INTRODUCTION
Most cytotoxic anticancer agents are water-insoluble and unstable and have low bioavailability (Liu et al., 2008). Their cancer therapy is limited by resistance, lack of specificity, and the harsh side effects of irradiation and chemotherapy (Mansoori et al., 2017). At the same time, nanomedicine improves hydrophobic drug aqueous solubility and reduces medication toxicity (Aljabali et al., 2022; Ferrari, 2005; Khater et al., 2021; Nsairat et al., 2021).
Mebendazole (MBZ; methyl N-(6-benzoyl-1H-benzimidazol-2-yl) carbamate) (Fig. 1A) is a first-line medication approved by the Food and Drug Administration for parasite infections (Keystone and Murdoch, 1979; Panic et al., 2014). MBZ (Vermox, generic name) is most often used for intestinal nematode infections (ascariasis, hookworm infections, trichuriasis, strongyloidiasis, and enterobiasis) and intestinal tapeworm infections (taeniases and hymenolepiasis) (Chai et al., 2021). MBZ’s low water solubility and class II status limit its therapeutic usage with decreased bioavailability (De La Torre-Iglesias et al., 2014). In vivo and in vitro cytotoxicity against human lung cancer cells was reported in 2002, where MBZ increased apoptosis dosage time dependently (Mukhopadhyay et al., 2002). MBZ’s antiproliferative efficacy against glioblastoma (Bai et al., 2011; De Witt et al., 2017), non-small cell lung cancer (NSCLC) cell line (Mudduluru et al., 2016), and gastric cancer (Pinto et al., 2015) is due to its capacity to disrupt microtubule polymerization and permanently decrease glucose absorption (Borgers et al., 1975; De Witt et al., 2017). Also, MBZ stopped tumors from growing and stopped them from spreading to the lungs in rats with thyroid cancer (Williamson et al., 2020).
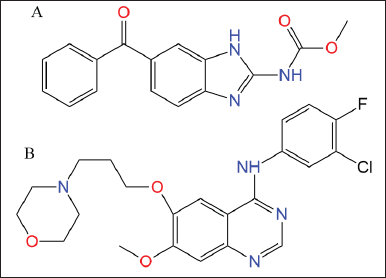 | Figure 1. Chemical structure of (A) MBZ and (B) GEF. [Click here to view] |
Repurposing MBZ as an anticancer drug may enhance cancer treatment (Guerini et al., 2019). MBZ has a favorable and safe profile, pharmacokinetics that enables therapeutic concentrations at the disease site, convenience of administration, and inexpensive pricing. MBZ is cytotoxic and synergizes with ionizing radiation and chemotherapy medicines to stimulate an antitumor immune response (Guerini et al., 2019). Hutchins et al. (2009) and Ren et al. (1994) found that MBZ blocks Ran GTPase, which is needed for the cell cycle to move forward, mitotic spindle formation, and cell growth.
Ran, a Ras-family G-protein, transfers macromolecules between the cytoplasm and nucleus (Rojas et al., 2012). Ran GTP is necessary for cell cycle, mitotic spindles, and cell proliferation (Hutchins et al., 2009; Ren et al., 1994). Matchett et al. (2014) found that Ran GTP overexpression promotes tumor invasion in ovarian, renal, and human lung carcinoma cells, as well as in a model of breast cancer. Ran GTP is also overexpressed in tumor-derived cell lines and tumor tissue (El-Tanani et al., 2016). By targeting Ran-binding proteins or silencing Ran, cancer cells can be eliminated (Boudhraa et al., 2020).
Gefitinib (GEF; N-(3-chloro-4-fluorophenyl)-7-methoxy-6-(3-morpholinopropoxy) quinazolin-4-amine) is an orally active first-generation tyrosine kinase inhibitor (Zhang, 2016). It showed slow oral absorption and was highly pH dependent (Nurwidya et al., 2016). After platinum- and docetaxel-based chemotherapies failed, GEF was approved as first-line treatment for locally advanced NSCLC (Cohen et al., 2003). GEF (ZD1839; AstraZeneca) is the first epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor for unresectable NSCLC (Araki et al., 2012). NSCLC patients treated with GEF exhibited resistance due to a secondary genetic mutation in exon 20 of the EGFR (Wu and Shih, 2018). This mutation activated the PI3K/Akt/mTORC1 and Ras/MEK/ERK pathways. Ran is a possible therapeutic target for malignancies whose mutations/expression alterations correspond with Ras/MEK/ERK pathway activation (Yuen et al., 2012). Ran silencing causes cancer cell death in cells with Ras/MEK/ERK mutations, and targeting Ran can be employed in tandem with other cancer treatments (Yuen et al., 2012).
Due to their biocompatibility, stability, ease of synthesis, and high drug loading and encapsulation efficiencies, liposomal nanocarriers were the first choice to improve lipophilic medicines’ pharmacological properties, water solubility, stability, and/or toxicity (Alshaer et al., 2019; Odeh et al., 2019). Nanoliposomes keep chemically active ingredients from breaking down too quickly in the body’s bloodstream (Nsairat et al., 2022; Odeh et al., 2020; Sercombe et al., 2015).
In this study, we synthesized nanoliposomal formulations encapsulating MBZ and GEFwhich were investigated against the A549 lung cancer cell line. We developed monodispersed nanoliposomes encapsulating MBZ and GEF with outstanding colloidal stability over 1 month and satisfactory encapsulation efficiencies. IC50 values, wound closure, and colony formation assays confirmed the in vitro cytotoxicity of drug-loaded nanoliposomes to block cancer cell proliferation and migration.
In this work, we succeeded in encapsulating MBZ and GEF in nanoliposomes. The new MBZ-loaded nanoliposomes demonstrated promising cytotoxicity against A549 cell lines.
MATERIALS AND METHODS
Chemicals
Sigma (St. Louis, MO) provided MBZ and GEF quinazolin-4-amine. 1,2-Dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and cholesterol (CHOL) were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL). Phosphate buffer saline (PBS) was obtained from Sigma-Aldrich (USA).
Methanol, ethanol, acetonitrile, and n-propanol (HPLC grade) and orthophosphoric acid (85%, AR grade) were purchased from Sigma-Aldrich (USA). Deionized water, ion-free water, and Millipore-0003 direct Q-5-UV from Millipore, USA. All other chemicals and solvents were of analytical grade. All reagents and chemicals were used without further treatment.
Cells
Human adenocarcinoma alveolar basal epithelial cell lines (A549) were acquired from the ECACC catalog no. 86012804 passage no. 18. The following materials and kits were used in this study: Promega kit for 3[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay (USA), dimethyl sulfoxide (DMSO) (GCC-UK), alcohol 70%, fetal bovine serum (FBS) (Euroclone, Italy), L-glutamine (Euroclone, Italy), penicillin/streptomycin (Euroclone, Italy), and Dulbecco’s Modified Eagle Medium-high glucose (DMEM-high glucose).
Preparation of drug-loaded liposomes
Nanoliposomes were prepared using the conventional thin film hydration extrusion technique (Alshaer et al., 2019; Lafi et al., 2021; Zhang, 2017). Briefly, the 65:35 molar ratio of DPPC and CHOL was mixed alone for blank nanoliposomes or with 1 mg of MBZ or GEF dissolved in an appropriate organic solvent. For MBZ, 3.0 ml of chloroform, ethanol, and methanol (2:1:1) were used with heating at 50°C until a clear solution formed, while only 3.0 ml of absolute ethanol (99%) was enough to dissolve the GEF.
The organic solvent(s) was then evaporated under reduced pressure, down to 100 mbar (BUCHI, R-300, Germany) at 30–40 rpm and 45°C–50°C water bath until a dried homogeneous thin film was obtained and stored at −20°C for 24 hours under N2. After 24 hours, the film was hydrated with 3 ml of deionized water and PBS (pH = 7.4) for MBZ and GEF experiments, respectively, with vigorous vertexing every 2–3 minutes (Wisd VM-10 orbital motion). The obtained MBZ and GEF nanoliposomes suspension were frozen and thawed for 12 cycles to enhance drug entrapment and encapsulation into unilamellar nanoliposome vesicles (Costa et al., 2014; Nsairat et al., 2022). The nanovesicles suspension was successively extruded through a polycarbonate membrane (100 nm, Whatman®) using Mini-Extruder (Avanti Polar Lipids, Inc., USA) at 50°C for 13 times to obtain the final nanoliposomes with low polydispersity and the desired size, which were stored at 4°C for further use (Nsairat et al., 2020).
HPLC quantification of MBZ and GEF
The quantification of MBZ and GEF was performed by the HPLC system (Shimadzu Prominence Liquid Chromatography equipped with LC-20AD quaternary solvent delivery system, auto-sampler having a universal loop injector of capacity 1–100 µl, and an SPD-M20A diode array detector monitored between 200 and 350 nm) using reversed phase C-18 column 150 × 4.6 mm, 5 μm (Shimadzu Corporation, Kyoto, Japan) as a stationary phase. The mobile phase for both drugs consists of 130 mM ammonium acetate and acetonitrile (63:37, v/v) adjusted to pH 6 for the MBZ and pH 5 for GEF using glacial acetic acid (Chandrashekara et al., 2014). The mobile phase was provided in an isocratic mode with a flow rate of 1 ml minute−1, detection by UV at 234 and 260 nm for MBZ and GEF, respectively, with an injection volume of 10 μl. All chromatographic conditions were performed at 30°C. The calibration curve was constructed using known concentrations of standard MBZ in the range 1.669–20.534 (Kumar et al., 2008) and 0.5–33.0 µg ml−1 for GEF, respectively (Faivre et al., 2011). The retention times for MBZ and GEF were 5.37 and 3.84 minutes, respectively. MBZ has been quantified using the linear regression equation of Y = 59,209X – 428.47 (n = 3, r2 = 0.9999). The GEF has been quantified using the linearity regression equation Y = 28,637X – 25,998 (n = 3, r2 = 0.9999).
Drug encapsulation efficiency (EE%) was expressed as the percentage of drug encapsulated inside the nanoliposomes with a total drug added and calculated according to
The unloaded drugs (free) were removed using Amicon® filters (cut-off point 10 kDa) (Millipore, Germany), in which 400 µl of liposomal suspension was placed followed by centrifugation at high speed (15,000 rpm, 4°C) using Microcentrifuge (SIGMA 1-16, 100–240 V, 50/60 Hz) for 3 minutes until 100 µl filtrate is reached. Then, the concentrated nanoliposomes were redispersed with 300 µl of deionized water up to the original 400 µl. Finally, the filtrated nanoliposome was placed in Eppendorf tubes and stored at 4°C for further use. The loaded drugs were quantified after liposome disruption using acetonitrile with vigorous stirring for 10 minutes followed by high-speed centrifugation for 30 minutes (Alshaer et al., 2019; Hasan et al., 2019).
Dynamic light scattering (DLS) measurements
The average particle size, polydispersity index (PDI), and zeta potential of drug-loaded liposomes and blank liposomes formulations were measured at 25°C by DLS using nano-ZS (Malvern Instruments, UK) zeta-sizer. An aliquot of 10 µl of liposomes formulation was diluted up to 1.0 ml of deionized water before measurement to yield an appropriate counting rate. All measurements were made in triplicate at room temperature.
Preliminary stability study: the average particle size, PDI, and zeta potential of the drug-loaded liposome and the blank liposome were evaluated immediately after preparation (time = 0) and after storage at 4°C for a period of 4 weeks.
In vitro cytotoxicity assay
The in vitro cytotoxic activities of MBZ and GEF nanoliposomes and nanoliposomes combination compared to their free drugs were investigated against A549 lung cancer cell lines.
A549 cells were cultured in DMEM growth medium (Euroclone®, Italy), which was supplemented with 10% (v/v) FBS and antibiotics (5 ml/500 ml penicillin/streptomycin). Cells were cultured at a confluence of 75%–95% in a humidified 5% CO2 incubator at 37°C. 10 × 103 cells per well were planted in 96-well plates (TPP, Switzerland) (Mosmann, 1983).
After 24 hours, cells were treated with various concentrations of MBZ and GEF nanoliposomes and nanoliposomes combination therapy, as well as with free MBZ (0.1–0.9 μM) and GEF (50–400 nM). After 24, 48, 72, and 96 hours, cell growth was evaluated using the tetrazolium dye (MTT) assay (Horiuchi et al., 1988; Mosmann, 1983). Untreated cells were used as a negative control and as a blank to calibrate the spectrophotometer. After a 1-hour incubation time, the absorbance at 570 nm was determined using a well-plate reader (Elisa reader) (BioTek). The IC50 values were determined using GraphPad Prism version 9 software (GraphPad Software Inc., USA). All experiments were performed in triplicates. Cell vitality % was calculated using Equation (2), where A is the absorbance.
In vitro wound healing assay
To evaluate the cancer cells migration, the in vitro scratch or wound healing assay was used as described by Grimmig et al. (2019). Scratches were created on a confluent cell monolayer, leading cells on the scratch’s edge to migrate toward the center, sealing the scratch and forming new cell-cell connections.
A549 cells were seeded in sterile 6-well cell culture plates (800,000 cells/6 well) and incubated for 24 hours at 37°C with 5% CO2. The next day, a vertical scratch was formed in the center of the A549 monolayer using a sterile 1,000 µl micropipette tip for either free drugs or nanoliposomes treatment. Then, each well was rinsed twice with sterile PBS. After 1 day, cells were treated with MBZ and GEF at concentrations of IC50, 0.5 × IC50, and 2 × IC50 for each free drug, as indicated by the MTT test. Finally, scratch images were taken before and during cell treatment using the phase contrast microscope (model P. MICRO-001, Nikon) equipped with a 4× magnification objective piece. The Motic Images Plus version 2.0 software was used to determine the wound closure area (µm2). DMSO and free media were used as a negative control.
A549 cells were seeded at 200,000 cells/6 wells for the liposome formula, and the vertical scratch was formed using a sterile 1,000 µl micropipette tip. The wound closure rate was observed on day 1 (before treatment) and day 4 after 72 hours of cell therapy (after 72 hours of treatment). Blank nanoliposomes and media were employed as a negative control. All experiments were performed in triplicates.
Colony formation assay
Colony-genic assays were performed in accordance with the published protocol by Franken et al. (2006). A549 cells were seeded on sterile 6-well cell culture plates (3,000 cells/6 well) and incubated overnight at 37°C to ensure adequate plate adhesion. On the next day, cells were treated with IC50, 0.5 × IC50, or 2 × IC50 of free MBZ, GEF doses, or a combination of the two drugs as indicated by the MTT test. Untreated cells were used as a negative control (Fig. 2).
After 24 hours, all treatments were removed and replaced with fresh media every 2 or 3 days over a 12-day period. The colony counting procedure was performed manually using an inverted microscope (Nikon) equipped with a 10× objective lens.
A549 cells were seeded in sterile 6-well cell culture plates (3,000 cells/6 well) for nanoliposomes encapsulated drugs and nanoliposomes combination therapy. Untreated cells and cells treated with blank nanoliposomes were used as a negative control. All treatments were withdrawn after 72 hours, and fresh media replacements were performed every 2 or 3 days for a 12-day period. All experiments were performed in triplicates.
Statistical analysis
All results were displaced as the mean ± standard deviation of three independent experiments. Statistical significance was determined using unpaired two-tailed Student’s t-test with GraphPad Prism version 6 (GraphPad Software Inc., USA). A value of p < 0.05 was considered to indicate a statistically significant difference.
RESULTS AND DISCUSSION
Characterization and stability of the liposomal formulations
DLS measurements revealed that blank nanoliposomes, MBZ and GEF nanoliposomes all had a comparable average particle size and zeta potential distribution (Fig. 3). Blank nanoliposomes have an average size of 153.72 ± 6.21 nm and a PDI of 0.18 ± 0.01. Drug loading into nanoliposomes slightly increased the average particle size up to 176.52 ± 8.98 and 188.32 ± 5.28 nm for MBZ and GEF, respectively, without significantly affecting PDI values (Table 1). The zeta potential of the blank nanoliposomes was −18.46 ± 1.0 mV, while it was −17.00 ± 0.15 and −17.16 ± 0.25 for MBZ and GEF, respectively. DLS measurements over a period of 4 weeks revealed that the nanoparticles were stable in terms of average particle size, zeta potential, and PDI (Table 1).
Encapsulation efficiency
EE% of each drug incorporated into nanoliposomes was determined using Equation (1) utilizing the HPLC calibration curves as indicated in Materials and Methods (Table 2). The EE% of MBZ and GEF was found to be 38.70% ± 1.98% and 55.06% ± 1.98%, respectively.
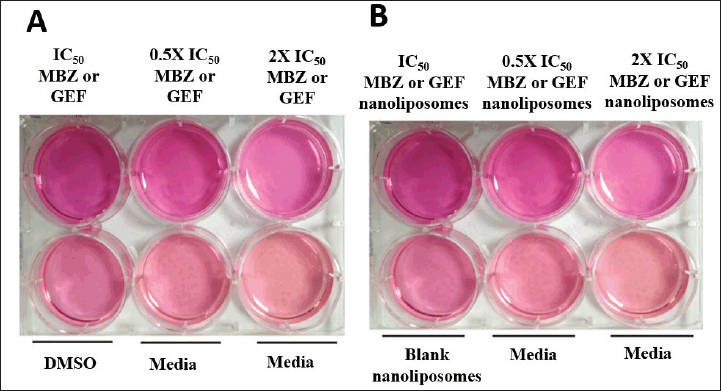 | Figure 2. Colony assay in a 6-well for A549 cell line for (A) free drug treatments and (B) drug-loaded liposomes. [Click here to view] |
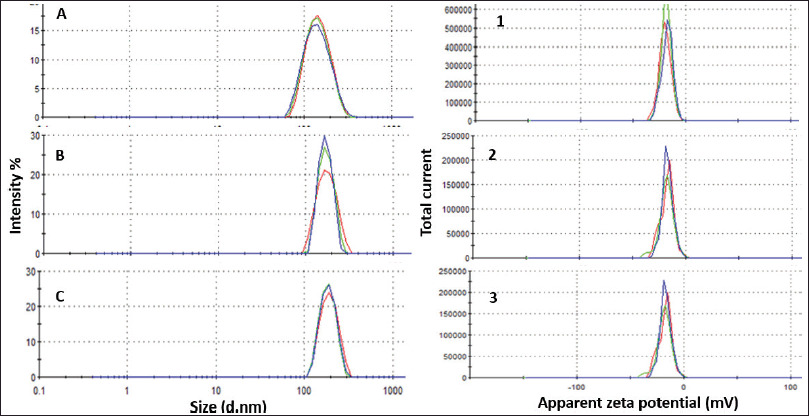 | Figure 3. Particle size distribution of (A) blank liposomes, (B) MBZ-liposomes, and (C) GEF liposomes. Zeta potential distribution for (1) blank liposomes, (2) MBZ-liposomes, and (3) GEF liposomes. [Click here to view] |
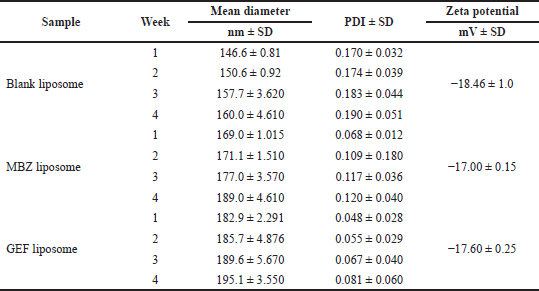 | Table 1. Liposomes characterization and stability: average diameter, PDI, and zeta potential, (mean ± SD, ,n = 3) over a 4-week period. [Click here to view] |
In vitro cytotoxicity assay
Cell viability assay was performed to investigate the cytotoxic activity of MBZ and GEF loaded nanoliposomes and the nanoliposomal combination of both drugs against A549 cells compared to the free drugs utilizing the MTT protocol. After incubation of different drug trreatments for 24 and/or 72 hours against A549 cells, the IC50 values were determined as indicated in Figure 4A for free MBZ and GEF, MBZ- and GEF-loaded nanoliposomes, and the nanoliposomal combination of both drugs. Figure 4B and C also represent the dose-response curves for each treatment against A549 cells.
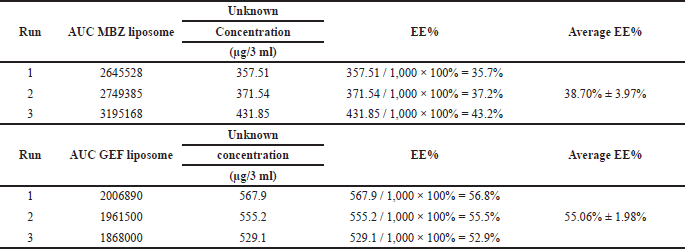 | Table 2. EE of MBZ and GEF calculated from the area-under-the-curve (AUC) measurements of the encapsulated drug using the standard calibration curves. [Click here to view] |
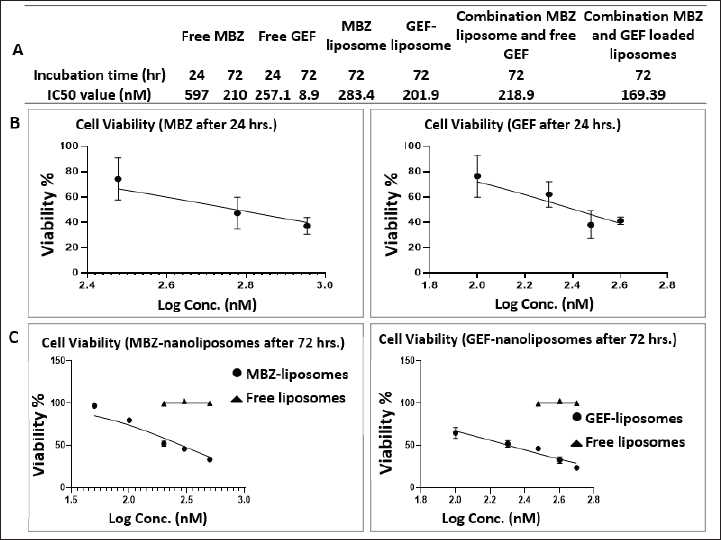 | Figure 4. (A) Table represents the IC50 values. (B and C) Dose-response curves after 24 and 72 hours incubation times for free drugs and drug-loaded nanoliposomes. [Click here to view] |
After 72-hour incubation time, the IC50 was 210, 8.9, 283, 201, and 169 nM for free MBZ and GEF, MBZ- and GEF-loaded nanoliposomes, and the nanoliposomal combination of both drugs, respectively. While the IC50 were 596 and 257 nM for free MBZ and GEF after 72 hours incubation time.
In vitro wound healing assay
The scratch assay is a well-established technique for evaluating cancer cell migration. It is based on the observation of the closure of the initial scratch after treating A549 cells with 0.5 × IC50, IC50, and 2 × IC50 values of free MBZ and GEF, free drugs combination therapy, and nanoliposomes formulations. Scratch images were visualized after 72 hours treatment incubation.
The wound closure percentage for free MBZ and GEF, liposome formulations, and combination therapies are summarized in Figure 5. Compared to free drugs and their combinations, drug-loaded nanoliposomes treatments dramatically inhibited scratch closure after 72 hours. Moreover, the wound closure rate of MBZ-loaded nanoliposomes was 12% for cells treated with the IC50 concentration (210 nM), which is lower than the wound closure rate of blank liposomes and medium. Free GEF represented the greatest wound closure rate of 30% at the IC50 (8.9 nM) concentration for free drug therapy. The nanoliposomes of both drug treatments displaced the lowest wound closure rate of 1.0% for nanoliposome treatment at the IC50 concentration.
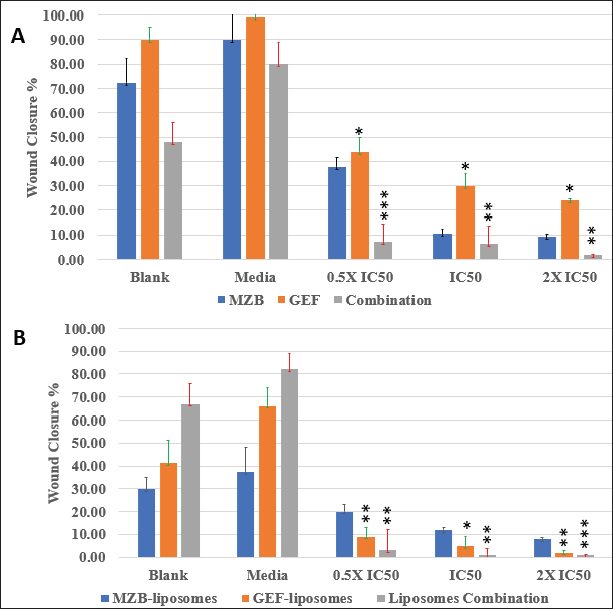 | Figure 5. Wound closure % for different types of drugs treatments against A549 cells after 72 hours: (A) free drug and (B) drug-loaded nanoliposomes. [Click here to view] |
Wound closure microscopic images and area (μm2) calculations of different types of free drug treatments and drug-loaded liposomes were available in the Supplementary Figures S1 and S2.
Colony assay
Colony assay is a frequently used in vitro technique for evaluating cancer cell growth. In addition, this test is utilized to determine the long-term effectiveness of anticancer agents (Franken et al., 2006). As indicated by the scratch assay, the nanoliposomal combination of both drugs was expected to displace the highest cytotoxicity against A549 cell lines with no colony “non-coloniality” was seen starting from the first day up to 12 days of cell treatment. Interestingly, the number of colonies created after cells were treated with either free MBZ or GEF-loaded liposome or free drugs which were almost similar (Supplementary Tables S1 and S2) with a smaller size compared to the cells treated with free drugs (Fig. 6).
DISCUSSION
This is the first study reporting the in vitro cytotoxic investigation of nanoliposomes that encapsulated MBZ alone or in combination with GEF against NSCLC. Average particle size, zeta potential, and PDI of the prepared nanoliposomes were determined using DLS technology. The mean diameter of unilamellar drug-loaded nanoliposomes was less than 200 nm which is small enough to allow passive diffusion into malignant tissue via the enhanced permeability and retention effect of the leaky and highly permeable tumor blood vessels. In addition, our nanoformulations were monodispersed with a PDI less than 0.2 and showed a suitable negative charge of −18.46 ± 1.0 mV, while it was −17.00 ± 0.15 and −17.16 ± 0.25 for MBZ and GEF, respectively, and mV maintains strong repulsion and enhanced nanoliposomes stability which is consistent with previously generated liposomes with high stability (Haeri et al., 2014; Rasmussen et al., 2020; Smith et al., 2017). Comparing the low EE% of most hydrophobic drugs into the small-sized (5–10 nm) nanoliposomes membrane bilayer, we reached a remarkable EE% of both MBZ and GEF of 38.70% ± 1.98% and 55.06% ± 1.98%, respectively, even with the bad solubility of MBZ in most of the low boiling point organic solvents.
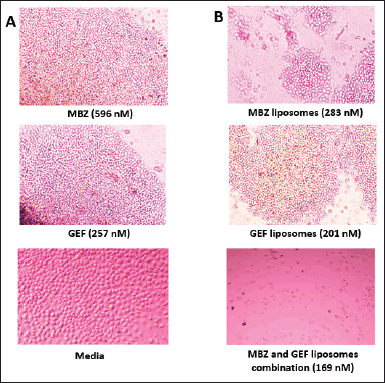 | Figure 6. Colony formation in A549 cells. (A) MBZ, GEF, and Media. (B) MBZ and GEF loaded liposomes and their liposomal combinations. [Click here to view] |
The cellular investigations indicated that our prepared blank nanoliposomes are biocompatible and safe compared to the drug-loaded nanoliposomes. Free MBZ and GEF were previously shown to be cytotoxic against A549 cell lines by Varbanov et al. (2017), with IC50 values of 0.65 and 0.4 M for MBZ and GEF, respectively (Ono et al., 2004). Our cell viability MTT assay demonstrated that the MBZ-loaded nanoliposome had a much greater cytotoxic impact against A549 lung cancer cells (IC50 = 283.4 nM after 72 hours incubation) than the free drug (IC50 = 596.5 nM after 24 hours incubation). The same result was obtained with free GEF (IC50 = 257.1 nM after 24 hours incubation) compared to the GEF-loaded nanoliposomes (IC50 = 201.9 nM after 72 hours incubation). Interestingly, both combinations of MBZ- and GEF-loaded nanoliposomes showed a higher cytotoxicity after 72 hours of therapy compared to the drug-loaded individual nanoliposomes.
The MTT results verified the enhanced antitumor effect and the synergy of the two drug combinations utilizing nanoliposomes as a suitable drug delivery system. Moreover, nanoliposomes usually mask the indesirable features of both drugs and may enhance their bioavailability and pharmacokinetics. Incubation for 24 hours may not represent the optimal effect time for the drug-loaded nanoliposomes due to the different cellular uptake mechanisms compared to the free drugs (Van Der Koog et al., 2022; Zaleskis et al., 2021).
The wound closure assay findings indicate the continuous action property of the drug-loaded nanoliposomes after 72 hours. The wound closure rate was much slower for loaded nanoliposomes of both drugs compared to free drugs, indicating that MBZ- and GEF-loaded nanoliposomes were more effective at preventing cancer cell migration than free drugs because of the enhanced physiochemical properties and improved cytotoxicity of the drug-loaded liposomes against A549 cell lines. The best wound closure percentage significant results were obtained after treatment A549 cell lines with IC50 and 2 × IC50 concentrations comparing the free drug with drug-loaded nanoliposomes. Utilizing these doses, a complete stop of cell migration with almost zero wound closure percentage was obtained by both MBZ and GEF drug-loaded nanoliposomes. These results were in good match with the MTT results and confirmed the improved cytotoxicity and synergistic effect of MBZ- and GEF-loaded liposomes against A549 cell lines to prevent lung cancer cell metastasis.
Furthermore, the colony formation assays also confirmed the effectiveness of MBZ- and GEF-loaded liposomes combinations in which no colony “non-coloniality” was observed starting from the first day up to 12 days of cell treatment. Despite the similarity of the number of counted colonies in both cells treated with either of the free drugs compared to the individual cases of drug-loaded nanoliposomes, still free MBZ and MBZ-loaded nanoliposomes showed significant decrease in the colony formation after 12 days compared to GEF experiments. This may be attributed to the effective of nanoliposomal formulations encapsulating MBZ alone or better if merged with proven GEF anticancer therapy that may lead to resensitization of the GEF effect against A549 cells.
CONCLUSION
In this study, we succeeded in encapsulating GEF, a first-line therapy for lung cancer, and MBZ, a new promising Ran GTPase inhibitor, inside conventional nanoliposomes to increase their anticancer cytotoxic impact against A549 and to resensitize the efficacy of GEF. The nanoliposomes were synthesized using the thin film hydration extrusion technique, and their average size, PDI, and zeta potential were determined and manufactured in a proper nanosized and high stable formula over 1-month storage period at 4°C (Hasan et al., 2014). The cellular investigation results demonstrated the effectiveness and the synergistics of the prepared drug-loaded nanoliposomes against A549 cell lines compared to free drug precursors after 72 hours with the superiority of the two drug combinations loaded into nanoliposomes. Both the wound closure test and the colony assay showed that the liposomal formulation is better than that of the free drugs.
Based on these data and the findings of our study, we hypothesize that the nanoliposomal form of MBZ may enhance the cytotoxic impact of GEF against lung cancer cell line and function in a synergistic way as an effective antimetastatic drug through MBZ inhibitory action against Ran GTPase. Further studies and tests may need to be conducted to evaluate the effectiveness of the role of MBZ-loaded liposomes in Ran GTPase inhibition.
ACKNOWLEDGMENTS
Special thanks and appreciation to Ms. Razan Madi and Ms. Rahmah Khirfan for their valuable help and support in the cell culture work.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Aljabali AAA, Obeid MA, Bakshi HA, Alshaer W, Ennab RM, Al-Trad B, Al Khateeb W, Al-Batayneh KM, Al-Kadash A, Alsotari S, Nsairat H, Tambuwala MM. Synthesis, characterization, and assessment of anti-cancer potential of ZnO nanoparticles in an in vitro model of breast cancer. Molecules, 2022; 27(6):1827.
Alshaer W, Zraikat M, Amer A, Nsairat H, Lafi Z, Alqudah DA, Al Qadi E, Alsheleh T, Odeh F, Alkaraki A, Zihlif M, Bustanji Y, Fattal E, Awidi A. Encapsulation of echinomycin in cyclodextrin inclusion complexes into liposomes: in vitro anti-proliferative and anti-invasive activity in glioblastoma. RSC Adv, 2019; 9(53):30976–88.
Araki T, Yashima H, Shimizu K, Aomori T, Hashita T, Kaira K, Nakamura T, Yamamoto K. Review of the treatment of non-small cell lung cancer with gefitinib. Clin Med Insights Oncol, 2012; 6:CMO.S7340.
Bai R-Y, Staedtke V, Aprhys CM, Gallia GL, Riggins GJ. Antiparasitic mebendazole shows survival benefit in 2 preclinical models of glioblastoma multiforme. Neur Oncol, 2011; 13(9):974–82.
Borgers M, De Nollin S, De Brabander M, Thienpont D. Influence of the anthelmintic mebendazole on microtubules and intracellular organelle movement in nematode intestinal cells. Am J Vet Res, 1975; 36(08):1153–66.
Boudhraa Z, Carmona E, Provencher D, Mes-Masson A-M. Ran GTPase: a key player in tumor progression and metastasis. Front Cell Dev Biol, 2020; 8:345.
Chai J-Y, Jung B-K, Hong S-J. Albendazole and mebendazole as anti-parasitic and anti-cancer agents: an update. Korean J Parasitol, 2021; 59(3):189–225.
Chandrashekara KA, Udupi A, Reddy CG. Separation and estimation of process-related impurities of gefitinib by reverse-phase high-performance liquid chromatography. J Chromatogr Sci, 2014; 52(8):799–805.
Cohen MH, Williams GA, Sridhara R, Chen G, Pazdur R. FDA drug approval summary: gefitinib (ZD1839) (Iressa) tablets. Oncologist, 2003; 8(4):303–6.
Costa AP, Xu X, Burgess DJ. Freeze-Anneal-Thaw cycling of unilamellar liposomes: effect on encapsulation efficiency. Pharm Res, 2014; 31(1):97–103.
De La Torre-Iglesias PM, García-Rodriguez JJ, Torrado G, Torrado S, Torrado-Santiago S, Bolás-Fernández F. Enhanced bioavailability and anthelmintic efficacy of mebendazole in redispersible microparticles with low-substituted hydroxypropylcellulose. Drug Des Devel Ther, 2014; 8:1467–79.
De Witt M, Gamble A, Hanson D, Markowitz D, Powell C, Al Dimassi S, Atlas M, Boockvar J, Ruggieri R, Symons M. Repurposing mebendazole as a replacement for vincristine for the treatment of brain tumors. Mol Med, 2017; 23:50–6.
El-Tanani M, Dakir El H, Raynor B, Morgan R. Mechanisms of nuclear export in cancer and resistance to chemotherapy. Cancers (Basel), 2016; 8:3.
Faivre L, Gomo C, Mir O, Taieb F, Schoemann-Thomas A, Ropert S, Vidal M, Dusser D, Dauphin A, Goldwasser F, Blanchet B. A simple HPLC-UV method for the simultaneous quantification of gefitinib and erlotinib in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci, 2011; 879(23):2345–50.
Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer, 2005; 5(3):161–71.
Franken NAP, Rodermond HM, Stap J, Haveman J, Van Bree C. Clonogenic assay of cells in vitro. Nat Protoc, 2006; 1(5):2315–9.
Grimmig R, Babczyk P, Gillemot P, Schmitz K-P, Schulze M, Tobiasch E. Development and evaluation of a prototype scratch apparatus for wound assays adjustable to different forces and substrates. Appl Sci, 2019; 9(20):4414.
Guerini AE, Triggiani L, Maddalo M, Bonù ML, Frassine F, Baiguini A, Alghisi A, Tomasini D, Borghetti P, Pasinetti N, Bresciani R, Magrini SM, Buglione M. Mebendazole as a candidate for drug repurposing in oncology: an extensive review of current literature. Cancers (Basel), 2019; 11(9):1284.
Haeri A, Alinaghian B, Daeihamed M, Dadashzadeh S. Preparation and characterization of stable nanoliposomal formulation of fluoxetine as a potential adjuvant therapy for drug-resistant tumors. Iran J Pharm Res, 2014; 13(Suppl):3–14.
Hasan M, Iqbal J, Awan U, Xin N, Dang H, Waryani B, Saeed Y, Ullah K, Rongji D, Deng Y. LX loaded nanoliposomes synthesis, characterization and cellular uptake studies in H2O2 stressed SH-SY5Y cells. J Nanosci Nanotechnol, 2014; 14(6):4066–71.
Hasan M, Zafar A, Yousaf M, Gulzar H, Mehmood K, Hassan SG, Saeed A, Yousaf A, Mazher A, Rongji D, Mahmood N. Synthesis of loureirin B-loaded nanoliposomes for pharmacokinetics in rat plasma. ACS Omega, 2019; 4(4):6914–22.
Horiuchi N, Nakagawa K, Sasaki Y, Minato K, Fujiwara Y, Nezu K, Ohe Y, Saijo N. In vitro antitumor activity of mitomycin C derivative (RM-49) and new anticancer antibiotics (FK973) against lung cancer cell lines determined by tetrazolium dye (MTT) assay. Cancer Chemother Pharmacol, 1988; 22(3):246–50.
Hutchins JRA, Moore WJ, Clarke PR. Dynamic localisation of Ran GTPase during the cell cycle. BMC Cell Biol, 2009; 10(1):66.
Keystone JS, Murdoch JK. Mebendazole. Ann Intern Med, 1979; 91(4):582–6.
Khater D, Nsairat H, Odeh F, Saleh M, Jaber A, Alshaer W, Al Bawab A, Mubarak MS. Design, preparation, and characterization of effective dermal and transdermal lipid nanoparticles: a review. Cosmetics, 2021; 8:39.
Kumar S, Chawla G, Sobhia ME, Bansal AK. Characterization of solid-state forms of mebendazole. Pharmazie, 2008; 63(2):136–43.
Lafi Z, Alshaer W, Hatmal MMM, Zihlif M, Alqudah DA, Nsairat H, Azzam H, Aburjai T, Bustanji Y, Awidi A. Aptamer-functionalized pH-sensitive liposomes for a selective delivery of echinomycin into cancer cells. RSC Adv, 2021; 11(47):29164–77.
Liu Z, Robinson JT, Sun X, Dai H. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J Am Chem Soc, 2008; 130(33):10876–7.
Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull, 2017; 7(3):339–48.
Matchett KB, Mcfarlane S, Hamilton SE, Eltuhamy YS, Davidson MA, Murray JT, Faheem AM, El-Tanani M. Ran GTPase in nuclear envelope formation and cancer metastasis. Adv Exp Med Biol, 2014; 773:323–51.
Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods, 1983; 65(1-2):55–63.
Mudduluru G, Walther W, Kobelt D, Dahlmann M, Treese C, Assaraf YG, Stein U. Repositioning of drugs for intervention in tumor progression and metastasis: old drugs for new targets. Drug Resist Updat, 2016; 26:10–27.
Mukhopadhyay T, Sasaki J, Ramesh R, Roth JA. Mebendazole elicits a potent antitumor effect on human cancer cell lines both in vitro and in vivo. Clin Cancer Res, 2002; 8(9):2963–9.
Nsairat H, Khater D, Odeh F, Al-Adaileh F, Al-Taher S, Jaber AM, Alshaer W, Al Bawab A, Mubarak MS. Lipid nanostructures for targeting brain cancer. Heliyon, 2021; 7(9):e07994.
Nsairat H, Khater D, Sayed U, Odeh F, Al Bawab A, Alshaer W. Liposomes: structure, composition, types, and clinical applications. Heliyon, 2022; 8(5):e09394.
Nsairat H, Mahmoud IS, Odeh F, Abuarqoub D, Al-Azzawi H, Zaza R, Qadri MI, Ismail S, Al Bawab A, Awidi A, Alshaer W. Grafting of anti-nucleolin aptamer into preformed and remotely loaded liposomes through aptamer-cholesterol post-insertion. RSC Adv, 2020; 10(59):36219–29.
Nurwidya F, Takahashi F, Takahashi K. Gefitinib in the treatment of nonsmall cell lung cancer with activating epidermal growth factor receptor mutation. J Nat Sci Biol Med, 2016; 7(2):119–23.
Odeh F, Nsairat H, Alshaer W, Alsotari S, Buqaien R, Ismail S, Awidi A, Al Bawab A. Remote loading of curcumin-in-modified β-cyclodextrins into liposomes using a transmembrane pH gradient. RSC Adv, 2019; 9(64):37148–61.
Odeh F, Nsairat H, Alshaer W, Ismail MA, Esawi E, Qaqish B, Bawab AA, Ismail SI. Aptamers chemistry: chemical modifications and conjugation strategies. Molecules, 2020; 25(1):3.
Ono M, Hirata A, Kometani T, Miyagawa M, Ueda S, Kinoshita H, Fujii T, Kuwano M. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol Cancer Ther, 2004; 3(4):465–72.
Panic G, Duthaler U, Speich B, Keiser J. Repurposing drugs for the treatment and control of helminth infections. Int J Parasitol Drugs Drug Resist, 2014; 4(3):185–200.
Pinto LC, Soares BM, Pinheiro Jde J, Riggins GJ, Assumpção PP, Burbano RM, Montenegro RC. The anthelmintic drug mebendazole inhibits growth, migration and invasion in gastric cancer cell model. Toxicol In Vitro, 2015; 29(8):2038–44.
Rasmussen MK, Pedersen JN, Marie R. Size and surface charge characterization of nanoparticles with a salt gradient. Nat Commun, 2020; 11(1):2337.
Ren M, Coutavas E, D’eustachio P, Rush MG. Effects of mutant Ran/TC4 proteins on cell cycle progression. Mol Cell Biol, 1994; 14(6):4216–24.
Rojas AM, Fuentes G, Rausell A, Valencia A. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J Cell Biol, 2012; 196(2):189–201.
Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK, Hua S. Advances and challenges of liposome assisted drug delivery. Front Pharmacol, 2015; 6:286.
Smith MC, Crist RM, Clogston JD, Mcneil SE. Zeta potential: a case study of cationic, anionic, and neutral liposomes. Anal Bioanal Chem, 2017; 409(24):5779–87.
Van Der Koog L, Gandek TB, Nagelkerke A. Liposomes and extracellular vesicles as drug delivery systems: a comparison of composition, pharmacokinetics, and functionalization. Adv Healthcare Mater, 2022; 11(5):2100639.
Varbanov HP, Kuttler F, Banfi D, Turcatti G, Dyson PJ. Repositioning approved drugs for the treatment of problematic cancers using a screening approach. PLoS One, 2017; 12(2):e0171052.
Williamson T, Mendes TB, Joe N, Cerutti JM, Riggins GJ. Mebendazole inhibits tumor growth and prevents lung metastasis in models of advanced thyroid cancer. Endocr Relat Cancer, 2020; 27(3):123–36.
Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer, 2018; 17(1):38.
Yuen HF, Chan KK, Grills C, Murray JT, Platt-Higgins A, Eldin OS, O’byrne K, Janne P, Fennell D, Johnston PG, Rudland PS, El-Tanani M. Ran is a potential therapeutic target for cancer cells with molecular changes associated with activation of the PI3K/Akt/mTORC1 and Ras/MEK/ERK pathways. Clin Cancer Res, 2012; 18(2):380–91.
Zaleskis G, Garberyte S, Pavliukeviciene B, Krasko JA, Skapas M, Talaikis M, Darinskas A, Zibutyte L, Pasukoniene V. Comparative evaluation of cellular uptake of free and liposomal doxorubicin following short term exposure. Anticancer Res, 2021; 41(5):2363–70.
Zhang H. Three generations of epidermal growth factor receptor tyrosine kinase inhibitors developed to revolutionize the therapy of lung cancer. Drug Des Devel Ther, 2016; 10:3867–72.
Zhang H. Thin-film hydration followed by extrusion method for liposome preparation. Methods Mol Biol, 2017; 1522:17–22.
SUPPLEMENTARY MATERIALS
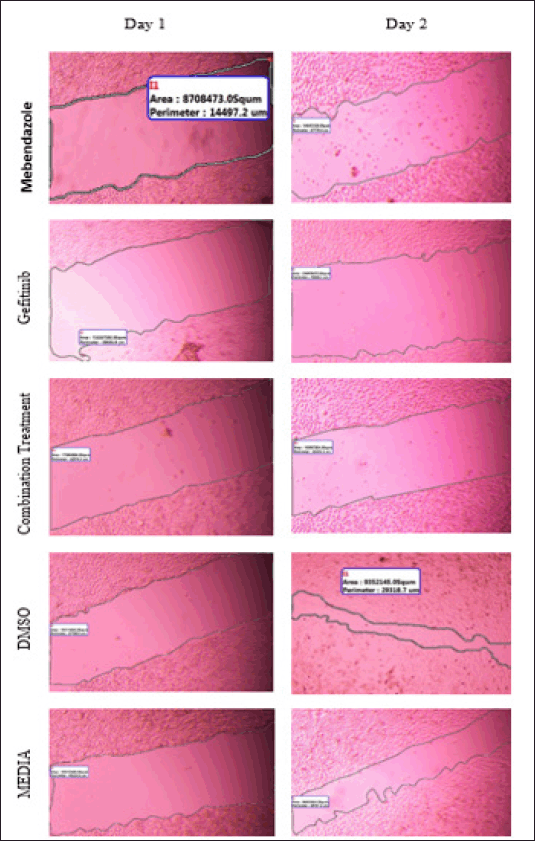 | Supplementary Figure S1. Wound closure area (μm2) for free MBZ, GEF and combination treatment. DMSO and media used as negative control. [Click here to view] |
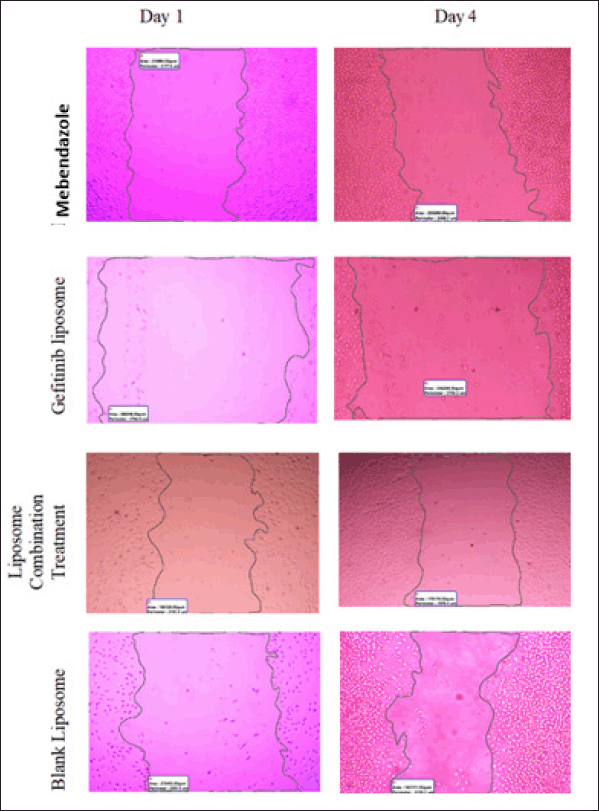 | Supplementary Figure S2. Wound closure area (μm2) for MBZ liposome, GEF liposome and combination treatments. Blank used as negative control. [Click here to view] |
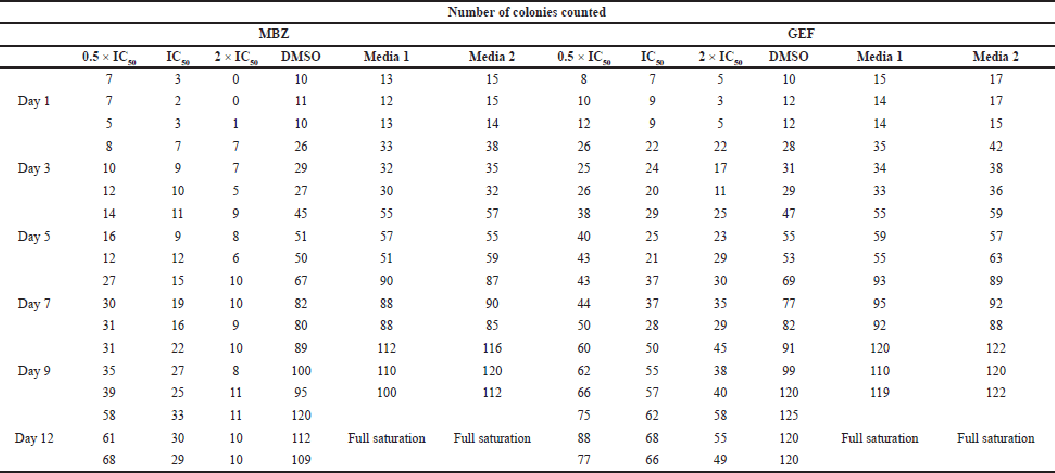 | Supplementary Table S1. Number of counted colonies for free MBZ and GEF. [Click here to view] |
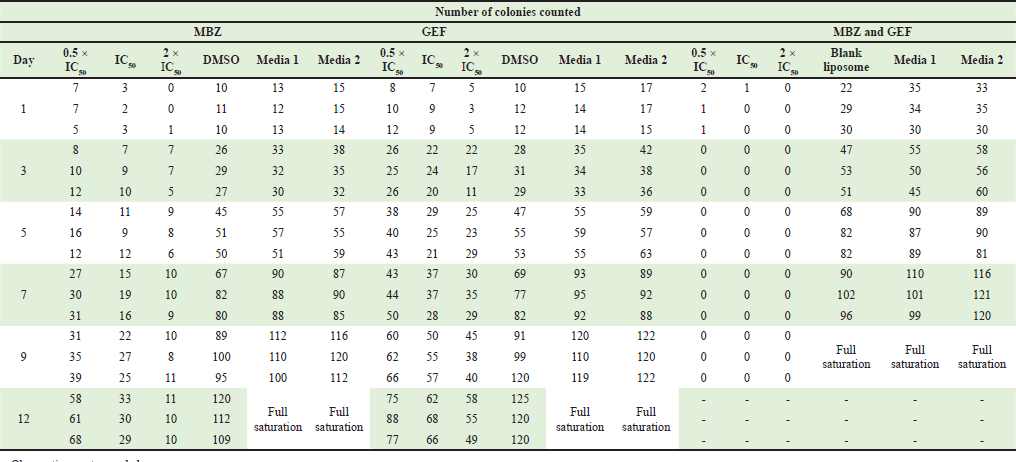 | Supplementary Table S2. Number of counted colonies for nanoliposomes loaded with MBZ, GEF and combination of MBZ and GEF. [Click here to view] |