1. INTRODUCTION
Bloodstream infection (BSI) is a significant cause of morbidity and mortality in hospitalized patients. The incidence numbers vary substantially between patients, as affected by intact or impaired defense mechanisms, pathological states, and pharmacological conditions (Viscoli, 2016). The European Antimicrobial Resistance Surveillance System reports an absolute increase of 47% in BSI between 2002 and 2008 (De Kraker et al., 2013). Other sources report more than 24,000 cases of BSI at 49 US hospitals over 7 years (Wisplinghoff et al., 2004). Deaths associated with sepsis have been estimated to be around 5 million cases in high-income countries (Fleischmann et al., 2016), and potentially higher numbers may be expected in low-income countries.
The most frequent pathogenic agents of bacteremia are Staphylococcus spp., Streptococcus spp., Enterobacter spp., Escherichia coli, Klebsiella pneumoniae, and Pseudomonas spp. (Al-Sweedan et al., 2012; Cisterna et al., 2001; Reimer et al., 1997). A study conducted in 2001 at Irbid, Jordan, documented that the predominant pathogens isolated from blood specimens were Gram-positive bacteria, mainly Streptococcus pneumoniae and coagulase-negative Staphylococcus spp. The Gram-negative bacteria were mainly E. coli, K. pneumoniae, Haemophilus influenzae, Neisseria meningitidis, and Yersinia spp. (Nimri et al., 2001).
Of great concern in the management of sepsis is the ever-growing resistance to antibiotic agents, predicting poor prognosis and posing a challenge on treatment. In Gram-negative bacteria, the production of carbapenemases and extended-spectrum ß-lactamases account for the most important causes of resistance, leading to treatment failure and further spread of resistance. For example, mortality rates of BSI caused by carbapenem-resistant K. pneumoniae can range between 30% and 60% and may approach 50% if colistin resistance is accompanied (Giacobbe et al., 2015). Carbapenem-resistant Pseudomonas aeruginosa (P. aeruginosa) and Acinetobacter baumannii complex (A. baumannii cpx) have been reported to show mortality rates of 72% and 70%, respectively (Lee et al., 2014, 2017). In Gram-positive agents, methicillin resistance represents the most concerning resistance factor that alters therapy. As many BSI are caused by Staphylococcus aureus (S. aureus), therapeutic choices are often reduced to vancomycin in methicillin-resistant S. aureus (MRSA), requiring additional monitoring to avoid toxic side effects. Although genetically coded resistance mechanisms are the main causes of antibiotic resistance, bacterial biofilm formation also shelters pathogens from therapeutic agents, making therapy regimens ineffective. Bacterial tolerance to antibiotics has been noticed to increase compared to the planktonic version of the cells (Spoering and Lewis, 2011). These are often formed on indwelling devices such as catheters or ventilators and might require the withdrawal of the device in case of being contaminated with bacterial biofilms, further complicating the therapy strategy.
As patterns of pathogens causing BSI change over the years, frequent monitoring is required for each region to provide up-to-date data to adjust infection control measures. In this work, we attempt to determine the current prevalence state of the bacteria causing BSI at King Abdullah University Hospital (KAUH), a private/referral clinical setting in Irbid, and representative for the northern region of Jordan. We assess their antimicrobial susceptibilities, along with the underlying genetic elements. We also assess their ability to form biofilms and determine minimum biofilm eradication concentration (MBEC) values in biofilm-forming strains.
MATERIALS AND METHODS
Bacterial isolation and identification
Two hundred bacterial isolates were collected in a 3-month period from positive blood cultures at the KAUH in Irbid, Jordan. The isolates were identified using the Vitek2 COMPACT BioMerieux system. Briefly, a 0.5 McFarland (MF) suspension of bacterial isolates was prepared using a 0.45% NaCl saline solvent. Bacterial suspensions were identified using the GN-ID and GP-ID cards to identify Gram-negative and Gram-positive isolates.
Antimicrobial susceptibility testing
Kirby-Bauer disc diffusion
As previously described, antimicrobial susceptibility testing was performed using the Kirby–Bauer disc diffusion method (Schwalbe et al., 2007). Briefly, Mueller–Hinton plates were seeded with 0.5 MF bacterial suspensions, followed by the addition of antimicrobial discs. Upon incubation at 37°C for 24 hours, the inhibition zones were evaluated according to the 2018 CLSI breakpoints (Wayne, 2018). Antimicrobial agents tested included erythromycin (15 μg), gentamycin (10 μg), levofloxacin (5 μg), penicillin (10 μg), oxacillin (1 μg), ampicillin (10 μg), piperacillin/tazobactam (100/10 μg), meropenem (10 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), clindamycin (2 μg), rifampicin (5 μg), doxycycline (30 μg), vancomycin (30 μg), cephazolin (30 μg), cefoxitin (30 μg), ceftazidime (30 μg), chloramphenicol (30 μg), azithromycin (15 μg), and colistin (10 μg) (Oxoid, Basingstoke, UK).
Minimum inhibitory concentration (MIC) via e-test
Isolates exhibiting strong biofilm formation in the quantitative method were included in this test. Antibiotic MICs were determined using graded antibiotic strips (e-tests), as previously described (Gontijo et al., 2017). Briefly, bacterial suspensions were prepared at a concentration of 0.5 MF and seeded on a Mueller–Hinton agar plate. The plates were dried for 15 minutes at room temperature, followed by the addition of the e-test. These were allowed to adsorb to the agar, and plates were incubated at 37°C for 24 hours. The MIC was determined as the lowest antibiotic concentration at which no bacterial growth was observed. Antibiotics tested included gentamycin, levofloxacin, piperacillin/tazobactam, meropenem, trimethoprim/sulfamethoxazole, rifampicin, doxycycline, vancomycin, chloramphenicol, and azithromycin.
MIC via broth microdilution
The MIC was determined using the microbroth dilution method described previously (Wiegand et al., 2008). The final reaction volume was set at 200 μl, and antibiotics tested include erythromycin, ceftazidime, cefazolin, and colistin.
Biofilm production
Qualitative biofilm formation testing using Congo red agar (CRA)
The isolates were tested for biofilm production using CRA (Mariana et al., 2009). Briefly, CRA was composed of 37% brain heart infusion broth, 50% sucrose, 10% agar, and 0.8% Congo red stain. The bacteria isolates were streaked onto CRA and incubated at 37°C for 24 hours. Isolates growing as black or Bordeaux-colored colonies were classified as biofilm formers, while those growing as red or pink colonies were classified as biofilm nonformers. Dark-colored colonies lacking dry crystalline morphology were classified as medium biofilm formers.
Quantitative biofilm formation testing using a microtiter plate assay
Quantitative evaluation of biofilm formation was determined using crystal violet as previously described (Mathur et al., 2006; O’Toole, 2011). Briefly, fresh bacterial suspensions were prepared in trypticase soy broth and 1% glucose. About 200 μl of a 0.5 MF dilution was added to a microtiter plate and incubated at 37°C for 24 hours. The uninoculated broth was used as a negative control. Upon incubation, plates were inverted and washed thoroughly with phosphate buffer saline (PBS). Biofilms were stabilized with 200 μl 30% acetic acid for 15 minutes and stained with 0.1% crystal violet for 30 minutes. On washing, the stain was solubilized with 200 μl 95% ethanol, and absorbance was read at OD550. The test was carried out in triplicates, and average readings were used to classify biofilm formation. The optical density cut-off value (ODc) was determined as three standard deviations above the mean optical density of the negative control. The interpretation of biofilm formation was according to the following scheme: OD ≤ ODc no biofilm formation, ODc< OD ≤ 2 × ODc weak biofilm formation, 2 × ODc< OD ≤ 4 × ODc moderate biofilm formation, and 4 × ODc< OD strong biofilm formation.
Minimum Biofilm Eradication Concentration (MBEC)
Strong biofilm formers were further evaluated for MBEC using various antimicrobial agents. The Calgary biofilm device was used for this assay (Feng et al., 2013; Schwering et al., 2013). Briefly, 150 μl of a new 0.5 MF bacterial suspension, prepared in Mueller–Hinton broth (MHB), were incubated in a microtiter plate with a peg lid at 37°C for 18–24 hours, in a humidified atmosphere. Uninoculated MHB was used as a negative control. Upon incubation, the peg lids were washed with PBS and challenged in 200 μl of antimicrobial agents at varying concentrations for 18–24 hours at 37°C. Upon incubation, peg lids were washed with PBS, and the remaining biofilms were allowed to recover in 200 μl MHB for 18–24 hours at 37°C. Plates were subjected to 30–40 kHz for 10 minutes in a sonicator upon recovery. The peg lids were removed, and microtiter plates were incubated at 37°C overnight. MBEC was the lowest antimicrobial concentration revealing no visible bacterial recovery growth (Ceri et al., 1999). The test was performed in triplicates. Antibiotics used to determine MBEC included piperacillin/tazobactam, meropenem, levofloxacin, gentamicin, aztreonam, vancomycin, rifampicin, erythromycin, doxycycline, chloramphenicol, cefazolin, and ceftazidime.
DNA extraction for PCR
Genomic DNA was extracted using the Promega Wizard Genomic DNA Purification Kit. Briefly, 1 ml of fresh bacterial culture was pelleted and resuspended in 480 μl 50 mM EDTA. Staphylococcus aureus strains were pretreated with 60 μl 10 mg/ml lysozyme and 60 μl 10 mg/ml lysostaphin and incubated at 37°C for 45 minutes. Upon pretreatment, extraction was performed according to the manufacturer’s protocol. The extracts were eluted at 100 μl for additional downstream applications.
PCR
Detection of carbapenemase genes
Gram-negative isolates were tested for the presence of KpC, NDM, and VIM carbapenemase genes, in a conventional multiplex PCR, using previously described primer sets (Poirel et al., 2011) (Supplementary 1). The reaction mixture was composed of 12.5 μl 2× i-MAX II master mix (iNtRON Biotechnology, Seongnam-Si, Gyeonggi-d, Korea), 2 μl DNA eluate, 1 μl of each primer at a final concentration of 6.25 pmol/μl, and 4.5 μl nuclease-free water. The PCR program was performed at the following conditions: 10 minutes of initial denaturation at 94°C, 36 cycles of 30 seconds at 94°C, 40 seconds at 52°C, and 50 seconds at 72°C, and a final extension step at 72°C for 5 minutes. Amplicons were detected in a 2% agarose gel electrophoresis at 130 V for 30 minutes.
Detection of extended-spectrum β-lactamase (ESBL) genes
Gram-negative isolates were tested for the presence of the CTX, TEM, and SHV ESBL genes, in a conventional PCR using previously described primer sets (Supplementary 2) (Moghaddam et al., 2012). The reaction mixture was composed of 12.5 μl 2× master mix, 3 μl DNA eluate, 1 μl of each primer at a final concentration of 6.25 pmol/μl, and 7.5 μl nuclease-free water. The PCR program was performed under the following conditions: 5 minutes of initial denaturation at 94°C, 35 cycles of 30 seconds at 94°C; 30 seconds at 50°C, 52°C, and 56°C for blaCTX, blaTEM, and blaSHV, respectively; 30 seconds for blaCTX and blaTEM and 60 seconds for blaSHV at 72°C; and a final extension step for 5 minutes at 72°C. Amplicons were detected in a 2% agarose gel electrophoresis at 130V for 30 minutes.
Detection of resistance genes in S. aureus
Staphylococcus aureus strains were tested for the presence of nuc, ermA, ermB, ermC, mrs A/B, and mecA, in a conventional multiplex PCR, with previously described primer sets (Supplementary 3) (Fasihi et al., 2017). The reaction mixture was composed of 12.5 μl 2× i-MAX II master mix (iNtRON Biotechnology, Seongnam-Si, Gyeonggi-d0, Korea), 2 μl DNA eluate, 0.5 μl of each primer at a final concentration of 10 pmol/μl, and 9.5 μl nuclease-free water. The PCR program was performed at the following conditions: 5 minutes of initial denaturation at 96°C, 30 cycles of 45 seconds at 95°C, 2 minutes at 72°C, and a final extension step at 72°C for 5 minutes. Amplicons were detected in a 2% agarose gel electrophoresis at 130V for 30 minutes.
Statistical analysis
A chi-square test was used to find statistically significant levels of resistance among different bacterial BSIs. A value of p < 0.05 was considered statistically significant.
RESULTS
Bacterial identification
Two hundred bacterial isolates from 200 positive blood cultures were included in this work. About 49.5% of isolates were Gram-positive and belonged to the genera Staphylococcus [6.5% MRSA and 43% coagulase-negative Staphylococci (CoNS)]. The remaining 101 isolates (51%) were Gram-negative bacteria, comprising the species E. coli (28%), K. pneumoniae (13%), P. aeruginosa 5%, Enterobacter cloacae complex (3%), and A. baumannii complex (1.5%) (Fig. 1).
Antimicrobial susceptibilities
While all isolates of P. aeruginosa were susceptible to all tested antibiotics, higher resistance rates were observed in A. baumannii strains, as all were nonsusceptible to gentamicin and ceftazidime (p < 0.05). About 67% of those were nonsusceptible to meropenem and piperacillin/tazobactam. The lactose-fermenting organisms were not susceptible to ampicillin but showed higher susceptibility percentages among the remaining antibiotics tested (Table 1) (Supplementary 4).
All isolated Gram-positive organisms were susceptible to vancomycin, doxycycline, and chloramphenicol but were tested nonsusceptible to penicillin. All S. aureus strains were tested as nonsusceptible to oxacillin in the double agar diffusion test, thus identifying MRSA (Table 2). CoNS were more resistant to erythromycin than MRSA (p < 0.05).
Multidrug resistance (MDR) was observed in 94 Gram-positive isolates (94.9%), testing as nonsusceptible to three or more antimicrobial agents belonging to different categories, compared to 65 Gram-negative isolates (50.5%).
Biofilm formation
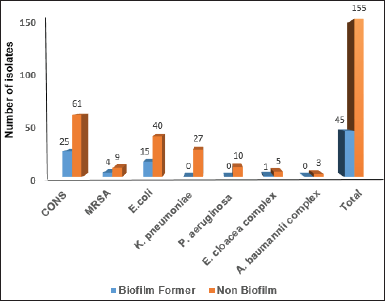
Biofilm formation was not observed in 155 (77.5%) isolates tested using the CRA screening method. Among the remaining 45 biofilm producers (22%), CoNS and E. coli shared the highest rates with 27% and 29%, respectively (Fig. 2).
More detailed results were seen in the quantitative measurements as 31 of all isolates (15.5%) were identified as strong, 32 (16%) as moderate, and 33 (16.5%) as weak biofilm formers. Among the remaining 104 isolates (52%) unable to form biofilms in quantitative testing were mainly CoNS 43.2%, followed by E. coli (36.5%) (Table 3).
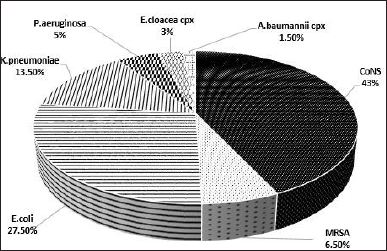 | Figure 1. Percentage of bacteria isolated from BSI. BSI, bloodstream infection; CONS, coagulase-negative Staphylococci; MRSA, methicillin-resistant Staphylococcus aureus. [Click here to view] |
MBEC
Among the 31 isolates, 14 CoNS, 1 S. aureus, 7 K. pneumoniae, 4 P. aeruginosa, 3 E. cloacea cpx, and 2 E. coli strains were quantitatively defined as strong biofilm formers, and assessed for their MBEC. Nine CoNS strains (64% of the CoNS tested) required a vancomycin MBEC ≥ 500 μg/ml while exhibiting susceptible MIC values. Similar results were noticed with chloramphenicol, rifampicin, doxycycline, gentamycin, and levofloxacin, requiring a similar concentration in 5 (35.7%), 3 (21.4%), 3 (21.4%), 1 (7.1%), and 1 (7.1%) isolates, respectively. Only two isolates (14.2%) resistant to gentamycin, required MBEC values ≥ 500 μg/ml. Similarly, high concentrations for biofilm eradication were only noticed with one strain nonsusceptible to levofloxacin (7.1%). All CoNS strains required an MBEC of ≤ 0.5 using erythromycin. All MBEC values ≥ 500 μg/ ml had susceptible MIC values to the corresponding antimicrobial agent (Table 4).
In 42 times, Gram-negative isolates required ≥ 500 μg/ml of piperacillin/tazobactam, meropenem, levofloxacin, gentamicin aztreonam, doxycycline, chloramphenicol, cefazolin, and ceftazidime to eradicate biofilms, while isolates grown as planktonic were susceptible to the corresponding antibiotic agent. Only in nine tests of 3 K. pneumoniae isolates, did a nonsusceptible MIC value corresponded to an MBEC value of ≥ 500 μg/ml using aztreonam and doxycycline (Table 5).
PCR
The prevalence of tested resistance genes in Gram-negative and Gram-positive bacteria is shown in Table 6 and Figure 3.
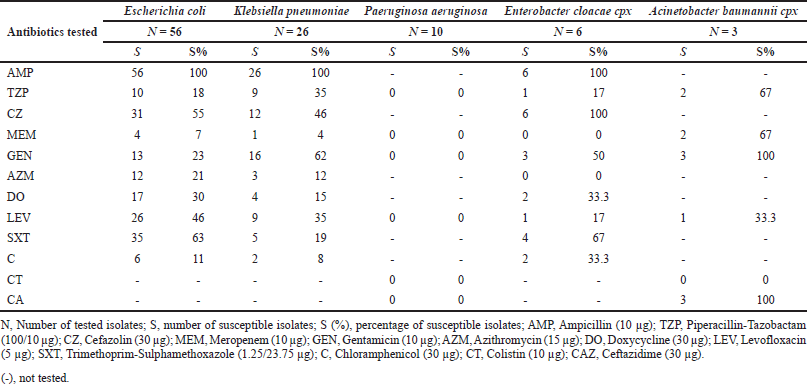 | Table 1. Antimicrobial susceptibilities of Gram-negative isolates. [Click here to view] |
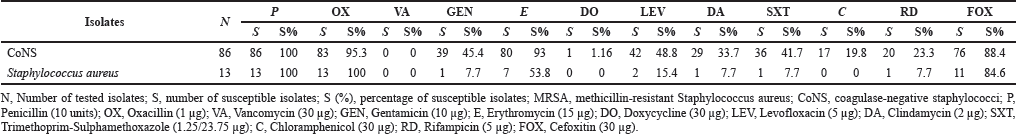 | Table 2. Antimicrobial susceptibility of Gram-positive bacteria isolated from blood. [Click here to view] |
 | Figure 2. Qualitative biofilm formation test using Congo red agar. [Click here to view] |
The ß-lactamase gene detected most frequently was SVH, found in 54 isolates (53.4%), mainly in K. pneumoniae and E. coli, in 21 (20.7%) and 23 (22.7%), respectively. Twenty-four isolates (23.7%) were positive for TEM mainly found in E. coli isolates, and 14 (13.8%) for CTX mainly found in E. cloacae. Carbapenemases were less frequently detected as only 15 isolates (14.8%) tested positive for VIM, mainly K. pneumoniae, followed by KPC and NDM, detected in 9 (8.9%) and 3 (2.9%), respectively. In CoNS, mecA, ermA, ermB, ermC, nuc, and mrsA/B were detected in 40 (46.5%), 11 (12.7%), 3 (3.4%), 24 (27.9%), 12 (13.9%), and 0 isolates, respectively. In MRSA, these were found in 7 (53.8%), 0, 0, 3 (23%), 1 (7.6%), and 0 isolates, respectively.
DISCUSSION
This work included a total sample volume of 200 bacterial species, isolated from positive blood cultures, derived from the KAUH. Gram-negative bacteria were isolated in 50.5% of the total sample size and Gram-positive ones in 49.5%. The most frequently isolated bacterial species were CoNS isolated in 43% of the times. This is consistent with previously described numbers in other regions (Gheybi et al., 2008; Kirchhoff and Sheagren, 1985) and with previous investigations conducted in the same clinical setting between 2003 and 2008, recovering CoNS from positive blood cultures as the most frequent species in 59% of the time (Al-Sweedan et al., 2012). CoNS comprise multiple Staphylococcus species, such as S. epidermidis, S. hominis, S. capitis, S. haemolyticus, and others, and are acquired during sample withdrawal from the skin. Their recovery from blood cultures is often considered a contamination, of no etiological importance than an actual infection, with no further identification and therapy conducted. While this is true for most CoNS species, more attention has to be paid to S. epidermidis, as it is reported to be the causative agent of various bacterial infections, including BSI, especially in patients carrying implant devices (Namvar et al., 2014). Therefore, further identification of S. epidermidis should be considered when isolating CoNS, accurately identifying the underlying cause of infection and allowing for appropriate therapy adjustment. Among the Gram-negative bacteria isolated was E. coli, the most identified species, comprising 27.5% of the total sample size, followed by K. pneumoniae with 13.5%. The most common Gram-negative bacteria recovered from blood cultures used to be K. pneumoniae in this region (Al-Sweedan et al., 2012).
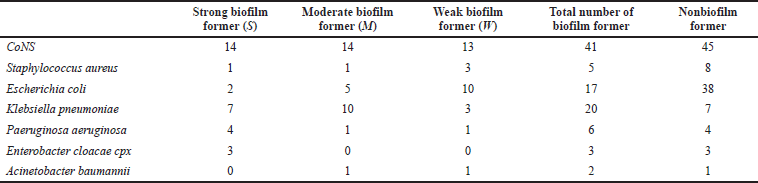 | Table 3. Quantitative biofilm formation test using microtiter plate. [Click here to view] |
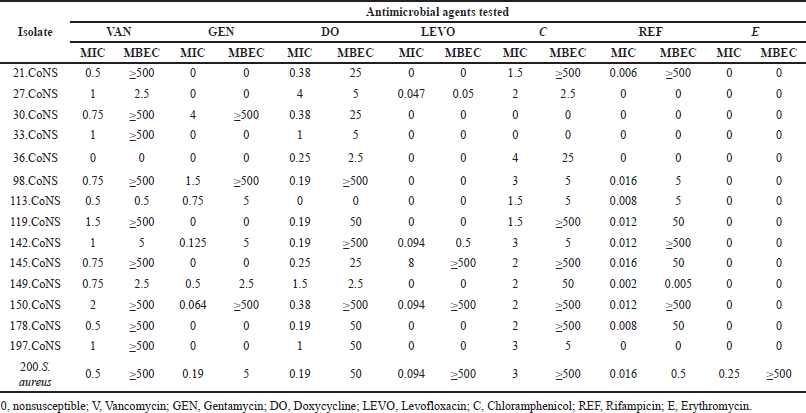 | Table 4. MIC and MBEC results for Gram-positive strong biofilm formers. [Click here to view] |
Antimicrobial susceptibility profiles revealed that piperacillin-tazobactam was effective in 64.3% of Gram-negative isolates. Chloramphenicol was effective in 69.3%, and azithromycin in 70.2% of these bacilli. Meropenem, empirical therapy for infections caused by Gram-negative bacteria, was effective in 92% of these isolates. Ampicillin, on the other hand, was ineffective in all lactose-fermenting isolates. Particular attention was paid to colistin, as global resistance concerns to this antibiotic have been reported in various regions (Zafer et al., 2019). In our sample size, no resistance to colistin was observed in any of the nonfermenting lactose isolates. An alarming concern was the observed resistance to methicillin in 100% of S. aureus strains since previous values had been 36% in the same hospital during the past decade (Al-Sweedan et al., 2012). The reason for optimism was given by the susceptibilities observed to vancomycin and doxycycline, as all MRSA isolates were susceptible to these agents. MDR, on the other hand, defined according to previously set criteria (Magiorakos et al., 2012), was observed in 64.3% of all Gram-negative isolates, along with 94.9% in Gram-positive isolates. Numbers that come in concordance with previous reports in which ESBL-producing Enterobacteriaceae were the most common MDR, followed by MRSA (Capsoni et al., 2019).
When examining the underlying resistance genes in MRSA, 53.8% harbored the mecA gene, responsible for the production of penicillin-binding protein 2a, conferring resistance to methicillin. Methicillin resistance in the remaining MRSA isolates could be driven by the commonly reported homolog of mecA and mecC (García-Álvarez et al., 2011; Shore et al., 2011), emphasizing the need to screen for both genetic elements when defining MRSA. Among the resistance genes found in Gram-negative isolates, ß-lactamases were more frequently found than carbapenemases, with blaSHV being the predominant gene found in 53.4% of tested species, followed by blaTEM in 23.7% of these isolates. An outcome achieved from other countries such as Saudi Arabia, South India, China, and Korea listed blaTEM as the predominant ß-lactamase gene found (Al Sheikh et al., 2014; Men et al., 2013; Rameshkumar et al., 2016; Shin et al., 2009). Among the carbapenemases detected was blaVIM, the most frequent gene detected in 14.8% of all Gram-negative isolates, followed by blaKPC seen in only 8.9% of these isolates. Although different from other regions where KPC accounts for one of the most carbapenemases found, the lower prevalence of this resistance gene could be explained by our lower incidences of K. pneumoniae, as KPC is found mainly in this species found, the lower prevalence of this resistance gene could be explained by our lower incidences of K. pneumoniae, as blaKPC is found mainly in this species (Han et al., 2020).
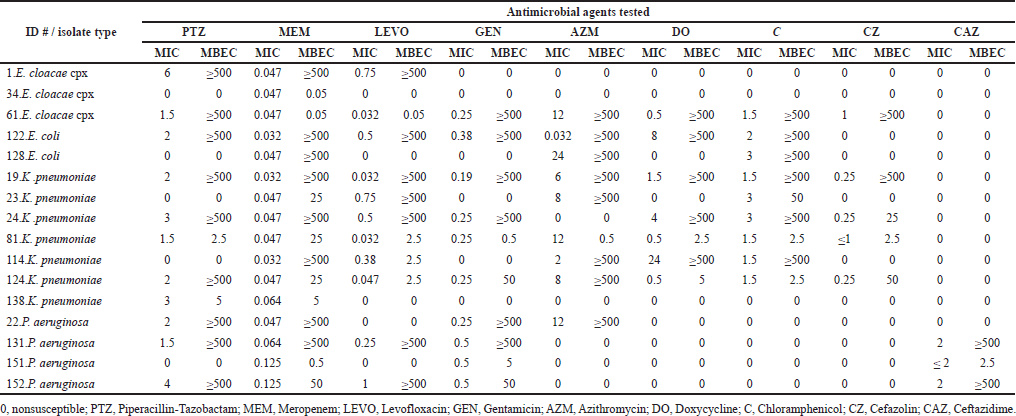 | Table 5. MIC and MBEC results for Gram-negative strong biofilm formers. [Click here to view] |
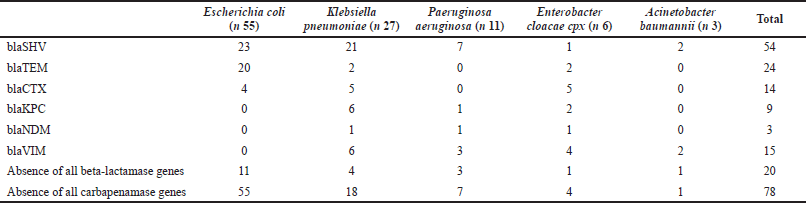 | Table 6. The frequency of beta-lactamase gene and carbapenamases genes among Gram-negative bacteria. [Click here to view] |
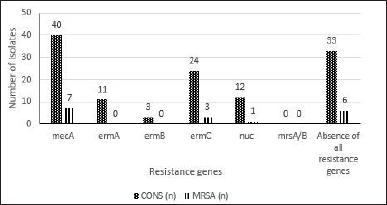 | Figure 3. The frequency of methicillin resistance S.aureus nuc, mecA and mrsA/B and erythromycin and clindamycin-resistant genes ermA, ermB and ermC. [Click here to view] |
Additional resistance due to biofilm production was assessed by determining the ability of all isolates to form biofilms, using quantitative and qualitative methods, with the subsequent measurement of antibiotic concentrations required to eradicate those biofilms. While only 22.5% of biofilm formers were detected quantitatively using CRA, more accurate results were achieved by the microtiter plate method. A result that might be contributed to the media’s nutritious conditions in the microtiter plate method as previously described (Reisner et al., 2006). About 48% were identified as either strong, moderate, or weak biofilm formers with almost equal proportions. CoNS were the predominant species that formed biofilms, followed by E. coli. When comparing the MIC values with MBEC values of biofilm formers, data indicate that concentrations required to eradicate biofilms are higher than the MIC. In 66% of all biofilm-forming Staphylococci, a vancomycin concentration >500 μg/ml was required, despite susceptible MIC values. A similar increase in MBEC values was noticed in Gram-negative isolates in all tested antibiotics. The result is not surprising, as some reports estimate a 10–100-fold concentration of antibiotics needed to eradicate bacteria in biofilms (Ceri et al., 2001).
CONCLUSION
This work provides an up-to-date state on the main causative agents for BSI, emphasizing the ever-growing issue of antibiotic resistance and the therapy challenge posed by biofilm-forming bacteria in a clinical setting sample in Irbid, Jordan. It highlights the need for novel therapy and disinfection strategies to eradicate these organisms and minimize the consequences posed by such pathogens. Although some of our results appeared to be unchanged compared to previous values, others showed some variation in prevalence in different regions and the same clinical setting a decade later. Again, constant monitoring of clinically relevant microorganisms and virulent characteristics is emphasized on, as infection control measures can only be successful with accurate numbers.
FUNDING
This work was supported by the Deanship of Research at Jordan University of Science and Technology, Jordan [grant number: 20190361].
AUTHOR CONTRIBUTIONS
Nid’a Alshraiedeh has substantially contributed to conceptualization, data curation, design of study, analysis and interpretation of the data, funding acquisition, project administration, resources, supervision, and writing the original draft and revising it critically for intellectual contents.
Majd Alzedan has substantially contributed to the design of the work, material preparation, acquisition of data, analysis, interpretation of data, and writing the original draft.
RashaBani Salameh has substantially contributed to analysis, interpretation of data, and writing the original draft.
Rawan Alsharedeh has substantially contributed to the conception, design of the work analysis, interpretation of data, and writing the original draft.
Majed Masadeh has substantially contributed to the conception and critical revision of draft for important intellectual content.
Enas Alrafayah, Toga Zghoul, and Farah Atawneh have substantially contributed to analysis, interpretation of data, and critical revision of the draft for important intellectual content.
All authors have reviewed and approved the final version of the manuscript. In addition, all authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest for this manuscript.
ETHICAL APPROVALS
The study was approved by the institutional review board (IRB) of the Jordan University of Science and Technology and KAUH. Informed consent form was waived by the IRB of the Jordan University of Science and Technology and KAUH (IRB ref: 24/124/2019, date 21.5.2.19). All methods were carried out in accordance with approved guidelines and regulations.
DATA AVAILABILITY
All data generated and analyzed during this study are included in this published article and its supplementary information files.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Al Sheikh YA, Marie MA, John J, Krishnappa LG, Dabwab KH. Prevalence of 16S rRNA methylase genes among β-lactamase-producing Enterobacteriaceae clinical isolates in Saudi Arabia. Libyan J Med, 2014; 9(1):24432.
Al-Sweedan SA, Hyajneh W, Al-Ostath A. Patterns of bacteremia in cancer patient with febrile neutropenia at King Abdullah University Hospital--Jordan 2003--2008. J Pediatr Infect Dis, 2012; 7(1):15–20.
Capsoni N, Bellone P, Aliberti S, Sotgiu G, Pavanello D, Visintin B, Callisto E, Saderi L, Soldini D, Lardera L, Monzani V, Brambilla AM. Prevalence, risk factors and outcomes of patients coming from the community with sepsis due to multidrug resistant bacteria. Multidiscip Respir Med, 2019; 14(1):1–11.
Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol, 1999; 37(6):1771–6.
Ceri H, Olson M, Morck D, Storey D, Read R, Buret A, Olson B. The MBEC assay system: Multiple equivalent biofilms for antibiotic and biocide susceptibility testing. Methods Enzymol, 2001; 337:377–85.
Cisterna R, Cabezas V, Gómez E, Busto C, Atutxa I, Ezpeleta C. 2001. Community-acquired bacteremia. Rev Esp Quimioter, 2001; 14:369–82.
De Kraker ME, Jarlier V, Monen JC, Heuer OE, van de Sande N, Grundmann H. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin Microbiol Infect, 2013; 19(9):860–8.
Fasihi Y, Saffari F, Ghahraman MR, Kalantar-Neyestanaki D. Molecular detection of macrolide and lincosamide-resistance genes in clinical methicillin-resistant Staphylococcus aureus isolates from Kerman, Iran. Arch Pediatr Infect Dis, 2017; 5(1) :e37761.
Feng X, Sambanthamoorthy K, Palys T, Paranavitana C. The human antimicrobial peptide LL-37 and its fragments possess both antimicrobial and antibiofilm activities against multidrug-resistant Acinetobacter baumannii. Peptides, 2013; 49:131–7.
Fleischmann C, Scherag A, Adhikari NK, Hartog CS, Tsaganos T, Schlattmann P, Angus DC, Reinhart K, International Forum of Acute Care Trialists. Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am J Respir Crit Care Med, 2016; 193(3):259–72.
García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RL, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis, 2011; 11(8):595–603.
Gheybi S, Fakour Z, Karamyar M, Khashabi J, Ilkhanizadeh B, Asghari SF, Mahmoudzadeh H, Majlesi AH. Coagulase negative Staphylococcus, the most common cause of neonatal septicemia in Urmia, Iran. Iran J Pediatr, 2008; 18(3):237–43.
Giacobbe DR, Del Bono V, Trecarichi EM, De Rosa FG, Giannella M, Bassetti M, Bartoloni A, Losito AR, Corcione S, Bartoletti M, Mantengoli E, Saffioti C, Pagani N, Tedeschi S, Spanu T, Rossolini GM, Marchese A, Ambretti S, Cauda R, Viale P, Viscoli C, Tumbarello M, ISGRI-SITA (Italian Study Group on Resistant Infections of the Società Italiana Terapia Antinfettiva). Risk factors for bloodstream infections due to colistin-resistant KPC-producing Klebsiella pneumoniae: results from a multicenter case–control–control study. Clin Microbiol Infect, 2015; 21(12):1106–e1.
Gontijo FD, de Melo AT, Pascon RC, Fernandes L, Paes HC, Alspaugh JA, Vallim MA. The role of Aspartyl aminopeptidase (Ape4) in Cryptococcus neoformans virulence and authophagy. PLoS One, 2017; 12(5):e0177461.
Han R, Shi Q, Wu S, Yin D, Peng M, Dong D, Zheng Y, Guo Y, Zhang R, Hu F, China Antimicrobial Surveillance Network (CHINET) Study Group. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol, 2020; 10:314.
Kirchhoff LV, Sheagren JN. Epidemiology and clinical significance of blood cultures positive for coagulase-negative Staphylococcus. Infect Control, 1985; 6(12):479–86.
Lee CH, Su TY, Ye JJ, Hsu PC, Kuo AJ, Chia JH, Lee MH. Risk factors and clinical significance of bacteremia caused by Pseudomonas aeruginosa resistant only to carbapenems. J Microbiol Immunol Infect, 2017; 50(5):677–83.
Lee HY, Chen CL, Wu SR, Huang CW, Chiu CH. Risk factors and outcome analysis of Acinetobacter baumannii complex bacteremia in critical patients. Crit Care Med, 2014; 42(5):1081–8.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect, 2012; 18(3):268–81.
Mariana NS, Salman SA, Neela V, Zamberi S. Evaluation of modified Congo red agar for detection of biofilm produced by clinical isolates of methicillinresistance Staphylococcus aureus. Afr J Microbiol Res, 2009; 3(6):330–8.
Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol, 2006; 24(1):25–9.
Men TY, Wang JN, Li H, Gu Y, Xing TH, Peng ZH, Zhong L. Prevalence of multidrug-resistant gram-negative bacilli producing extended-spectrum β-lactamases (ESBL s) and ESBL genes in solid organ transplant recipients. Transpl Infect Dis, 2013; 15(1):14–21.
Moghaddam MN, Forghanifard MM, Moshrefi S. Prevalence and molecular characterization of plasmid-mediated extended-spectrum β-lactamase genes (balaTEM, blaCTX and blASHV) among urinary Escherichia coli clinical isolates in Mashhad, Iran. Iran J Basic Med Sci, 2012; 15(3):833–9.
Namvar AE, Bastarahang S, Abbasi N, Ghehi GS, Farhadbakhtiarian S, Arezi P, Hosseini M, Baravati SZ, Jokar Z, Chermahin SG. Clinical characteristics of Staphylococcus epidermidis: a systematic review. GMS Hyg Infect Control, 2014; 9(3):Doc23.
Nimri LF, Rawashdeh M, Meqdam MM. Bacteremia in children: etiologic agents, focal sites, and risk factors. J Trop Pediatr, 2001; 47:356–60.
O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp, 2011; (47):2437.
Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis, 2011; 70(1):119–23.
Rameshkumar G, Ramakrishnan R, Shivkumar C, Meenakshi R, Anitha V, Venugopal Reddy YC, Maneksha V. Prevalence and antibacterial resistance patterns of extended-spectrum beta-lactamase producing Gram-negative bacteria isolated from ocular infections. Indian J Ophthalmol, 2016; 64(4):303.
Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J Bacteriol, 2006; 188(10):3572–81.
Reimer LG, Wilson ML, Weinstein MP. Update on detection of bacteremia and fungemia. Clin Microbiol Rev, 1997; 10:444–65.
Schwalbe R, Steele-Moore L, Goodwin AC. Antimicrobial susceptibility testing protocols. CRC Press, Boca Raton, FL, 2007.
Schwering M, Song J, Louie M, Turner RJ, Ceri H. Multi-species biofilms defined from drinking water microorganisms provide increased protection against chlorine disinfection. Biofouling, 2013; 29(8):917–28.
Shin SY, Kwon KC, Park JW, Song JH, Ko YH, Sung JY, Shin HW, Koo SH. Characteristics of aac (6’)-Ib-cr gene in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from Chungnam area. Korean J Lab Med, 2009; 29(6):541–50.
Shore AC, Deasy EC, Slickers P, Brennan G, O’Connell B, Monecke S, Ehricht R, Coleman DC. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother, 2011; 55(8):3765–73.
Spoering AL, Lewis KI. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol, 2001; 183(23):6746–51.
Viscoli C. Bloodstream infections: the peak of the iceberg. Virulence, 2016; 7(3):248–51.
Wayne PA. Clinical and laboratory standards institute, performance standards for antimicrobial susceptibility testing , Clinical and Laboratory Standards Institute, Wayne, PA, 2018.
Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc, 2008; 3(2):163–75.
Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis, 2004; 39(3):309–17.
Zafer MM, El-Mahallawy HA, Abdulhak A, Amin MA, Al-Agamy MH, Radwan HH. Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann Clin Microbiol Antimicrob, 2019; 18(1):1–8.
SUPPLEMENTARY MATERIALS
 | Table 1. Primers used for amplification of carbapenemase genes s (Poirel et al., 2011). [Click here to view] |
 | Table 2. Primers used for amplification of ESBL genes (Moghaddam et al., 2012). [Click here to view] |
 | Table 3. Primers used for amplification of resistance genes in Gram-positive isolates (Fasihi et al., 2017 ). [Click here to view] |