo-Morales J, Jiménez IA, Piñero JE. Sesquiterpenoids and flavonoids from Inula viscosa induce programmed cell death in kinetoplastids. Biomed Pharmacother, 2020; 130: 110518. CrossRef
Zhang WH, Williams ID, Che CT. Chabrolols A, B and C, three new norditerpenes from the soft coral Nephthea chabroli. Tetrahedron Lett, 2001; 42(28):4681–5. CrossRef