INTRODUCTION
Coronavirus disease 2019 (COVID-19) refers to a respiratory infection brought on by the SARS-CoV-2 virus and has emerged as a pandemic since 2020. The discovery of effective therapy with accessible raw materials for patients with COVID-19 is a critical attempt that is being intensified. Utilizing molecular databases to investigate different compounds that may have therapeutic effects on the virus is one of the methods explored by researchers to find new medications to combat SARS-CoV-2 (Wu et al., 2020).
Through the latest advances in computational biology and molecular bioinformatics, a number of natural phytocompounds have been published to have antiviral potential against several specific targets of SARS-CoV-2 including angiotensin-converting enzyme-2 (ACE-2) and several viral important proteins, such as spike protein, which contains S1 and S2 domains, 3-chymotrypsin-like protease (3-CLpro), papain-like protease (PLpro), helicases, and RNA-dependent RNA polymerase (RdRp) (Khare et al., 2020). The in-silico prediction of possible phytochemical compounds on key SARS-CoV-2 proteins has been extensively investigated. Andrographis paniculata (Burm. F.) Nees or Justicia paniculata Burm. F. is a medicinal plant whose natural compounds have been shown to have potential as SARS-CoV-2 antivirals (Murugan et al., 2021; Vijayakumar et al., 2022).
The main phytocompounds of A. paniculata are diterpene compounds including andrographolide, 14-deoxyandrographolide, 14-deoxy-11,12-didehydroandrographolide, 14-deoxy-11-oxoandrographolide, neoandrographolide, andrographiside, deoxyandrographoside, andrograpanin, deoxyandrographolide-19-D-glucosi-de, 14-deoxy-12-methoxyandrographolide, and flavonoid compounds (Dwivedi et al., 2021). The outcomes of in-silico testing on the main compounds of A. paniculata have varied according to several researches. These take into account the importance of affinity energy, bond conformation stability, and comparisons to currently available reference medications like remdesivir, lopinavir, and hydroxychloroquine. This systematic review’s objective was to determine the phytochemical compounds of A. paniculata that may be active against SARS-CoV-2 using an in-silico technique as a reference and scientific support for prospective herbal COVID-19 treatment options.
METHODS
This systematic review included data from two databases, PubMed and Google Scholar, covering original English language articles which applied in-silico methods for A. paniculata phytochemical compounds on SARS-CoV-2 protein. A systematic search was conducted using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Protocols 2020 starting from August to September 2022. Keywords for the search included “Andrographis paniculata,” “in silico,” and “COVID-19.” The inclusion criteria were applied to a total of 38 scientific articles, and 29 articles were chosen for review. Three authors independently evaluated the quality of each article using Duffy’s Research Appraisal Checklist Approach. Nine articles were excluded because the target proteins used in the in-silico assays were not derived from proteins in SARS-CoV-2 and the intervention utilized derivatives of A. paniculata phytochemical compounds (Fig. 1).
The extracted data were analyzed to determine the in-silico method used, the A. paniculata compounds tested, the SARS-CoV-2 protein target, and the authors’ conclusions (Table 1). The data were synthesized according to the Synthesis Without Meta-analysis guide to determine A. paniculata compounds as potential protein inhibitors in SARS-CoV-2. Afterward, the potential antiviral phytochemical compounds of A. paniculata are further elaborated according to their protein targets in the form of highly conserved sites from changes in the evolutionary phylogenetic tree, such as 3CLpro (main protease), PLpro, RdRp, and spike proteins.
RESULTS
Literature evidence showed that 107 phytochemical compounds of A. paniculata were in silico tested on SARS-CoV-2 protein. Fifty of these compounds showed their potential as inhibitors of the SARS-CoV-2 protein target. The main parameter used to assess the docking results is the affinity binding energy, which in some studies is amplified by the results of molecular dynamic simulations and Molecular Mechanics Poisson-Boltzmann Surface Area. Additionally, many in-silico studies also assessed the Absorption, Distribution, Metabolism, and Excretion (ADME) and toxicity predictions of A. paniculata compounds. Overall, the determination of a compound’s potential as a SARS-CoV-2 inhibitor was carried out in three ways, including the determination of binding affinity energy value parameters, comparison with reference drugs, and consideration of bond conformations at protein amino acid residues. The articles reviewed showed several variations of in-silico tests, particularly molecular docking in terms of methods, software, targets, and ligand modeling. The two most popular docking programs were AutoDock (37.93%) and AutoDock Vina (24.14%). In-silico experiments on a number of SARS-CoV-2 protein targets were carried out in most of the study (58.62%). The majority of these protein targets were found in the NCBI GenBank and RCSB Protein Bank Database.
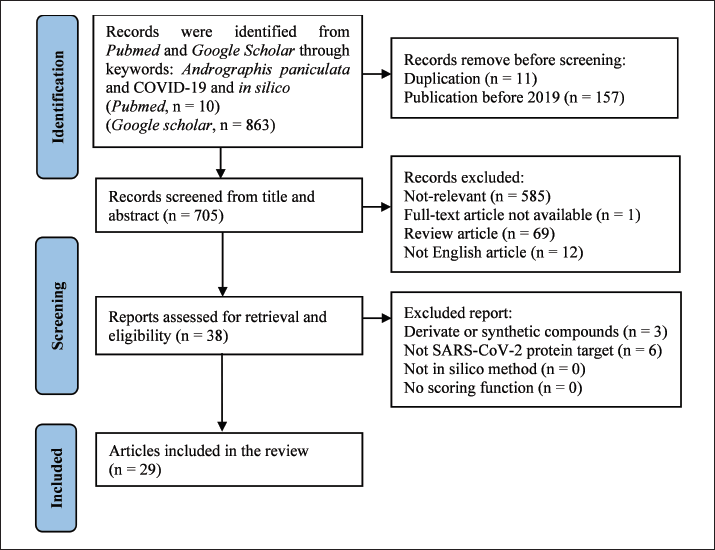 | Figure 1. Flowchart on search strategies. [Click here to view] |
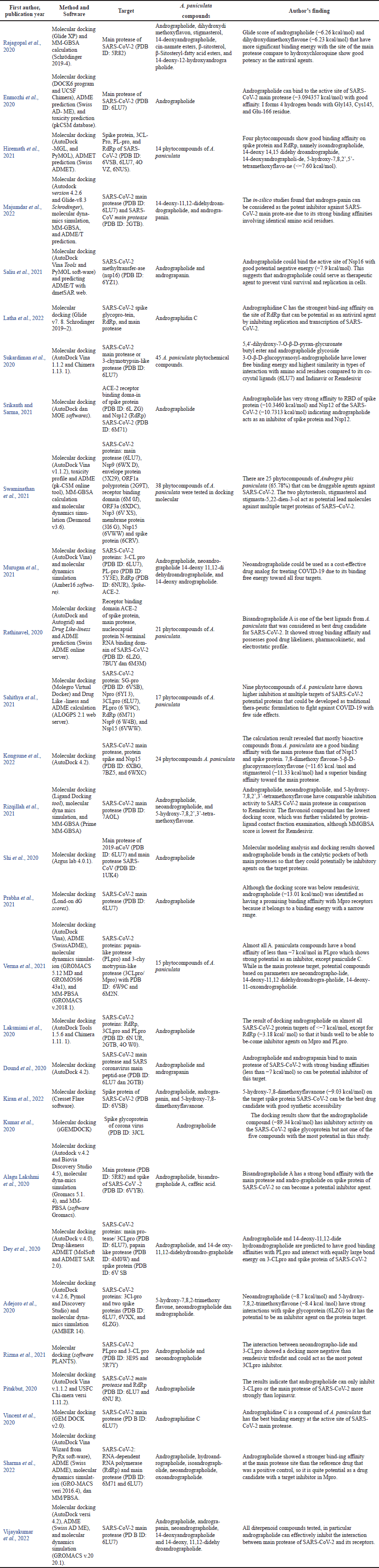 | Table 1. Characteristicsics and the main finding of studies included in the systematic review. [Click here to view] |
The reviewed in-silico studies of A. paniculata phytochemical compounds used a total of 15 SARS-CoV-2 proteins as targets, namely, 3-CLpro (Mpro), PLpro (Nsp3), RdRp (Nsp12), spike protein, non-structural protein 15 (Nsp15), Nsp9, Nsp16, envelope protein, ORF1a, ORF3a, membrane protein, nucleocapsid N-terminal RNA-binding domain, SGpro, Npro, and main peptidase. Main protease (Mpro) was the most widely used target protein, with 25 articles. Spike protein (11 articles), RdRp (8 articles), PLpro (7 articles), Nsp15 (3 articles), and Nsp9 (2 articles) followed sequentially. Only one article of the remaining nine SARS-CoV-2 proteins was focused on. The five most used SARS-CoV-2 proteins were conserved structures from phylogenetic changes and therefore were highly potential antiviral drug targets, namely, Mpro, PLpro, RdRp, nsp15, and spike protein. Furthermore, the potential of compounds will be reviewed based on these five potential targets.
There are 22 diterpenoid compounds, 18 flavonoids, four phytosterols, three sesquiterpene compounds, and one compound each from the triterpene and phenylpropanoid classes among the 50 A. paniculata compounds reported as potential inhibitors of various potential protein targets of SARS-CoV-2. Interestingly, several studies involving many A. paniculata compounds as populations for molecular docking tests showed that andrographolide, recognized as the main herbal compound, performed lower in binding affinity energy, indicating that other compounds from this plant also have better potential as antivirals on several SARS-CoV-2 protein targets. These compounds include neoandrographolide, andrographidin C, or stigmasterol and stigmasta-5,22-dien-3-ol from the phytosterol class which are considered to be multi-target antiviral drug agents (Swaminathan et al., 2021) (Table 2).
Based on the protein target, the majority of A. paniculata phytochemical compounds were found as inhibitors of spike proteins, namely, 37 compounds, followed by PLpro with 32 compounds, Mpro (main protease) with 30 compounds, Nsp15 with 30 compounds, and RdRp with 11 compounds (Fig. 2). This finding is in line with the results of Kongsune et al. (2022), which showed that most of the A. paniculata compounds have greater binding energy values to the main protease, followed by spike protein and Nsp15. However, more hydrogen bonds were found at the active site of Nsp15, then spike protein and Mpro sequentially. This is due to the docking results that also consider other interactions, such as Van der Waals and electrostatic bonds (Kongsune et al., 2022).
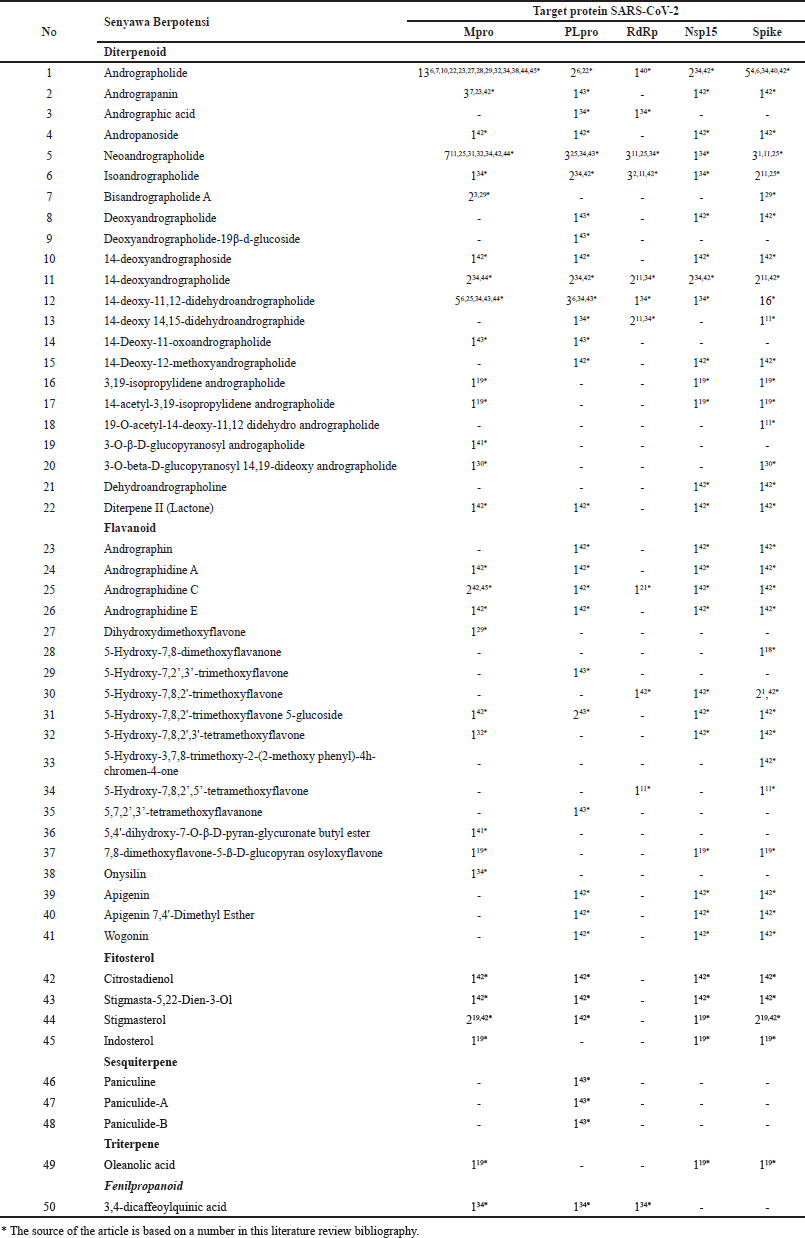 | Table 2. Literature evidence of multi-target A. paniculata phytocompounds for COVID-19. [Click here to view] |
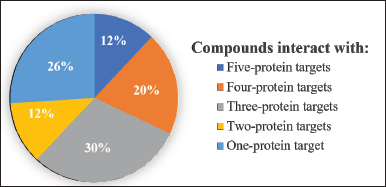
Scientific evidence from in-silico studies shows the potential of several A. paniculata compounds being multi-target inhibitors of SARS-CoV-2 (Fig. 3). Andrographolide, neoandrographolide, isoandrographolide, 14-deoxyandrographolide, 14-deoxy-11,12 didehydroandrographolide, and andrographidine C are the six compounds that have inhibitory activity on all five protein targets. A total of 10 compounds interact with four protein targets, 15 compounds interact with three protein targets, 6 compounds interact with two protein targets, and 13 other phytochemical compounds interact only with one potential protein of SARS-CoV-2.
DISCUSSION
An in-vivo study revealed that 14-deoxy-11,12-didehydroandrographolide, the primary diterpenoid compound of A. paniculata, significantly reduced lung titers in infected mice when administered at non-toxic concentrations of 1,000 or 500 mg/kg/day 4 or 48 hours prior to H5N1 influenza A virus infection (Cai et al., 2016). Several in-vitro studies also showed the potency of A. paniculata phytocompounds as an antiviral. In Calu-3 cells infected with SARS-CoV-2, post-infection therapy with A. paniculata extract and andrographolide dramatically decreased virion generation with an IC50 of 0.036 µg/ml and 0.034 µm, respectively, limiting the transmission of COVID-19 (Sa-Ngiamsuntorn et al., 2021). Andrographolide also suppressed the main protease activity of SARS-CoV-2 in the cleavage assay by producing an IC50 value of 15.05 ± 1.58 M (Shi et al., 2020). Furthermore, exploration of potential phytochemical compounds in essential proteins of SARS-CoV-2 with in-silico predictions has been carried out. Based on existing literature evidence, A. paniculata phytochemical compounds are capable of inhibiting the activity of several potential protein targets in SARS-CoV-2. In addition, below is a more detailed discussion of the mechanism of inhibition based on the protein targets from existing in-silico studies.
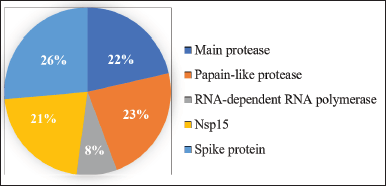 | Figure 2. Percentage of target-based A. paniculata compounds. [Click here to view] |
 | Figure 3. Percentage of multi-target A. paniculata compounds. [Click here to view] |
Main protease
The main protease is a highly conserved cysteine hydrolase class protein in β-coronavirus (Hu et al., 2022). This enzyme mediates the cleavage of 11 non-structural proteins that are necessary for viral replication and transcription. Inhibition of viral proteases will reduce the formation of mature functional proteins (Mengist et al., 2021). According to research, the most alluring residue for SARS-CoV-2 Mpro to make hydrogen bonds with diverse ligands is Gly143, followed by Glu166, Cys145, and His163 (Nguyen et al., 2020). Some of the amino acid residues with crucial roles in the main protease are targeted by A. paniculata phytochemical compounds through hydrogen bonding. Andrographolide forms four hydrogen bonds to the main protease through residues Gly143, Cys145, and Glu166 (Enmozhi et al., 2020). This electrophilic compound inhibits the main protease activity of SARS-CoV-2 by entering the catalytic site through hydrogen bonds on amino acid residues and several water molecules (Rajagopal et al., 2020; Shi et al., 2020). The following list of molecular mechanisms that andrographolide possesses as antiviral characteristics was shown by in-silico investigations to be possible inhibitors of the SARS-CoV-2 main protease: 1) improved H1N1 virus-I which caused cell death by blocking virally induced activation of the retinoic acid-inducible gene-I- like receptors signaling pathway and reduced lung virus titer through its immune modulatory action, 2) modification of the virus replication route caused by Endoplasmic Reticulum stress-mediated Unfolded Protein Response, 3) induction of heme oxygenase-1 expression, 4) involvement of multiple pathways including Nuclear Factor kappa B and Janus kinase/signal transducers and activators of transcription, 5) inhibition of protease activity, 6) limiting of antigen expression, 7) inhibition of glycoprotein expression, and 8) suppression of lytic protein expression (Banerjee et al., 2020).
The non-covalent bond interactions of the four diterpenoid compounds studied by Majumdar et al. (2022) on amino acids of main protease proteins are hydrogen, pi-alkyl, alkyl, carbon-hydrogen, hydrophobic, and polar bonds. The in-silico study carried out by Sukardiman et al. (2020) presented another finding stating that the glycoside form of andrographolide, 3-O-β-D-glucopyranosyl andrographolide, produces more negative binding energy than its aglycone form. The physicochemical properties of the glycoside affect the solubility and polarity of the compound thus inducing the receptor site of main protease (Sukardiman et al., 2020). The study conducted by Rajagopal et al. (2020) demonstrated that dihydroxy dimethoxy flavone compounds have the same excellent affinity as andrographolide on the SARS-CoV-2 main protease receptor due to lipophilic properties and hydrogen bond formation. Stigmasterol from the phytosterol class of A. paniculata forms hydrogen bonds, ionic interactions, and strong hydrophobicity on various amino acid residues of the main protease and exerts a stable configuration on the spike and Mpro protein targets (Swaminathan et al., 2021).
However, different results were also reported for some major diterpene compounds such as andrographolide, andrograpanin, 14-deoxyandrographolide, and 14-deoxy-11,12-didehydroandrographolide where these compounds were reported to have no stable binding to the main protease target. Research by Sukardiman et al. (2020) assessed the potential of compounds with standard reference ligand bond affinity values, where the main diterpene compound is found to have a weaker binding affinity. This was also confirmed by the results of Swaminathan et al. (2021) concerning the lack of potential of several main diterpene compounds as main protease inhibitors with an affinity energy assessment greater than −7 kcal/mol.
Papain-like protease
PLpro is a crucial coronavirus enzyme necessary to process viral polyproteins to activate the replication complex and facilitate virus spread. This protein in SARS-CoV-2 shares 83% sequence identity making it highly conserved (Shin et al., 2020). The antiviral interferon pathway can be maintained with the help of PLpro inhibition, which can also lessen viral replication in infected cells and interfere with virus-induced cytopathogenic effects. The reduction in the replication of the active virus (subgenomic RNA4-encoding E gene in SARS-CoV-2), as determined by genetic surveillance of intracellular viral RNA synthesis, is the outcome of the suppression of PLpro. As a result, the released virus particles from the infected cell are reduced (Shin et al., 2020). In order to reduce SARS-CoV-2 infection and improve antiviral immunity, PLpro should be targeted. Flexible binding sites at residues Tyr269 and Gln270 for small molecules present in SARS-CoV PLpro are also found in SARS-CoV-2 PLpro. Research by Ibrahim et al. (2020) showed that the key residues that appear on SARS-CoV-2 PLpro are Tyr268 and Gln 269.
The compound reported to bind most extensively to Plpro is 14-deoxy-11,12-didehydroandrographolide with stable binding to the active site of this protein through hydrogen (Gln269), carbon-hydrogen (Gly160), and Van der Waals bonds on other residues (Verma et al., 2021). Meanwhile, neoandrographolide binds to the active site through hydrogen bonds (Asn109, Gln269, and Va159) and carbon-hydrogen bonds (Gly160 and Thr158). This is in line with the results by Sahithya et al. (2021), which showed that A. paniculata compounds interact with the main amino acid residue of SARS-CoV-2 PLpro, namely, Tyr268 and several other residues. Andrographolide, as the main compound, was found to bind toward the active site of PLpro through hydrogen bonds at the Tyr274 residue (Dey et al., 2020). Van der Waals interactions and hydrogen bonds are the main factors in the inhibitory bonding process of flavonoid phytochemical compounds from A. paniculata on the target protein PLpr (Rajagopal et al., 2020). The two main compounds of the phytosterol class, stigmasterol and stigmasta-5,22-dien-ol, display hydrophobic interactions at Arg131, Thr199, Tyr239, and Leu286 residues with high affinity, making them potential inhibitors of the SARS-CoV-2 PLpro protein (Swaminathan et al., 2021).
However, in-silico research conducted by Rizma et al. (2021) suggested that the two main diterpenoid compounds of A. paniculata, andrographolide and neoandrographolide, have no potential as PLpro inhibitors. The study assessed the potential of the compound based on the comparison of the estimated affinity energy of the docking results with the natural ligand PLpro and the reference drug remdesivir (Rizma et al., 2021). The study by Dey et al. (2020) also found that the compound 14-deoxy-11,12-didehydroandrographolide lacked a hydrogen bond to the amino residue PLpro despite having a fairly high-affinity value.
RNA-dependent RNA polymerase
RdRp is a highly conserved protein in three coronaviruses, namely, SARS-CoV, SARS-CoV-2, and MERS-CoV. This protein is suggested to be a broad-spectrum antiviral target in coronaviruses. RdRp is predicted to be the pivotal enzyme responsible for viral replication and transcription complexes (Jiang et al., 2021). Aside from that, these proteins also play a role in aiding viral escape from host defense mechanisms which has important implications during viral evolution. Potential treatment strategies to prevent viral replication include antiviral medications that specifically target the active sites of RdRp, Asp760, and Asp761 (Aftab et al., 2020). The FDA has approved the anti-RdRp medications, remdesivir and ribavirin.
The major diterpenoid compound of A. paniculata, andrographolide, inhibits RdRp activity by binding to residue Thr556 through the formation of four hydrogen bonds (Srikanth and Sarma, 2021). Neoandrographolide and isoandrographolide are the two compounds reported to mostly interact with RdRp. A study by Hiremath et al. (2021) reported that neoandrographolide interacts with the active site of this protein through hydrogen bonds at four residues, namely, Ala125, Asp126, Arg132, and His133. This finding is supported by the results from Murugan et al.’s (2021) study, which suggested that neondrographolide is a compound with the potential as an inhibitor of RdRp even though its binding affinity value is greater than that of remdesivir drugs, which is due to the stability of its binding to the Asp336, Thr440, and Asp507 residues of the target protein. The study by Latha et al. (2022) showed that andrographidin C, being one of the major diterpenoid compounds of A. paniculata, interacts with RdRp similarly to nucleotide analog drugs, that is, through non-obligate RNA chain breaks. Such interaction is stable due to the presence of pi-pi bonds from uracil bases and the affinity is also strengthened by hydrogen bonds at amino residues Glu811 and Lys551 (Dey et al., 2020). The phytochemical compound was also discovered to interact with the major site of RdRp, Asp761, making it a potential inhibitor of this protein (Sahithya et al., 2021).
However, in-silico research conducted by Laksmiani et al. (2020) concluded that andrographolide has less potential as an RdRp inhibitor by having a more positive affinity value than remdesivir drugs. This is supported by the results of Sharma et al. (2022) which showed that some of the main diterpenoid compounds of A. paniculata, including andrographolide and neoandrographolide, have no interaction with amino acid residues of the RdRp target protein. Although reported as having good binding affinity, isoandrographolide does not have hydrogen bonding with the amino residue RdRp (Hiremath et al., 2021).
Non-structural protein 15
Coronaviruses use the uridine-specific endoribonuclease known as Nsp15 to break viral RNA and circumvent the host immune system (Saramago et al., 2022). This protein features a conserved active site across Coronaviridae. Nsp15 is crucial for the pathogenesis of coronaviruses. According to research, this RNase is in charge of destroying viral dsRNA intermediates that would otherwise hinder host cells from recognizing them. Additionally, this protein dramatically slows down the type I interferon response (Saramago et al., 2022). As a result, developing antiviral drug candidates that target these proteins in particular could be a possible treatment for COVID-19.
The active site of Nsp15 shares six essential residues that are conserved among SARS-CoV-2, SARS-CoV, and MERS-CoV proteins, namely, His235, His250, Lys290, Thr341, Tyr343, and Ser294 (Kim et al., 2020). Based on the results by Sahithya et al. (2021), A. paniculata phytochemical compounds bind to the Nsp15 active site more efficiently than hydroxychloroquine at the amino acid residues Thr (167), Lys (71, 90), Glu (203), Asn (200), Arg (199), Thr (196, 275), Lys (277), Val (295), Tyr (89, 279), Ser (198, 274), Glu (69), Asp (268, 273, 297), Gly (165), and Met (272). Although it does not bind to the key residue of Nsp15, the compound has a stronger affinity than the antiviral drugs nelvinafir and hydroxychloroquine to many other residues (Sahithya et al., 2021). Andrographidin C is reported to have high binding affinity energy and interacts with amino residues Asn29, Asn30, and Pro51 (Swaminathan et al., 2021). However, some of the main compounds of A. paniculata have also been reported to have less potential due to having a more positive binding affinity energy than existing reference drugs (Kongsune et al., 2022).
Spike protein
Spike glycoproteins have an essential role in the attachment, fusion, and entrance of the virus into the host cell (Duan et al., 2019). A spike protein that binds to the ACE-2 receptor enables SARS-CoV-2 to enter human cells and cause infection (Banerjee et al., 2022). The spike protein, which is involved in the process of membrane fusion and receptor recognition, is made up of the S1 and S2 subunits. The coronavirus must bind to the specific receptor ACE-2 in order to enter the target cell. This is accomplished by the S1 subunit of the spike protein, while the S2 subunit facilitates the fusing of viral membrane cells by establishing a six-helix bond via a two-heptad repetitive domain (Huang et al., 2020). Located on the surface of the virus, the spike glycoprotein is targeted by the host immune response as the main target by neutralizing antibodies. Therefore, targeting this protein as an ativirus drug target may reduce the potential for SARS-CoV-2 infection in humans.
Results by Srikanth and Sarma (2021) indicated that the main compound of the diterpenoid group, andrographolide, binds to the spike glycoprotein at the tyrosine kinase phosphorylation site suggesting inactivation of the target protein. Andrographolide creates a hydrogen bond interaction with the amino residue Lys807 on the spike protein. The diterpene lactone compound from A. paniculata binds to the core site located under the surface area of the spike protein complex and ACE-2 thereby indirectly modulating the interaction between the two proteins (Murugan et al., 2021). Research by Kiran et al. (2022) stated that the compound 5-hydroxy-7,8-dimethoxyflanone, one of the main flavonoid compounds of A. paniculata, has moderate affinity energy on spike protein targets through hydrogen bonding interactions with amino residues Asp 364 and Gly339.
CONCLUSION
The potential proteins of the SARS-CoV-2 have been discovered to be inhibited by a number of phytocompounds from the plant A. paniculata using in-silico tests. The most studied and highly conserved SARS-CoV-2 potential proteins from phylogenetic changes for inhibitory targets of phytochemical compounds of A. paniculata are the main protease, PLpr, RdRp, Nsp15, and spike protein. A number of six phytochemical compounds of A. paniculata showed inhibitory activity on the five SARS-CoV-2 target proteins including andrographolide, neoandrographolide, isoandrographolide, 14-deoxyandrographolide, 14-deoxy 11,12-didehydroandrographolide, and andrographidine C which are the main diterpenoid compounds of A. paniculata.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Adejoro IA, Babatunde DD, Tolufashe GF. Molecular docking and dynamic simulations of some medicinal plants compounds against SARS-CoV-2: an in silico study. J Taibah Univ Sci, 2020; 14(1):1563–70. CrossRef
Aftab SO, Ghouri MZ, Masood MU, Haider Z, Khan Z, Ahmad A, Munawar N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J Transl Med, 2020; 18(1):1–15. CrossRef
Alagu Lakshmi S, Shafreen RMB, Priya A, Shunmugiah KP. Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: using structure-based drug discovery approach. J Biomol Struct Dyn, 2021; 39(13):4594–609. CrossRef
Banerjee A, Czinn SJ, Reiter RJ, Blanchard TG. Crosstalk between endoplasmic reticulum stress and anti-viral activities: a novel therapeutic target for COVID-19. Life Sci, 2020; 255:117842; doi:10.1016/j.lfs.2020.117842. CrossRef
Banerjee S, Wang X, Du S, Zhu C, Jia Y, Wang Y, Cai Q. Comprehensive role of SARS-CoV-2 spike glycoprotein in regulating host signaling pathway. J Med Virol, 2022; 94:4071–87. CrossRef
Cai W, Chen S, Li Y, Zhang A, Zhou H, Chen H, Jin M. 14-Deoxy-11,12-didehydroandrographolide attenuates excessive inflammatory responses and protects mice lethally challenged with highly pathogenic A(H5N1) influenza viruses. Antiviral Res, 2016; 133:95–105. CrossRef
Dey YN, Khanal P, Patil BM, Wanjari MM, Srivastava B, Gurav SS. The role of andrographolide and its derivative in COVID-19 associated proteins and immune system 2020. Available via https://doi.org/10.21203/rs.3.rs-35800/v1 CrossRef
Dound YA, Chaudhary S, Sehgal R, Chaudhary SS, Dound BA. Plant based molecules for the management of Covid-19 2020. Available via https://pubchem.ncbi.nlm.nih.gov/
Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front Immunol, 2019; (11):576622. CrossRef
Dwivedi MK, Sonter S, Mishra S, Singh P, Singh PK. Secondary metabolite profiling and characterization of diterpenes and flavones from the methanolic extract of Andrographis paniculata using HPLC-LC-MS/MS. Futur J Pharm Sci, 2021; (1):1–28.. CrossRef
Enmozhi SK, Raja K, Sebastine I, Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J Biomol Struct Dyn, 2020; 39:1–7. CrossRef
Hiremath S, Kumar HDV, Nandan M, Mantesh M, Shankarappa KS, Venkataravanappa V, Basha CRJ, Reddy CNL. In silico docking analysis revealed the potential of phytochemicals present in Phyllanthus amarus and Andrographis paniculata, used in ayurveda medicine in inhibiting SARS-CoV-2. 3 Biotech, 2021; 11(2):44. CrossRef
Hu Q, Xiong Y, Zhu GH, Zhang YN, Zhang YW, Huang P, Ge GB. The SARS-CoV-2 main protease (Mpro): structure, function, and emerging therapies for COVID-19. MedComm (Beijing), 2022; 3(3):e151. CrossRef
Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin, 2020; 41:1141–9. CrossRef
Ibrahim TM, Ismail MI, Bauer MR, Bekhit AA, Boeckler FM. Supporting SARS-CoV-2 papain-like protease drug discovery: in silico methods and benchmarking. Front Chem, 2020; 8:996. CrossRef
Jiang Y, Yin W, Xu HE. RNA-dependent RNA polymerase: structure, mechanism, and drug discovery for COVID-19. Biochem Biophys Res Commun, 2021; 538:47. CrossRef
Khare P, Sahu U, Pandey SC, Samant M. Current approaches for target-specific drug discovery using natural compounds against SARS-CoV-2 infection. Virus Res, 2020; 290:98169. CrossRef
Kim Y, Jedrzejczak R, Maltseva NI, Wilamowski M, Endres M, Godzik A, Michalska K, Joachimiak A. Struktur kristal Nsp15 endoribonuklease NendoU dari SARS-CoV-2. Protein Sci, 2020; 29(7):1596–605. CrossRef
Kiran G, Karthik L, Shree Devi MS, Sathiyarajeswaran P, Kanakavalli K, Kumar KM. In silico computational screening of kabasura kudineer - official siddha formulation and JACOM against SARS-CoV-2 spike protein. J Ayurveda Integr Med, 2022; 13(1): 100324. CrossRef
Kongsune P, Innok W, Rungrotmongkol T. In silico screening of potential inhibitor from Andrographis paniculata constituents against three targets of SARS-CoV-2: main protease, spike protein, and Nsp15. ASEAN J Sci Technol Rep, 2022; 25(1):69–76. CrossRef
Kumar MP, Sundaram KM, Ramasamy MS. Coronavirus spike (S) glycoprotein (2019-Ncov) targeted siddha medicines kabasura kudineer and thonthasura kudineer in silico evidence for corona viral drug. Asian J Pharm Res Health Care, 2020; 12(1):20–7. CrossRef
Latha D, Hrishikesh D, Shiban G, Chandrashekar C, Bharath BR. In silico, in vitro screening of plant extracts for anti-SARS-CoV-2 activity and evaluation of their acute and sub-acute toxicity. Phytomedicine Plus, 2022; 2(2):100233. CrossRef
Laksmiani NP, Larasanty LP, Santika AA, Prayoga PA, Dewi AA, Dewi NP. Active compounds activity from the medicinal plants against SARS-CoV-2 using in silico assay. Biomed Pharmacol J, 2020; 13(2):873–81. CrossRef
Majumdar M, Singh V, Misra TK, Roy DN. In silico studies on structural inhibition of SARS-CoV-2 main protease Mpro by major secondary metabolites of Andrographis paniculata and Cinchona officinalis. Biologia (Bratisl), 2022; 77(5):1373–89. CrossRef
Mengist HM, Dilnessa T, Jin T. Structural Basis of Potential Inhibitors Targeting SARS-CoV-2 Main Protease. Front Chem, 2021; 9:7. CrossRef
Murugan NA, Pandian CJ, Jeyakanthan J. Computational investigation on Andrographis paniculata phytochemicals to evaluate their potency against SARS-CoV-2 in comparison to known antiviral compounds in drug trials. J Biomol Struct Dyn, 2021; 39(12):4415–26. CrossRef
Nguyen DD, Gao K, Chen J, Wang R, Wei GW. Unveiling the molecular mechanism of SARS-CoV-2 main protease inhibition from 137 crystal structures using algebraic topology and deep learning. Chem Sci, 2020; 11(44):12036–46. CrossRef
Pitakbut T. The Antiviral activity of andrographolide, the active metabolite from Andrographis paniculata (Burm. f.) Wall. ex Nees. against SARS-CoV-2 by using bio-and chemoinformatic tools. Walailak J Sci Technol, 2020; 17(8):851–66. CrossRef
Prabha T, Kapoor VK, Selvamani P, Latha S, Sivakumar T, Jubie S. Dual modulators of selected plant secondary metabolites targeting COVID-19 main protease and interleukin-2: an in-silico approach based novel hypothesis. Coronaviruses, 2020; 2(2):223–34. CrossRef
Rajagopal K, Varakumar P, Baliwada A, Byran G. Activity of phytochemical constituents of curcuma longa (turmeric) and Andrographis paniculata against coronavirus (COVID-19): an in silico approach. Futur J Pharm Sci, 2020; 6(1):104. CrossRef
Rathinavel T. Virtual screening of COVID-19 drug from three Indian traditional medicinal plants through in silico approach antioxidant assay of phytoconstituents from Eclipta alba view project hyaluronic acid-drug conjugation technology view project [Internet]. Article Res J Biotechnol, 2020. Available via www.rcsb.org
Rizma BRP, Ananto AD, Sunarwidhi AL. The study of potential antiviral compounds from Indonesian medicinal plants as anti-COVID-19 with molecular docking approach. J Mol Docking, 2021; 1(1):32–9. CrossRef
Rizqillah RK, Fatriansyah JF, Fadilah F, Sulhadi S, Wahyuni S, Sudirman MA, Nafisah HC, Lestari SD. In silico molecular docking and molecular dynamics examination of Andrographis paniculata compounds of Andrographolide, Neoandrogra-pholide, and 5-hydroxy-7,8,2’,3’-tetramethoxyflavone inhibition activity to SARS-CoV-2 main protease. BIO Web Conf, 2021; 41:07002. CrossRef
Sa-Ngiamsuntorn K, Suksatu A, Pewkliang Y, Thongsri P, Kanjanasirirat P, Manopwisedjaroen S, Charoensutthivarakul S, Wongtrakoongate P, Pitiporn S, Chaopreecha J, Kongsomros S, Jearawuttanakul K, Wannalo W, Khemawoot P, Chutipongtanate S, Borwornpinyo S, Thitithanyanont A, Hongeng S. Anti-SARS-CoV-2 activity of Andrographis paniculata extract and its major component andrographolide in human lung epithelial cells and cytotoxicity evaluation in major organ cell representatives. J Nat Prod, 2021; 84(4):1261–70. CrossRef
Sahithya MD, Srinath M, Kumar BC, Giri CC. Molecular in silico docking interventions in various SARS-CoV-2 receptor targets involving bioactive herbal compounds of Andrographis paniculata (Burm. f.) Nees against COVID-19. Ann Phytomed Int J, 2021; 10(1) (COVID-19):S4–12. CrossRef
Saliu TP, Umar HI, Ogunsile OJ, Okpara MO, Yanaka N, Elekofehinti OO. Molecular docking and pharmacokinetic studies of phytocompounds from Nigerian medicinal plants as promising inhibitory agents against SARS-CoV-2 methyltransferase (nsp16). J Genet Eng Biotechnol, 2021; 19(1):172. CrossRef
Saramago M, Costa VG, Souza CS, Bárria C, Domingues S, Viegas SC, Lousa D, Soares CM, Arraiano CM, Matos RG. The nsp15 nuclease as a good target to combat SARS-CoV-2: mechanism of action and its inactivation with FDA-approved drugs. Microorganisms, 2022; 10(2):342. CrossRef
Sharma A, Vora J, Patel D, Sinha S, Jha PC, Shrivastava N. Identification of natural inhibitors against prime targets of SARS-CoV-2 using molecular docking, molecular dynamics simulation and MM-PBSA approaches. J Biomol Struct Dyn, 2022; 40(7):3296–11. CrossRef
Shi TH, Huang YL, Chen CC, Pi WC, Hsu YL, Lo LC, Chen WY, Fu SL, Lin CH. Andrographolide and its fluorescent derivative inhibit the main proteases of 2019-nCoV and SARS-CoV through covalent linkage. Biochem Biophys Res Commun, 2020; 533(3):467–73. CrossRef
Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, Mehdipour AR, Tascher G, Geurink PP, Wilhelm A, van der Heden van Noort GJ, Ovaa H, Müller S, Knobeloch KP, Rajalingam K, Schulman BA, Cinatl J, Hummer G, Ciesek S, Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature, 2020; 587(7835):657–2. CrossRef
Srikanth L, Sarma PVGK. Andrographolide binds to spike glycoprotein and RNA-dependent RNA polymerase (NSP12) of SARS-CoV-2 by in silico approach: a probable molecule in the development of anti-coronaviral drug. J Genet Eng Biotechnol, 2021; 19(1). CrossRef
Sukardiman, Ervina M, Fadhil Pratama MR, Poerwono H, Siswodihardjo S. The coronavirus disease 2019 main protease inhibitor from Andrographis paniculata (Burm. f) Ness. J Adv Pharm Technol Res. 2020; 11(4):157–62. CrossRef
Swaminathan K, Karunakaran KN, Vidyalakshmi S. SARS-CoV2 multiple target inhibitors from Andrographis paniculata: an in-silico report. Eur J Mol Clin Med, 2021; 08(03):1653–85.
Verma D, Mitra D, Paul M, Chaudhary P, Kamboj A, Thatoi H, Janmeda P, Jain D, Panneerselvam P, Shrivastav R, Pant K, Das Mohapatra PK. Potential inhibitors of SARS-CoV-2 (COVID 19) proteases PLpro and Mpro/ 3CLpro: molecular docking and simulation studies of three pertinent medicinal plant natural components. Curr Res Pharmacol Drug Discov, 2021; 2:100038. CrossRef
Vijayakumar M, Janani B, Kannappan P, Renganathan S, Al-Ghamdi S, Alsaidan M, Abdelaziz MA, Peer Mohideen A, Shahid M, Ramesh T. In silico identification of potential inhibitors against main protease of SARS-CoV-2 6LU7 from Andrographis panniculata via molecular docking, binding energy calculations and molecular dynamics simulation studies. Saudi J Biol Sci, 2022; 29(1):18–29. CrossRef
Vincent S, Arokiyaraj S, Saravanan M, Dhanraj M. Molecular docking studies on the anti-viral effects of compounds from Kabasura Kudineer on SARS-CoV-2 3CLpro. Front Mol Biosci, 2020; 7:613401. CrossRef
Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B, 2020; 10(5):766–88. CrossRef