INTRODUCTION
Resistance of bacterial strains to the existing antibiotics causes many problems in the therapy of diseases of bacterial aetiology (Ventola, 2015); the situation can be highly dangerous in the case of such pathogens as Pseudomonas aeruginosa (Khan et al., 2020; Pang et al., 2019; Yakout and Abdelwahab, 2022). It is known that the composition of antibiotics (Jones et al., 2022; Olsson et al., 2020) or their protection from deactivating enzymes using corresponding inhibitors (Brown and Wobst, 2022) is the possible way to solve this problem. The selection of more stable molecules in the existing groups of antibiotics is also a promising strategy to improve the therapy; Plazomicin (Wagenlehner et al., 2019), dalbavancin (Cercenado, 2017), oritavancin (Mitra et al., 2015), and meropenem (El-Gamal, 2011) serve as good examples of such molecules.
One more strategy of antimicrobial drug development, with some problems and limitations (Tommasi et al., 2015), is the development of inhibitors of bacterial protein targets, which could even form a new class of antibiotic or antibacterial agents. One of these attractive targets is bacterial TrmD, which is crucially different from its ortholog of eukaryotes and archaea (Goto-Ito et al., 2017). Blockage of this enzyme leads to +1 frameshifting at the ribosomal translation stage of protein synthesis (Gamper et al., 2015). The disruption of protein synthesis in its turn negatively influences the production of bacterial membrane proteins, such as efflux drugs pumps (Masuda et al., 2019), which leads to the misfunctioning of the outer membrane of Gram-negative bacterial cells (Zhang et al., 2013), can favorize the accumulation of antibiotics and increases their activity (Richter et al., 2017).
Small molecules draw the attention of researchers as candidates for bacterial TrmD inhibitors. Among the most promising heterocyclic systems which are present in the structure of highly active inhibitors, the assemblies of pyrazole with indole were reported (Whitehouse et al., 2019). Compounds with pyrazole heterocyclic pattern modified with 3-indolyl or 2-thienylcaboxamide substituents were identified as leaders in HTS assays aiming at the development of TrmD P. aeruginosa inhibitors (Zhong et al., 2019a). The derivatives of thieno[2,3-d]pyrimidine are one the effective classes of the well-known TrmD inhibitors (Hill et al., 2013), which allowed to disclose differences in inhibitors’ binding mechanisms between Gram-negative tRNA (Guanine37-N1)-methyltransferases from mycobacterial and also their Gram-positive counterparts. The study of thienopyrimidine derivatives complexes with different TrmDs also helped to get a deeper insight into the molecular mechanism of their action (Zhong et al., 2019b). One of the newest results concerning libraries potential of thieno[2,3-d]pyrimidines as TrmD inhibitors is the study that revealed the derivative of 3,5,6,8-tetrahydropyrido[4′,3′:4,5]thieno[2,3-d]pyrimidine modified at position three with 3-indolylthioacetamide substituent as the most active compound (Malasala et al., 2022). Besides, the activity of thienopyrimidines as TrmD inhibitors has also been reported (Badiger et al., 2015; Triloknadh et al., 2021). It is also interesting that thieno[2,3-d]pyrimidin-6-carboxylic acids derivatives are inhibitors of N-acetyltransferases of bacterial polysaccharides, which showed nanomolar inhibitory activity and were active against Campylobacter jejuni PglD (De Schutter et al., 2017).
MATERIALS AND METHODS
Chemistry
All the convenient solvents and commercially available reagents including chromatography grades we used with no additional purification; for additional purification of dimethylformamide molecular sieves 4Å were applied for 72 hours. For the structural analysis by NMR 1H and 13C the devices Bruker 170 Avance 500 (500 MHz for 1H and 125 MHz for 13C nuclei) and Varian Mercury-400 (400 MHz for 1H and 100 MHz for 13C nuclei) were used with reference to tetramethylsilane. The solvents for NMR studies were deuterated (DMSO-d6 or CDCl3). LC-MS spectra were obtained on Shimadzu 10-AV LC, Gilson-215, equipped with an autosampler, with the following detectors: UV (215 and 254 nm), electrospray ionization mass spectrometer (API 150EX), and ELS detectors. For the chromatographical part of the device Luna-C18 column, Phenomenex, 5 cm × 2 mm, was used. Euro Vector EA-3000 CHNS apparatus was used for elemental analyses. Kofler hot-bench device was used for measurements of the melting points.
Ethyl 4-chloro-5-methylthieno[2,3-d]pyrimidine-6-carboxylate (1)
It was prepared according to the previously reported method (Triloknadh et al., 2018) and chromatographically purified. Eluent CHCl3:iPrOH (97:3).
Ethyl 4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxylate (2)
To the three-neck round-bottomed flask (1 l) 50 g (0.195 mol) of ethyl 4-chloro-5-methylthieno[2,3-d]pyrimidine-6-carboxylate (1), 17.5 g (0.29 mol) of methylboronic acid, 3.11 g (0.0043 mol) of Pd(dppf)Cl2·CH2Cl2, and 44.3 g (0.76 mol) of potassium fluoride were placed under the argon atmosphere. The amount of 1,4-dioxane taken was enough for the free stirring of the reaction mixture. The reaction mixture was heated in the silicone bath at 90°? for 24 hours. The reaction progress was monitored by TLC in the system ethyl acetate-hexane 1:2. After the full conversion of the starting chloride the solvent was evaporated (reduced pressure) and the crude mixture of organic products was dissolved in a minimal amount of ethyl acetate. The product was isolated chromatographically using the pad of silica and ethyl acetate-hexane (1:2) as the eluent. The main product was collected in the second fraction. Yield 38%, yellowish crystals; mp 113°C–114°C. 1H NMR (400 MHz, DMSO-d6): δ 1.33 (t, 3?, J = 7.1 Hz, CH3), 2.88–2.98 (m, 6?, 2CH3), 4.35 (q, 2?, J = 7.1 Hz, CH2), and 8.96 (s, 1?, CH); 13C NMR (100 MHz, DMSO-d6): δ 14.5, 16.3, 24.8, 62.1, 125.5, 130.4, 140.8, 155.2, 162.4, 165.9, and 166.9. LC-MS, m/z: 237 [M+H]+. Anal. Calcd. for C11H12N2O2S: C, 55.91; H, 5.12; N, 11.86. Found: C, 56.04; H, 5.17; N, 11.93.
4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxylic Acid (3)
To a conical flask (100 ml) 5 g (0.021 mol) of ethyl ester of 4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxylic acid (2) and 2,5 g (0.063 mol) sodium hydroxide were placed. Water (20 ml) was added to the reaction mixture and the content of the flask was gradually heated to boiling. The reaction mixture in a flask was boiled for 3 hours and then cooled and acidified with 85% orthophosphoric acid. Then the formed precipitate of compound 3 was drained. Plenty of water was used to remove inorganic admixtures by washing the filter cake. The precipitate was dried to constant mass at 60°C. Yield: 53%, a white powder; mp > 300°C; 1H NMR (500 MHz, DMSO-d6): δ 2.88–2.92 (m, 6?, 2CH3), 8.91 (s, 1?, CH), and 13.57 (br. s, 1?, COOH); 13C NMR (125 MHz, DMSO-d6): δ 15.5, 24.2, 126.9, 130.1, 139.0, 154.4, 163.6, 164.9, and 166.3. LC-MS, m/z: 209 [M+H]+. Anal. Calcd. for C9H8N2O2S: C, 51.91; H, 3.87; N, 13.45. Found: C, 52.10; H, 3.89; N, 13.59.
General method of 4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxamides synthesis
To 0.15 g (0.73 mmol) of 4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxylic acid (3), 1,1′-carbonyldiimidazole (CDI) 0.13 g (0.8 mmol) and 1.5 ml of anhydrous DMF were added. The reaction mixture was heated with vigorous stirring (70°?–80°?) then 0.0008 moles of the corresponding amine was added and the temperature was adjusted to 100°C and stirred more for 4–5 hours. After the mixture was cooled and 20 ml of water was added, the precipitate formed was drained. An analytical sample was dried to constant mass (ethanol was used as a solvent for crystallization).
N-benzyl-4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxamide (4a)
Yield 65%, a white powder; mp 155°C–156°C; 1H NMR (400 MHz, DMSO-d6): δ 2.75 (s, 3?, CH3), 2.91 (s, 3?, CH3), 4.49 (d, 1?, J = 5.7 Hz, CH2), 7.23–7.30 (m, 1?, H Ar), 7.31–7.40 (m, 4H, H Ar), 8.92 (s, 1?, CH), and 9.10 (m, 1?, NH); 13C NMR (125 MHz, DMSO-d6): δ 16.6, 24.5, 43.4, 127.4, 127.7, 128.8, 130.2, 131.4, 132.8, 139.4, 154.2, 162.6, 164.5, and 166.4. LC-MS, m/z: 298 [M+H]+. Anal. Calcd. for C16H15N3OS: C, 64.62; H, 5.08; N, 14.13. Found: C, 64.70; H, 5.10; N, 14.21.
4,5-dimethyl-N-(4-methylbenzyl)thieno[2,3-d]pyrimidine-6-carboxamide (4b)
Yield 53%, a white powder; mp 176°C–177°C; 1H NMR (400 MHz, DMSO-d6): δ 2.30 (s, 3?, CH3), 2.75 (s, 3?, CH3), 2.91 (s, 3?, CH3), 4.45 (d, 1?, J = 5.6 Hz, CH2), 7.16 (d, 1?, J = 7.2 Hz, H Ar), 7.25 (d, 1?, J = 7.2 Hz, H Ar), 8.86 (br s, 1?, NH), and 8.90 (s, 1?, CH); 13C NMR (100 MHz, DMSO-d6): δ 16.6, 21.1, 24.5, 43.2, 127.8, 129.4, 130.2, 131.5, 132.7, 136.4, 136.5, 154.2, 162.6, 164.4, and 166.4. LC-MS, m/z: 312 [M+H]+. Anal. Calcd. for C17H17N3OS: C, 65.57; H, 5.50; N, 13.49. Found: C, 65.59; H, 5.57; N, 13.57.
N-(4-chlorobenzyl)-4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxamide (4c)
Yield 92%, a white powder; mp 200°C–201°C; 1H NMR (400 MHz, DMSO-d6): δ 2.76 (s, 3?, CH3), 2.92 (s, 3?, CH3), 4.48 (d, 1?, J = 5.6 Hz, CH2), 7.31–7.45 (m, 4H, H Ar), 8.91 (s, 1?, CH), and 8.98 (br. s, 1?, NH); 13C NMR (125 MHz, DMSO-d6): δ 16.1, 24.0, 42.3, 128.0, 128.3, 128.7, 129.2, 129.7, 130.7, 131.5, 132.5, 138.0, 153.8, 162.2, 164.0, and 165.9. LC-MS, m/z: 332 [M+H]+. Anal. Calcd. for C16H14ClN3OS: C, 57.92; H, 4.25; N, 12.66. Found: C, 57.99; H, 4.36; N, 12.73.
N-(4-fluorobenzyl)-4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxamide (4d)
Yield 68%, a white powder; mp 175°C–176°C; 1H NMR (400 MHz, DMSO-d6): δ 2.75 (s, 3?, CH3), 2.91 (s, 3?, CH3), 4.48 (d, 1?, J = 6.5 Hz, CH2), 7.15 (t, 2H, J = 8.3 Hz, ? Ar), 7.41 (m, 2H, H Ar), 8.90 (s, 1?, H-4), and 8.97 (br. s, 1?, NH); 13C NMR (100 MHz, DMSO-d6): δ 16.6, 24.5, 42.8, 115.4, 115.7, 129.8, 129.8, 130.2, 131.3, 132.9, 135.6, 135.6, 154.2, 160.5, 162.6, 162.9, 164.4, and 166.4. LC-MS, m/z: 316 [M+H]+. Anal. Calcd. for C16H14FN3OS: C, 60.94; H, 4.47; N, 13.32. Found: C, 61.11; H, 4.53; N, 13.34.
N-(4-methoxybenzyl)-4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxamide (4e)
Yield 59%, a white powder; mp 159°C–160°C; 1H NMR (400 MHz, DMSO-d6): δ 2.74 (s, 3?, CH3), 2.91 (s, 3?, CH3), 3.75 (s, 3?, OCH3), 4.43 (d, 1?, J = 6.5 Hz, CH2), 6.92 (d, 1?, J = 8.3 Hz, H Ar), 7.29 (d, 1?, J = 8.3 Hz, H Ar), 8.89 (br s, 1?, NH), and 8.90 (s, 1?, CH); 13C NMR (125 MHz, DMSO-d6): δ 16.1, 24.0, 42.4, 55.0, 113.7, 128.7, 129.7, 130.9, 131.1, 132.1, 153.7, 158.3, 162.0, 163.9, and 165.9. LC-MS, m/z: 328 [M+H]+. Anal. Calcd for C17H17N3O2S: C, 62.37; H, 5.23; N, 12.83. Found: C, 62.48; H, 5.31; N, 12.85.
(4,5-dimethylthieno[2,3-d]pyrimidin-6-yl)(morpholin-4-yl)methanone (4f)
Yield 43%, a white crystalline powder; mp 195°C–196°C; 1H NMR (400 MHz, DMSO-d6): δ 2.55 (s, 3?, CH3), 2.90 (s, 3?, CH3), 3.19–3.69 (m, 8?, H morpholine), and 8.90 (s, 1?, CH); 13C NMR (125 MHz, DMSO-d6): δ 16.4, 24.1, 66.5, 129.3, 129.7, 129.8, 153.8, 162.6, 163.9, 167.2, and 168.7. LC-MS, m/z: 278 [M+H]+. Anal. Calcd for C13H15N3O2S: C, 56.30; H, 5.45; N, 15.15. Found: C, 56.38; H, 5.46; N, 15.23.
6-(1H-benzimidazol-2-yl)-4,5-dimethylthieno[2,3-d]pyrimidine (5)
To 3.0 g (0.0144 mol) of 4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxylic acid (3) 2.53 g (0.0156 mol) of 1,1’-carbonyldiimidazole and 25 ml of anhydrous DMF were added. The reaction mixture was stirred and heated at 70°?–80°? before the addition of 1.7 g (0.0156 mol) of o-phenylenediamine and the heating continued at 130°?–140°C for 3–5 hours. After cooling to the reaction mixture 100 ml of purified water was added and the precipitate formed was filtered off and washed with plenty of water. An analytical sample was purified by boiling in ethanol. Yield 62%, a beige powder; mp 239°C–240°C. 1H NMR (500 MHz, DMSO-d6): δ 2.94 (s, 3?, CH3), 2.96 (s, 3?, CH3), 7.25 (br. s, 2?, ? Ar), 7.64 (br. s, 2?, ? Ar), 8.90 (s, 1?, CH), and 12.91 (br. s, 1?, NH); 13C NMR (100 MHz, DMSO-d6): δ 16.8, 24.6, 126.9, 131.0, 131.3, 146.0, 153.8, 163.8, and 167.1. LC-MS, m/z: 281 [M+H]+. Anal. Calcd. for C15H12N4S: C, 64.26; H, 4.31; N, 19.98. Found: C, 64.29; H, 4.39; N, 20.06.
General method of N-Aryl-2-[2-(4,5-dimethylthieno[2,3-d]pyrimidin-6-yl)-1H-benzimidazol-1-yl]acetamides (6) synthesis via alkylation of 6-(1H-benzimidazol-2-yl)-4,5-dimethylthieno[2,3-d]pyrimidine (5)
To 0.28 g (0.001 mol) of 6-(1H-benzimidazol-2-yl)-4,5-dimethylthieno[2,3-d]pyrimidine (5) 0.14 g (0.001 mol), 0.001 mol of corresponding alkylating agent and 3 ml of dried dimethylformamide were added, and after the content of the flask was heated at 120°C for 5 hours, then chilled reaction mixture was quenched with water and the precipitate formed was drained and dried at raised temperature (65°C). All of compound 6 were additionally purified by boiling in ethanol.
2-[2-(4,5-dimethylthieno[2,3-d]pyrimidin-6-yl)-1H-benzimidazol-1-yl]-N-phenylacetamide (6a)
Yield 78%, a white powder; Mp >300°C; 1H NMR (500 MHz, DMSO-d6): δ 2.58 (s, 3?, CH3), 2.92 (s, 3?, CH3), 5.13 (s, 2?, CH2), 7.04 (t, 1?, J = 6.5 Hz, ? Ar), 7.24–7.41 (m, 4?, ? Ar), 7.48 (d, 2?, J = 7.1 Hz, H Ar), 7.61 (d, 1H, J = 7.1 Hz, H Ar), 7.78 (d, 1H, J = 7.1 Hz, H Ar), 8.95 (s, 1?, CH), and 10.34 (br. s, 1?, NH); 13C NMR (125 MHz, DMSO-d6): δ 16.7, 24.2, 47.8, 111.5, 119.6, 119.9, 120.2, 123.0, 123.9, 124.1, 124.5, 129.3, 129.7, 134.1, 136.4, 138.7, 143.0, 146.7, 153.9, 164.1, 165.4, and 167.9. LC-MS, m/z: 414 [M+H]+. Anal. Calcd for C23H19N5OS: C, 66.81; H, 4.63; N, 16.94. Found: C, 66.96; H, 4.65; N, 17.01.
N-(4-methylphenyl)-2-[2-(4,5-dimethylthieno[2,3-d]pyrimidin-6-yl)-1H-benzimidazol-1-yl]acetamide (6b)
Yield 68%, a white powder; Mp >300°C; 1H NMR (400 MHz, DMSO-d6): δ 2.22 (s, 3?, CH3), 2.58 (s, 3?, CH3), 2.93 (s, 3?, CH3), 5.10 (s, 2?, CH2), 7.08 (d, 2?, J = 7.8 Hz, ? Ar), 7.29–7.40 (m, 4?, ? Ar), 7.62 (d, 1H, J = 6.8 Hz, H Ar), 7.78 (d, 1H, J = 6.8 Hz, H Ar), 8.96 (s, 1?, CH), and 10.26 (br. s, 1?, NH); 13C NMR (125 MHz, DMSO-d6): δ 16.7, 20.8, 24.2, 47.7, 111.5, 119.6, 119.9, 123.0, 123.8, 124.6 129.6, 129.7, 133.1, 134.1, 136.2, 136.4, 143.0, 146.7, 153.9, 164.1, 165.1, and 167.9. LC-MS, m/z: 428 [M+H]+. Anal. Calcd for C24H21N5OS: C, 67.43; H, 4.95; N, 16.38. Found: C, 67.54; H, 5.06; N, 16.48.
N-(3,5-dimethoxyphenyl)-2-[2-(4,5-dimethylthieno[2,3-d]pyrimidin-6-yl)-1H-benzimidazol-1-yl]acetamide (6c)
Yield 81% yield, a white powder; mp 282°C–283°C. 1H NMR (400 MHz, DMSO-d6): δ 2.58 (s, 3?, CH3), 2.93 (s, 3?, CH3), 3.67 (s, 6?, 2 OCH3), 5.10 (s, 2?, CH2), 6.22 (s, 1?, H Ar), 6.70 (s, 2?, H Ar), 7.29–7.41 (m, 2?, ? Ar), 7.62 (d, 1H, J = 7.6 Hz, H Ar), 7.78 (d, 1H, J = 7.6 Hz, H Ar), 8.96 (s, 1?, CH), and 10.31 (br. s, 1?, NH); 13C NMR (100 MHz, DMSO-d6): δ 16.7, 24.3, 47.8, 55.5, 96.2, 97.9, 111.5, 119.9, 123.0, 123.9, 124.5, 129.8, 134.1, 136.5, 140.4, 143.0, 146.7, 154.0, 161.0, 164.2, 165.5, and 167.9. LC-MS, m/z: 474 [M+H]+. Anal. Calcd for C25H23N5O3S: C, 63.41; H, 4.90; N, 14.79. Found: C, 63.51; H, 4.97; N, 14.88.
Microbiology
The antimicrobial and antifungal activities of the obtained substances were estimated according to the recommendation of WHO and the national standard of Ukraine (Coyle, 2005; Nekrasova, 2007) using Staphylococcus aureus ???? 25923 and Bacillus subtilis ???? 6633 as two Gram-positive bacteria and the set of three Gram-negative strains, which were Escherichia coli ???? 25922, P. aeruginosa ???? 27853, and Proteus vulgaris ATCC 4636 strains. The fungal strain Candida albicans ???? 885/653 was used for antifungal activity studies. For the “agar well” diffusion method the microbial load was 107 CFU in 1 ml of the nutrient media and it was determined according to the MacFarland standard (McFarland, 1907). According to the national recommendations (Ministry of Public Health of Ukraine, 2001), each microorganism culture of 18–24-hour growth was used for the test. For antimicrobial activity studies, Muller–Hinton (HIMedia Laboratories Pvt. Ltd., India) agar was used, while Sabouraud agar (HIMedia Laboratories Pvt. Ltd., India) was used to study fungi. The administration of each tested sample including standards (Streptomycin) was performed in form of dimethylsulfoxide solutions with 100 μg/ml concentrations in the form of 0.3-ml aliquots (Magaldi et al., 2004). The activity of every tested compound sample was estimated three times experimentally. The assessment of antibacterial activity against each microorganism was performed according to the measured value of growth inhibition zones.
The activity of the compounds was assessed by the following criteria (Coyle, 2005; Nekrasova, 2007): absence of the growth inhibition zone or its diameter less than 10 mm, very low sensitivity of a microorganism to the tested compound in this concentration; the diameter near 10–15 mm, low sensitivity of a microorganism to the tested compound in this concentration; the diameter near 15–25 mm, good sensitivity of a microorganism to the tested compound in this concentration; the diameter exceeded 25 mm, high sensitivity of a microorganism to the tested compound in this concentration.
Docking studies
For the docking studies, we used the same set of computer programs as previously reported (Severina et al., 2020; Vlasov et al., 2021); namely AutoDockTools 1.5.6rc3 and AutoDock Vina, BIOVIA Draw 2019, Chem3D software, and Discovery Studio V17.2.0.16349. The macromolecule of the enzyme tRNA (Guanine37-N1)-methyltransferase in its conformation in crystal with the native ligand was downloaded from Protein Data Bank; the ID of the structure is 5ZHN (PDB, 2022). The docking procedure obeyed all the conventional algorithms (Torres et al., 2016). The output formats of the structures were transformed to a PDBQT format. All the docking parameters, validation techniques for the algorithm, and RSMD values were reported previously by us (Vlasov et al., 2021).
RESULTS AND DISCUSSION
Although some compounds with 4-O-alkylthieno[2,3-d]pyrimidine core were reported to be inactive as TrmD inhibitors, the most active compound I with oxo group at position four has been reported in the same article (Zhong et al., 2019b) and 4-N-alkyl derivatives showed good activity as inhibitors to the acetyltransferase of bacterial amino-sugar II (De Schutter et al., 2017). Antimicrobial properties were also revealed earlier for thieno[2,3-d]pyrimidine-4-carboxamides, which are also the compounds with aromatic pyrimidine ring, and notably benzylamide III was the most active against P. aeruginosa (Vlasova et al., 2019). The problem of the aromatic pyrimidine ring to antibacterial activity remained unclear. We found it interesting to obtain the derivatives with a small alkyl substituent at position four of thienopyrimidine attached to the heterocyclic system without any additional atoms linker O or N atoms.
For the further preparation of attractive antimicrobial substances, we additionally considered two possible main ways of modifying the carboxylic function at position six of the thienopyrimidine cycle: one of them is the preparation of amides and the other one is the use of heterocyclization methods which showed their effectiveness for construction of thieno[2,3-d]pyrimidines bearing 6-heteryl substituent with antimicrobial properties (Hill et al., 2013; Vlasov et al., 2021; Zhong et al., 2019b).
The examples of 4-alkylthieno[2,3-d]pyrimidines are quite rare, although recently some 4-methylthieno[2,3-d]pyrimidines with different heterocycles at position six were reported as ones with antiproliferative dose-dependent activity according to NCI-H460 xenograft model and their safety profile was found acceptable (Lin et al., 2018). The building blocks for these structures were obtained by reaction of 4-chlorothieno[2,3-d]pyrimidine with methylboronic acid. We found this approach promising and decided to test an opportunity of synthesis of 4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxylic acid as a perfect building block for the synthesis of amides and the construction of benzimidazole substituent at position six of the core heterocyclic system.
The first preparation of 4-alkyl derivatives of thieno[2,3-d]pyrimidine was reported in 1971 (Manhas and Sharma, 1971). The authors used 4-chloride of 2-phenyl-5,6,7,8-tetrahydro[1]-benzothieno[2,3-d]pyrimidine as the starting compound for the preparation of 4-methylsulphonylmethyl-2-phenyl-5,6,7,8-tetrahydro[1]-benzothieno[2,3-d]pyrimidine, which was further reduced with aluminum amalgam in aqueous tetrahydrofuran. The example of incidental preparation of 4-alkyl thieno[2,3-d]pyrimidine occurred during the experimental selection of metalation reagent (Therkelsen et al., 2004). Preparation of 4-methyl derivatives by cyclization of 4-chloro-5-(trimethylsilylethynyl)pyrimidines with the addition of sodium hydrosulfide was also suggested (Sakamoto et al., 1986). Preparation of 4-alkylthieno[2,3-d]pyrimidines by Suzuki coupling was first reported for the preparation of 4-butyl derivative as the result of the interaction of Pd(PPh3)4-mediated reaction of the corresponding chloride with butylboronic acid (Fernandez-Mato et al., 2011); similar transformations for methylboronic acid were reported recently (Lin et al., 2018).
 | Figure 1. The derivatives of thieno[2,3-d]pyrimidine with antibacterial properties. [Click here to view] |
In view of the high effectiveness of methylation in the reaction of different aryl and hetaryl halides in reaction with methylboronic acid (Falb et al., 2017; Karroum et al., 2018), we studied the Pd-catalyzed reaction of previously reported ethyl 4-chloro-5-methylthieno[2,3-d]pyrimidine-6-carboxylate (Triloknadh et al., 2018) with methylboronic acid (Fig. 2). It is interesting that the reaction appeared very sensitive to the quality of the starting chloride. For the crude chloride in the reaction with boronic acid, we saw the traces of the by-product with quasimolecular ion peak 459 (m/z), which probably resulted from Pd-catalyzed side N-arylation reaction of the chloride 1 with its oxo-derivative, which might have been as admixture. Therefore, for the scaling up of the reaction, the starting chloride 1 was purified chromatographically.
The reaction of pure compound 1 with methylboronic acid catalyzed by PdCl2 dppf allowed the preparation of 4-methyl derivative 2 in satisfactory yield (Fig. 2). At the next step, the ester 2 was hydrolyzed in alkaline conditions to give acid 3 with an excellent yield. Synthesis of amides 4 was performed by reaction of the acid 3 with the series of primary benzyl amines and morpholine in DMF, which was promoted by CDI. All of the amides 4 were obtained with good yields 43%–92%. In the 1H NMR spectra of the compounds 4 two signals of methyl groups at positions 4 and 5 of thienopyrimidine are observed at 2.54–2.75 and 2.90–2.92 ppm; the position of the down-field signal is not much influenced by the amide substituent which confirms that this is the signal of CH3 group at more distant form carboxamide substituent position four. For all of the compound 4, the signal of a proton of thieno[2,3-d]pyrimidine at position two is observed as a singlet at 8.90–8.98 ppm. For benzylamides 4a-4e the doublet signals of the methylene group are observed at 4.43–4.49 ppm while the broaden signal of NH is obviously seen at 8.86–9.10 ppm; the signals of aromatic protons are observed at 6.92–7.40 ppm. For morpholyl amide 4f the signals of methylene groups are at 3.19–3.69 ppm. 13C NMR spectra of the synthesized derivatives 4 have signals of both CH3 groups of thieno[2,3-d]pyrimidine at 16.1–16.6 ppm and 24.0–24.6 ppm.
We also studied the reaction of acid 3 in conditions similar to amides 4 synthesis using o-phenylenediamine as an amine. It was found that the reaction at 120°C–130°C during 2–3 hours results in the product of benzimidazole ring closure with the formation of 6-(1H-benzimidazol-2-yl)-4,5-dimethylthieno[2,3-d]pyrimidine 5. In the 1H NMR spectrum of the derivative 6, the signals of thienopyrimidine CH3 groups are observed as two singlets at 2.94 ?? 2.96 ppm; the proton at position two of the core heterocyclic system shows its signal at 8.90 ppm. The signals of the benzimidazole system in the spectrum are observed as two groups at 7.25 (br. s, 2?, ? Ar) and 7.64 (br. s, 2?, ? Ar), while the signal of benzimidazole NH is at 12.91 ppm.
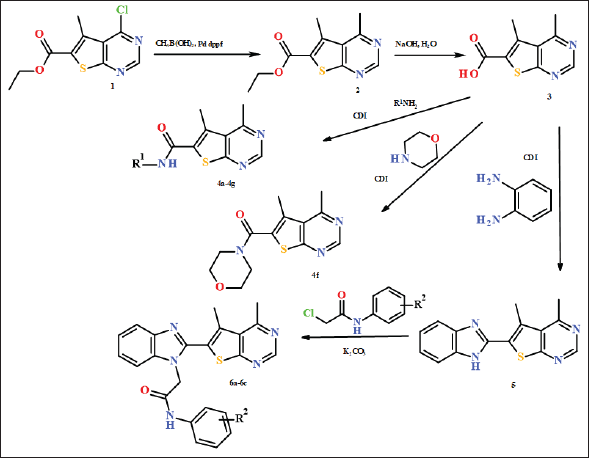 | Figure 2. Preparation of 4-methylthieno[2,3-d]pyrimidine derivatives 4a-f, 5, and 6a-c. [Click here to view] |
Compound 5 with its benzimidazole fragment was found suitable for alkylation with chloroacetamides. The alkylation was performed in dimethylformamide using potassium carbonate as the base. 2-[2-(4,5-dimethylthieno[2,3-d]pyrimidin-6-yl)-1H-benzimidazol-1-yl]-N-arylacetamides 6?-6? were isolated with good yields and crystallized from ethanol. 1H NMR spectra of the compounds 6 in comparison with the compound 5 have additional signals of protons of the CH2 group of acetamide in the region 5.10–5.13 ppm, while the signals of NH groups are observed in the region 10.26–10.34 ppm. The 13C NMR spectra of 6?-6? differ from the spectrum of 5 by additional signals of CH2 at 47.7–47.8 ppm and C=O signals at 165.1–165.5 ppm.
The results of antimicrobial activity screening, which was performed by agar “well” method for all of the synthesized compounds, showed that all of the compounds with the fragment of 4-methylthieno[2,3-d]pyrimidine were active against most bacteria and Candida fungi (Table 1). Among the Gram-positive bacteria B. subtilis ???? 6633 was the most sensitive; the compounds also most effectively inhibited the growth of P. aeruginosa ???? 27853 among the Gram-negative strains. Ester 2 has been found more active than the corresponding acid 3. The introduction of the benzylamide fragment also increased the activity of the compounds 4c, 4d, and 4e correspondingly 4-Cl, 4-F, and 4-OCH3 against B. subtilis and P. aeruginosa. Amide 4f with morpholine fragment did not display high antimicrobial activity regardless of its better solubility. The results of the antimicrobial activity study also showed the introduction of a benzimidazole ring instead of a carboxyl group. The antimicrobial activity of the compounds synthesized was found to be a little less than the activity of the reference drug Streptomycin, which was selected as the reference drug which is an aminoglycoside antibiotic with a wide spectrum of activity against both Gram-positive and Gram-negative bacteria; it is also active against Mycobacterium tuberculosis; this drug is also widely used in clinics (Ahmed et al., 2013; Jansen et al., 2016; Tereshko et al., 2003). Unfortunately, not of the TrmD inhibitors, for now, has been approved for medical use.
To suggest the possible mechanism of antibacterial activity for all the compounds which were studied for the antimicrobial activity we performed the docking study to the active site of TrmD selective inhibitors. The TrmD which was isolated from P. aeruginosa (Zhong et al., 2019b) was used as a protein model. The validation of docking methodology and RMSD calculation of the reference ligand pose to the native ligand were reported earlier (Vlasov et al., 2021, 2022). According to the docking studies, the obtained compounds showed different affinity to the active site TrmD inhibitor (Table 2).
The lowest level of affinity was predicted for both ester 2 and acid 3 (−7.4 kcal/mole against −8.2 kcal/mole affinity for the native ligand). Transformation of the carboxyl group to the benzimidazole cycle improves the binding energy for derivative 5 (−8.0 kcal/mole). Alkylation of the benzimidazole cycle increases the affinity; the scoring function for arylacetamides 6?-? reaches −8.7 kcal/mole value. The affinity results were predicted for amides 4, especially benzylamides 4a-e, which showed the values of scoring function from −9.3 to −9.9 kcal/mole. The calculated scoring function values are in good correlation with in vitro experimental results.
Detailed analysis of the interaction of the ligands with amino acids of the active site shows many hydrophobic interactions. Among them, there was the interaction of methyl group at position four of thieno[2,3-d]pyrimidine with the pyrrolidine cycle of proline Pro94, which can be a sign of the existence of stable ligand-enzyme conformations. All the studied ligands interact with the residues of glutamic acid Gly145, 146, and/or glutamine Gln95, which were proven to be the site of TrmD cofactor S-adenosylmethionine (SAM) binding. This determines the deep immersion of 4-methylthieno[2,3-d]pyrimidines in the cavity of the active site competing with SAM and thus inhibiting bacterial TrmDs like the one isolated from P. aeruginosa. As to the results of the docking, the best parameters of antibacterial activity via inhibition of TrmD were shown by benzyl amides 4?-? (Fig. 3).
 | Table 1. Antimicrobial activity screening results for 4-methylthieno[2,3-d]pyrimidine derivatives 2,3, 4a-f, 5, 6a-c. [Click here to view] |
 | Table 2. The resuls of the docking studies of 4-methylthieno[2,3-d]pyrimidine derivatives 2, 3, 4a-f, 5, 6a-c to the active site of selective TrmD P. aeruginosa inhibito rs. [Click here to view] |
 | Figure 3. 3D visualization of the ligand 4b and the amino acids of the active site of selective TrmD inhibitors (a) conformation with reference inhibitor (yellow structure) (Zong et al., 2019 ) in the active site of P. aeruginosa TrmD. [Click here to view] |
CONCLUSION
As the result of modification of position four of thieno[2,3-d]pyrimidine using Suzuki reaction of methyl boronic acid and readily available ethyl 4-chloro-5-methylthieno[2,3-d]pyrimidine-6-carboxylate the ester and 4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxylic acid resulted from hydrolysis of the ester were obtained. Modification of 4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxylic acid gave the series of amides and the derivative with benzimidazole fused system at position six. All of the synthesized compounds showed antimicrobial properties, especially against the strains of B. subtilis and P. aeruginosa. The most promising antimicrobial properties were determined for benzyl amides of 4,5-dimethylthieno[2,3-d]pyrimidine-6-carboxylic acid. The docking studies of the obtained compounds to the active site of selective TrmD inhibitors revealed that benzyl amides also had the best binding parameters to the tRNA (Guanine37-N1)-methyltransferase of P. aeruginosa. The docking studies confirmed the suggested mechanism of antibacterial action as in silico results are in correlation with the in vitro experiment.
ACKNOWLEDGMENTS
All the authors and contributors are truly grateful to Enamine Ltd. Company for providing analytical data like 1H, 13C NMR, and LC-MS spectra. They acknowledge the kind help of Dr. Tatyana P. Osolodchenko, Ph.D. in Biology, and her colleagues from the Mechnikov Institute of Microbiology and Immunology of the NAMS of Ukraine (Kharkiv) for the performance of antibacterial and antifungal studies.
AUTHOR CONTRIBUTIONS
Concept and design were contributed by S.V.V., V.A.G., O.D.V., and H.I.S.; data acquisition and data analysis were contributed by K.Y.K., V.S.V, P.E.S., H.I.S., O.D.V., O.V.B., and S.V.V.; preparation and submission of the manuscript were done by S.V.V., H.I.S., K.Y.K., and O.V.B.; supervision and final approval were contributed by V.A.G., S.V.V., H.I.S., and O.V.B. All the authors have made a significant contribution to the content of the submitted manuscript; they adjusted it and agree with its publication.
FINANCIAL SUPPORT
The research was funded by the Ministry of Health Care of Ukraine at the expense of the State Budget in framework # 2301020 “Scientific and scientific-technical activity in the field of health protection” on the topic “Synthesis and study of new thienopyrimidines for the detection of antimicrobial and related types of pharmacological activity” (State registration number: 0121U109472; Order of the Ministry of Health of Ukraine of November 17, 2020, ? 2651).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Ahmed Z, Saeed Khan S, Khan M. In vitro trials of some antimicrobial combinations against Staphylococcus aureus and Pseudomonas aeruginosa. Saudi J Biol Sci, 2013; 20(1):79–83. CrossRef
Badiger NP, Gaonkar SL, Shetty NS. Synthesis of some new thienopyrimidines and triazole fused thienopyrimidines and their antimicrobial activities. Internat J Chem Pharm Sci, 2015; 6(1):58–62.
Brown DG, Wobst HJ. A decade of FDA-approved drugs (2010-2019): trends and future directions. J Med Chem, 2021; 64(5):2312–38. CrossRef
Cercenado E. Espectro antimicrobiano de dalbavancina. Mecanismo de accion y actividad in vitro frente a microorganismos Gram positivos. Enferm Infecc Microbiol Clin, 2017; 35(1):9–14. CrossRef
Coyle MB. Manual of antimicrobial susceptibility testing. American Society for Microbiology, Washington, DC, 241 c p, 2005.
De Schutter JW, Morrison JP, Morrison MJ, Ciulli A, Imperiali B. Targeting Bacillosamine biosynthesis in bacterial pathogens: development of inhibitors to a bacterial amino-sugar acetyltransferase from Campylobacter jejuni. J Med Chem, 2017; 60(5):2099–118. CrossRef
El-Gamal M, Kim S, Oh CH. Synthesis and in vitro antibacterial activity of new meropenem analogs. J Antibiot, 2011; 64:687–8. CrossRef
Falb E, Ulanenko K, Tor A, Gottesfeld R, Weitman M, Afri M, Gottlieb HE, Hassner A. A highly efficient Suzuki–Miyaura methylation of pyridines leading to the drug pirfenidone and its CD3 version (SD-560). Green Chem, 2017; 19:5046–53. CrossRef
Fernandez-Mato A, Peinador C, Quintal JM. Convenient one-pot synthesis of functionalized thieno[3,2-d]pyrimidine and thieno[2,3-d]pyrimidine derivatives. Synthesis, 2011; (20):3323–31. CrossRef
Gamper HB, Masuda I, Frenkel-Morgenstern M, Hou YM. Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat Commun, 2015; 6:7226. CrossRef
Goto-Ito S, Ito T, Yokoyama S. Trm5 and TrmD: two enzymes from distinct origins catalyze the iIdentical tRNA modification, m¹G37. Biomolecules, 2017; 7(1):32. CrossRef
Hill PJ, Abibi A, Albert R, Andrews B, Gagnon MM, Gao N, Grebe T, Hajec LI, Huang J, Livchak S, Lahiri SD, McKinney DC, Thresher J, Wang H, Olivier N, Buurman ET. Selective inhibitors of bacterial t-RNA-(N(1)G37) methyltransferase (TrmD) that demonstrate novel ordering of the lid domain. J Med Chem, 2013; 56(18):7278–88. CrossRef
Jansen G, Mahrt N, Tueffers L, Barbosa C, Harjes M, Adolph G, Friedrichs A, Krenz-Weinreich A, Rosenstiel P, Schulenburg H. Association between clinical antibiotic resistance and susceptibility of Pseudomonas in the cystic fibrosis lung. Evol Med Public Health, 2016; 2016(1):182–94. CrossRef
Jones F, Hu Y, Coates A. The efficacy of using combination therapy against multi-drug and extensively drug-resistant Pseudomonas aeruginosa in clinical settings. Antibiotics (Basel), 2022; 11(3):323. CrossRef
Karroum NB, Patinote C, Deleuze-Masquefa C, Moarbess G, Diab-Assaf M, Cuq P, Kassab I. Methylation of imidazopyrazine, imidazoquinoxaline, and pyrazoloquinoxaline through Suzuki–Miyaura cross coupling. Chem Heterocyc Comp, 2018; 54(2):183–7. CrossRef
Khan M, Stapleton F, Summers S, Rice SA, Willcox MDP. Antibiotic resistance characteristics of Pseudomonas aeruginosa isolated from keratitis in Australia and India. Antibiotics (Basel), 2020; 9(9):600. CrossRef
Lin S, Wang C, Ji M, Wu D, Lv Y, Sheng L, Han F, Dong Y, Zhang K, Yang Y, Li Y, Chen X, Xu H. Discovery of new thienopyrimidine derivatives as potent and orally efficacious phosphoinositide 3-kinase inhibitors. Bioorg Med Chem, 2018; 26(3):637–46. CrossRef
Magaldi S, Mata-Essayag S, Hartung de Capriles C, Perez C, Colella MT, Olaizola C, Ontiveros Y. Well diffusion for antifungal susceptibility testing. Int J Infect Dis, 2004; 8(1)39–45. CrossRef
Malasala S, Polomoni A, Ahmad Md N, Shukla M, Kaul G, Dasgupta A, Chopra S, Nanduri S. Structure based design, synthesis and evaluation of new thienopyrimidine derivatives as anti-bacterial agents. J Molec Struct, 2021; 1234:130168. CrossRef
Manhas MS, Sharma SD. Hetcrocyclic compounds III. Synthesis of some substituted thienopyrimidines. J Heterocyc Chem, 1971; 8:1051–3. CrossRef
Masuda I, Matsubara R, Christian T, Rojas ER, Yadavalli SS, Zhang L, Goulian M, Foster LJ, Huang KC, Hou YM. tRNA methylation Is a global determinant of dacterial multi-drug resistance. Cell Syst, 2019; 8(4):302–14.e8. CrossRef
McFarland J. The nephelometer: an instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. J A Med Assoc, 1907; 49(14):1176–8. CrossRef
Ministry of Public Health of Ukraine (MHU). Bacteriological control of culture media; Newsletter ? 05.4.1/1670. Ministry of Public Health of Ukraine, Kiev, Ukraine, 45 p, 2001.
Mitra S, Saeed U, Havlichek DH, Stein GE. Profile of oritavancin and its potential in the treatment of acute bacterial skin structure infections. Infect Drug Resist, 2015; 8:189–97. CrossRef
Nekrasova LS, Svita VM, Glushkevich TG, Tomchuk VV, Zherebko NM, Yanovs’ka VV. Methodological guidelines “Determination of the Sensitivity of Microorganisms to Antibiotics”; ? MB 9.9.5-143-2007. Ministry of Public Health of Ukraine, Kiev, Ukraine, 24 p, 2007.
Olsson A, Wistrand-Yuen P, Nielsen EI, Friberg LE, Sandegren L, Lagerbäck P, Tängdén T. Efficacy of antibiotic combinations against multidrug-resistant Pseudomonas aeruginosa in automated time-lapse microscopy and static time-kill experiments. Antimicrob Agents Chemother, 2020; 64(6):e02111–9. CrossRef
Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv, 2019; 37(1):177–92; doi:10.1016/j.biotechadv.2018.11.013 CrossRef
Protein Data Bank. Available via http://www.rcsb.org/pdb/home/home.do (Accessed 10 April 2022).
Richter MF, Drown BS, Riley AP, Garcia A, Shirai T, Svec RL, Hergenrother PJ. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature, 2017; 545(7654):299–304. CrossRef
Sakamoto T, Kondo Y, Watanabe R, Yamanaka H. Condensed heteroaromatic ring systems. VII: synthesis of thienopyridines, thienopyrimidines, and furopyridines from o-substituted N-heteroarycetylenes. Chem Pharm Bull, 1986; 34(7):2719–24. CrossRef
Severina HI, Skupa OO, Voloshchuk NI, Georgiyants VA. Synthesis, docking study, and pharmacological evaluation of S-acetamide derivatives of 4,6-dimethyl-2-thiopyrimidine as anticonvulsant agents. J Appl Pharm S, 2020; 10(07):001–8. CrossRef
Tereshko V, Skripkin E, Patel DJ. Encapsulating streptomycin within a small 40-mer RNA. Chem Biol, 2003; 10(2):175–87. CrossRef
Therkelsen FD, Rottlaender M, Thorup N, Bjerregaard PE. 4-Metalated condensed pyrimidines: their preparation and reaction with aldehydes under barbier-type conditions. Org Lett, 2004; 6(12):1991–4. CrossRef
Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov, 2015; 14(8):529–42. CrossRef
Torres PHM, Sodero ACR, Jofily P, Silva-Jr FP. Key topics in molecular docking for drug design. Int J Mol Sci, 2019; 20(18):4574–603. CrossRef
Triloknadh S, Venkata Rao C, Nagaraju K, Hari Krishna N, Venkata Ramaiah C, Rajendra W, Trinath D, Suneetha Y. Design, synthesis, neuroprotective, antibacterial activities and docking studies of novel thieno[2,3-d]pyrimidine-alkyne Mannich base and oxadiazole hybrids. Bioorg Med Chem Lett, 2018; 28(9):1663–9. CrossRef
Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P & T, 2015; 40(4):277–83.
Vlasov SV, Vlasova OD, Severina HI, Krolenko KY, Borysov OV, Abu Sharkh AIM., Vlasov VS, Georgiyants VA. Design, synthesis and in vitro antimicrobial activity of 6-(1H-Benzimidazol-2-yl)-3,5-dimethyl-4-oxo-2-thio-3,4-dihydrothieno[2,3-d]pyrimidines. Sci Pharm, 2021; 89(4):49. CrossRef
Vlasov SV, Severina HI, Borysov OV, Krolenko KY, Shynkarenko PE, Saidov NB, Vlasov VS, Georgiyants VA. Synthesis and antimicrobial evaluation of 2-(6-Imidazo[1,2-a]pyridin-2-yl-5-methyl-2,4-dioxo-3-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-1(2H)-yl)-N-arylacetamide derivatives. Molbank, 2022; 1:M1331. CrossRef
Vlasova OD, Krolenko KYu, Nechayev MA, Shynkarenko PE, Kabachnyy VI, Vlasov SV. Efficient method for the synthesis of novel substituted thieno[2,3-d]pyrimidine-4-carboxylic acids, their derivatization, and antimicrobial activity. Chem Heterocycl Comp, 2019; 55(2):184–8. CrossRef
Wagenlehner FME, Cloutier DJ, Komirenko AS, Cebrik DS, Krause KM, Keepers TR, Connolly LE, Miller LG, Friedland I, Dwyer JP. Once-daily Plazomicin for complicated urinary tract infections. New Engl J Med, 2019; 380(8):729–40. CrossRef
Whitehouse AJ, Thomas SE, Brown KP, Fanourakis A, Chan DS, Libardo MDJ, Mendes V, Boshoff HIM, Floto RA, Abell C, Blundell TL, Coyne AG. Development of inhibitors against Mycobacterium abscessus tRNA (m1G37) methyltransferase (TrmD) using fragment-based approaches. J Med Chem, 2019; 62(15):7210–32. CrossRef
Yakout MA, Abdelwahab IA. Diabetic foot ulcer infections and Pseudomonas aeruginosa biofilm production during the COVID-19 pandemic. J Pure Appl Microbiol, 2022; 16(1):138–46. CrossRef
Zhang G, Meredith, TC, Kahne D. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr Opin Microbiol, 2013; 16(6):779–85. CrossRef
ZhongW, Koay A, Ngo A, Li Y, Nah Q, Wong YH, Chionh YH, Ng HQ, Koh-Stenta X, Poulsen A, Foo K, McBee M, Choong ML, El Sahili A, Kang C, Matter A, Lescar J, Hill J, Dedon P. Targeting the bacterial epitranscriptome for antibiotic development: discovery of novel tRNA-(N1G37) methyltransferase (TrmD) inhibitors. ACS Infect Dis, 2019a; 5(3):326–35. CrossRef