INTRODUCTION
Sir Alexander Fleming’s discovery of antibiotics in 1928 was one of the most significant milestones in human history (Kourkouta et al., 2018). Antibiotics are now commonly used in the food industry for disease prevention (McDermott et al., 2002), in addition to their use in the treatment of infectious diseases (Gelband et al., 2015). On the other hand, antibiotic abuse and misuse are increasing, which is why phrases like preantibiotic era have emerged to highlight the severity of the increase in antibiotic resistance, along with superbug emergence of multidrug-resistant bacteria (MDR) including vancomycin-resistant Enterococcus and methicillin-resistant Staphylococcus aureus (MRSA) (Albsoul-Younes et al., 2010). The high prevalence of MDR bacterial infections poses a challenge to the healthcare systems, not only because these bacteria are resistant to the majority of traditional antibiotics, but also because of their ability to form biofilms, which act as massive barriers to antibiotics passing through (Ventola, 2015).
Increase in the number of recent antibiotics presented for use in medicine rose considerably during the mid-20th century. For instance, from 1935 to 1968, 12 new classes were introduced. Even so, the new classes plummeted precipitously after that, introducing just two new classes between 1969 and 2003 (Mohr, 2016). This decline in the newly introduced antibiotics to the market was caused by the high toxicity, poor stability, hydrophobicity, and increased research and development costs (Lewis, 2013). The COVID-19 epidemic, with its catastrophic effects on people and economies, mainly caused by an infection with no available curing antibiotic or antiviral agent, should be enough incentive to act before it is too late; thus, new effective antibiotics with novel mechanisms of action are needed (Cama et al., 2021).
In nature and all types of life, bacteria, vertebrates, insects, and plants, antimicrobial peptides (AMPs) are widely propagated. These peptides are known to be a large part of the innate immune system and play a significant role in providing the host organism with first-line defenses against invasion or attack by bacteria, viruses, and fungi. These actions are achieved either directly by outer membrane attachment and subsequent destruction of the microbial membrane or through indirect activation of the immune system (Liévin-Le Moal and Servin, 2006). Most AMPs have a net positive charge to promote electrostatic interaction with the bacterial membrane, which is needed for peptide activity (Epand and Epand, 2009; Harding et al., 2018; Lundstedt et al., 2021). Furthermore, their structure contains around 40% hydrophobic residue, which interacts with the lipid core in the targeted membrane and facilitates membrane permeabilization (Fjell et al., 2012; Giangaspero et al., 2001; Huang et al., 2010; Toke, 2005).
The AMPs have a broad spectrum of activity against various microorganisms and rapid-killing kinetics (Huan et al., 2020). Moreover, they can also inhibit biofilm formation (Klubthawee et al., 2020). Finally, the nonspecific multitarget mode of action is responsible for their low resistance levels (Kumar et al., 2018). These characteristics paved the way for AMPs as a potential future replacement for traditional antibiotics.
On the other hand, source limitations, instability, toxicity, and bioavailability hampered the commercial development of these natural peptides for even the most basic applications (Azmi et al., 2016). Therefore, attempts are continuously made toward modifying these peptides to enhance their physicochemical characteristics. These changes are made rationally based on an understanding of the structure–activity relationship of AMPs. In this regard, various bioinformatics tools, online libraries, and databases are accessible online to assist the researchers in studying the influence of each attribute on the activity and selectivity of the modified peptides.
Few of the approaches utilized to enhance AMPs properties include sequence alteration of natural peptides and hybridization of different AMPs (Masadeh et al., 2022). In the current study, we are employing these two ways to design HEA-9, a novel peptide having enhanced activity and selectivity compared to the parent peptides, cecropin A and BMAP-27. Cecropin A is a naturally occurring AMP that insects produce as part of their innate defense mechanism. Cecropin A works as a bactericide by enhancing membrane permeabilization. It is also effective against Gram-negative bacteria; however,it is ineffective against S. aureus (Moore et al., 1996). BMAP-27, on the other hand, is an alpha-helical cathelicidins-derived peptide with a significant antibacterial action against a broad spectrum of pathogens. However, its high toxicity and hemolytic activity toward human blood cells made it unsuitable for clinical applications (Gennaro and Zanetti, 2000; Lee et al., 2011). This research focuses on the development of a novel modified hybrid peptide and its application in the prevention and treatment of infections caused by MDR S. aureus and MDR Escherichia coli. Moreover, the activity of the antibiofilm of this peptide was assessed versus the same strains, through the use of two complementary methods that were carried out sequentially: in silico and in vitro studies. In addition, our strategy focuses on measuring the added value of the combination of this novel peptide in small concentrations with traditional antibiotics aiming to increase effectiveness and combat the problem of MDR pathogens.
MATERIALS AND METHODS
Materials
The strains of the bacteria employed in the current work were acquired from the American Type Tissue Culture Collection (ATCC). They included two Gram-positive bacteria, S. aureus (ATCC29213), as the control strain and the MDR strain MRSA (ATCC BAA-41), as well as two Gram-negative bacteria, the control strain E. coli (ATCC25922) and MDR strain E. coli (ATCC BAA-2452). All the bacteria were cultivated on Muller-Hinton (MH) agar purchased from Scharlab, S.L. (Spain). The antibiotics’ pure formulas, ciprofloxacin, rifampicin, and doxycycline, were acquired from Sigma-Aldrich (USA). Additionally, ampicillin was purchased from Duchefa Biochemie. The peptide powder was synthesized and acquired from BIOMATIK (Cambridge, Canada). MH broth (Bio LAB) was used to dissolve all the antibacterial compounds and prepare the bacterial suspension. Phosphate-buffered saline (PBS) and dimethyl sulfoxide (DMSO) were purchased from Capricorn Scientific and Thermo Scientific, respectively. Triton X-100 was purchased from Sigma-Aldrich. Amphotericin B solution, trypsin-EDTA 1X, and penicillin and streptomycin were purchased from HiMedia. Fetal bovine serum and Roswell Park Memorial Institute (RPMI) with L-glutamine were also bought from Capricorn Scientific. MTT and Trypan Blue were acquired from Sigma-Aldrich and Atom Scientific, respectively.
Peptide design, molecular modeling, and in silico analyses
Firstly, the network protein sequence analysis (NPS) HNN secondary structure prediction software was utilized to calculate the helicity of the modified hybrid peptide (Combet et al., 2000). The HydroMCale program from the HELIQUEST service was then used to compute the hydrophobicity (H) and the HEA-9 peptide hydrophobic moment (μH) (Gautier et al., 2008). Next, the isoelectric point, water solubility, molecular weight, and net charge at neutral pH of the parent peptides, hybrid, and HEA-9 peptides were calculated using Innovagen’s peptide calculator. Next, the protein-binding potential (Boman index) of the parent and hybrid peptides was estimated using the AMP calculator and prediction tool from the AMP database (APD3) (Wang et al., 2016a). Next, EXPASY’s ProtParam program was used to determine the physicochemical properties of the HEA-9 peptide (Gasteiger et al., 2005). Finally, the I-TASSER software was used to predict the three-dimensional structure from the primary amino acid sequence of the HEA-9 peptide (Zhang, 2008).
Peptide synthesis and purification
The HEA-9 peptide was produced utilizing the solid-phase method and fluorenylmethyloxycarbonyl (Fmoc) chemistry and purified using reverse-phase high-performance liquid chromatography (HPLC) with an Inertsil ODS-SP 4.6 mm * 250 mm column and gradients of acetonitrile-TFA/H20-TFA as a mobile phase at 1.0 ml/minute. The peptide’s identification was validated using electrospray ionization mass spectrometry (ESI-MS). The peptide was obtained from Biomatik (Cambridge, Canada).
Antimicrobial susceptibility test by evaluating the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC)
This study evaluated the antibacterial activity of the HEA-9 peptide, four antibiotics, and the combination of HEA-9 peptide and traditional antibiotics against all bacterial strains utilizing the broth microdilution technique published in the Clinical and Laboratory Standard Institute (CLSI) (CLSI, 2014). Briefly, the HEA-9 peptide stock solution was made by dissolving the peptide powder in 5% DMSO and MH broth. The peptide concentration was freshly prepared by diluting the stock solution with the MH broth twofold serially. Firstly, 50 μl of the peptide concentration was transported into a 96-well plate, followed by another 50 μl of freshly prepared bacterial suspension with a 106 CFU/ml cell density. The positive control was made with 50 μl of bacterial suspension and 50 μl of MH broth. On the other hand, the negative control consisted of only 100 μl MH broth. The plates were then incubated for 18–24 hours at 37°C in a humidified environment (Binder incubator, type B53). Optical density (OD) at 600 nm was measured using an enzyme-linked immunosorbent assay (Epoch, BioTek) microplate reader after incubation to determine bacterial growth.
The MBC was also computed by transferring 20 μl aliquots from the MIC well and two additional higher concentrations to a fresh 96-well plate containing 80 μl presterilized PBS to prepare 8 dilutions of each concentration. Then, 10 μl of each dilution was transferred to a presterilized MH agar plate and incubated for 24 hours at 37°C in a humidified incubator in accordance with the recommendations of the CLSI. The minimal effective concentration (MBC) of a peptide was determined to be the concentration at which less than 0.1% bacterial subculture survives.
Determination of the fractional inhibitory concentration (FIC)
The FIC index was measured by dividing the lowest inhibitory concentration of every antibiotic in combination with that of antibiotics alone. The microdilution checkerboard technique was used to estimate the antibacterial activity of HEA-9 antibiotic combinations.
The FIC index for the combinations was calculated as follows (Masadeh et al., 2022):
The FIC values were interpreted as follows:
≤0.5: synergistic effect,
0.5 to ≤1: additive effect,
1 to <4: indifference,
FIC ≥ 4: antagonistic effect.
Determination of the antibiofilm activity
The antibiofilm activity of the HEA-9 peptide was tested utilizing the producer’s guidelines and as reported by Ceri et al. (2001). The biofilm was formed by adopting the procedure described by Ceri et al. (2001) and using the Calgary Biofilm Device (Innovotech Inc., Edmonton, Canada). Using Mueller- Hinton broth media, 107 CFU/ml bacterial suspensions of Gram-positive MDR S. aureus (ATCC BAA-41) and Gram-negative MDR E. coli (ATCC BAA-2452) bacteria were prepared by diluting a fresh bacterial culture. Then, 96-well plates containing 150 μl of the bacterial inoculum were then covered with a 96-peg lid for the biofilm to grow on. Plates’ incubation was done using an orbital shaker incubator (JSR shaking incubator) for 24 hours at 37°C with agitation at 110 rpm.
Following the formation of the biofilms, the 96-pig lids were washed three times with 200 μl of PBS to eliminate additional nonadherent cells (planktonic bacteria), followed by air-drying for 1 minute. The 96-pig lids were then placed over 96-well plates including 200 μl of 8 concentrations of the HEA-9 peptide through diluting a stock solution exploiting Molar Hinton broth as a solvent. The positive and negative controls for this challenge plate were prepared by filling the last two column wells with 200 μl of broth; after that, the plate was incubated for 4 hours in an orbital shaker (JSR shaking incubator) at 37°C with agitation at 110 rpm. The 96-pig lids were washed three times after the biofilm treatment using 200 μl PBS and air-dried for 1 minute. After that, the 96-pig lids were placed over a 96-well plate containing 200 μl PBS and sealed and then sonicated in a water bath (Clifton digital ultrasonic cleaner) for 20 minutes for the biofilm to detach from the pig lids. After sonication, 50 μl from each well of the recovery plate was moved to a fresh 96-well plate having 100 μl of broth and then incubated for 18–20 hours. The OD of the biofilms was calculated at = 600 utilizing a plate reader to estimate the minimal biofilm eradication concentration (MBEC). The MBEC was defined as the minimal concentration of HEA-9 peptide required to inhibit biofilm regrowth. In addition, 20 μl aliquots were removed from the recovery plate after sonication into a fresh 96-well plate, including 80 μl of PBS. Then, each aliquot was twofold serially diluted 8 times to measure the biofilm viable cell count following peptide treatment. The aliquots of the bacterial suspension were plated on an MH agar for counting. The minimum bactericidal concentration on biofilm (MBCb) was the least peptide concentration required to kill 99.9% of bacteria.
Cytotoxicity assays
Two tests were used to assess the toxicity of HEA-9 in vitro: the 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) cell proliferation test and the erythrocyte hemolysis assay.
MTT cell proliferation assay
The African green monkey kidney epithelial cell-derived Vero cell line was utilized in this experiment. The cells were grown in a PRMI medium, which was prepared by adding 10% fetal bovine serum (50 ml), 1% penicillin and streptomycin (5 ml), and 1% amphotericin B solution (5 ml). Firstly, the frozen Vero cell stock was melted in a water bath, and then 2 ml of cell suspension was placed into a cell culture flask (T75, Korea) containing 25 ml of ready RPMI media. The flask was then incubated at 37°C for 24 hours in a 5% CO2 incubator (Euroclone CO2 incubator) for cell attachment. The media was changed every 24 hours for 2 weeks until the confluency reached 70%–80%. Thereafter, the medium was discarded, and the cells were harvested and counted. In a flat-bottomed 96-well plate, the Vero cells were seeded at a density of 25,000 cells per well. The plates were incubated for 18–24 hours at 37°C in a 5% CO2 incubator. The next day, the medium in the wells was removed. The HEA-9 peptide was made into seven different concentrations by diluting the stock solution with RPMI culture media. Then, 100 μl of all concentrations was added to the cells in the wells. The plates were then incubated for 20 hours at 37°C in a 5% CO2 incubator. Following the incubation period, the drug solution was discarded, and 20 μl of MTT (5 mg/ml in PBS) solution was added to all treated and the positive control wells. After that, the plates were incubated for 4–6 hours under the same conditions. Finally, the MTT/peptide solution was exchanged with 100 μl of DMSO in all wells. The solution was pipetted up and down until the formazan crystals dissolved in DMSO and the purple color showed. Plate absorbance was computed at λ = 595 nm through a microplate reader.
Erythrocyte hemolytic assay
The hemolysis testing was performed for determining the toxicity of the HEA-9 peptides toward erythrocyte cells (Almaaytah et al., 2012). Briefly, 2 ml of horse blood was suspended in 48 ml of presterilized PBS and centrifuged at 2,000 rpm for 5 minutes. Then, the supernatant was discarded and substituted with another 48 ml of PBS. This step was repeated three times, and the pellet was eventually suspended in 48 ml of PBS and vortexed very well to prepare a 4% RBC suspension. After that, 2 ml of the blood suspension was mixed with 8 concentrations of the HEA-9 peptide that had already been prepared (ranging from 1.56 to 200 μM). Then, 100% hemolysis was induced in the positive control through adding 10 μl of 1% Triton X-100 to 2 ml of 4% blood suspension, and the negative control was prepared by adding 2 ml of PBS to 2 ml of 4% blood suspension. Following that, the samples were incubated at 37°C for 60 minutes. Following incubation, the samples were gently mixed before being moved to an Eppendorf test tube and centrifuged at 2,000 rpm for 5 minutes. Then, a 96-well plate was used to collect the supernatant. Finally, the percentage of hemolysis was determined by measuring the absorbance at 450 nm in a microplate reader.
RESULTS
Peptide design, molecular modeling, and in silico analyses
The HEA-9 peptide is made up of 21 amino acids. It was composed of two parts: the N-terminus was derived from the helix N-terminus of cecropin A (3–9) amino acids, and the C-terminus was derived from the helical N-terminus of BMAP-27 (3–14) amino acids (these residues are underlined in Table 1). Additionally, the first lysine amino acid in the hybrid peptide was changed into glutamic acid (the substituted amino acid is bold and underlined in Table 1) to enhance the physicochemical characteristics of the AMP by lowering its net positive charge without compromising its hydrophobicity (Bauer, 1989;Janocha, 2011). HNN was used to predict the helicity percentages (Masadeh, 2022). The secondary structure of HEA-9 was predicted to have 90.48% alpha helices and 9.52% random coils. These values differed from the parent peptides, which used less helicity and more random coils, as indicated in Table 1.
The ProtParam analysis software and APD3 were used to examine the physicochemical properties of the parents, hybrid, and HEA-9 peptides (Kardani and Bolhassani, 2021; Roy et al., 2011). The results are shown in Table 2. HEA-9 has an estimated molecular weight of 2,788.51 g/mol and an isoelectric point of 10.79. The instability index is 12.37. This value reflects the peptide’s stability in a test tube, which has to be <30 {Sahay, 2020 #32}. In addition, the aliphatic index value shows the thermostability of the substance. The result for HEA-9 indicates that the HEA-9 peptide is thermostable. The GRAVY score is the grand average of hydropathy −0.805, which is negative and near zero for the HEA-9 peptide, indicating the peptide’s hydrophilicity (Baeumlisberger et al., 2010). Finally, the Boman index is computed from APD to evaluate the peptide’s protein binding potential (Table 2).
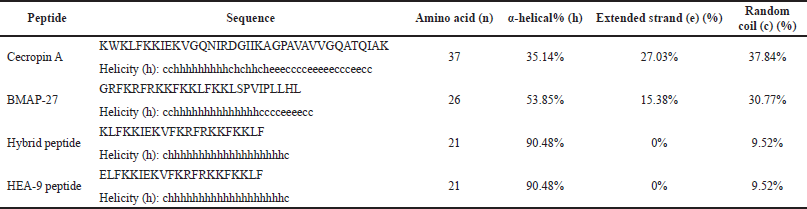 | Table 1. The results of NPS HNN secondary structure analysis. The h in the peptides sequence represents the helical portions of the peptides. The underlined amino acids in parent peptides represent those used in the hybrid peptide. The underlined and bolded amino acid represents the substituted one when preparing the final HEA-9 peptide from the hybrid peptide. [Click here to view] |
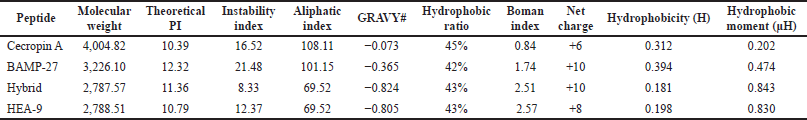 | Table 2. The physicochemical properties, the net charge, the mean hydrophobicity (H), and hydrophobic moment (μ?) for the parent, hybrid, and HEA-9 peptides using the ProtParam software and APD3, Innovagen’s peptide calculator, and the HydroMCale software, respectively. [Click here to view] |
Also, the net charge for the parent, hybrid, and HEA-9 peptides was calculated using Innovagen’s peptide calculator software (Gupta et al., 2013); HEA-9 (+8) has a greater positive charge than cecropin A (+6) and a smaller net positive charge than both the hybrid and BMAP-27 (+10) (Table 2). Additionally, the HEA-9 peptide exhibited lower hydrophobicity than the parent peptides, cecropin A and BAMP-2, as shown in Table 2. This can influence its ability to be partitioned into the lipid bilayer and hence its efficacy and toxicity (Ciumac et al., 2019). Increasing or lowering the hydrophobic percentage outside of its optimal range may decrease antimicrobial activity due to increased self-association caused by the increased hydrophobicity, reducing the peptide concentration required for bacterial membrane action (Chen et al., 2007). Additionally, the HEA-9 peptide showed a greater hydrophobic moment than the parent peptides (Table 2), which is a quantitative indication of amphipathicity (Ron?evi? et al., 2019). The amphipathicity of a peptide sequence relates to its topographic distribution of hydrophobic (binds to the lipid bilayer) and polar (attaches to the phospholipids) residues, which leads to pronounced spatial separation in the active AMP structure (Jureti? et al., 2018). Besides that, amphipathicity amplifies helical peptide activity by allowing them to sink their hydrophobic faces into the membrane bilayer, which is a necessary stage in membrane depolarization (Jiang et al., 2021) .
Moreover, the HEA-9’s three-dimensional structural model was created using the I-TASSER software (Beaufays et al., 2012). The best model was chosen, and it showed a continuous, unbroken alpha-helix conformation of the HEA-9 peptide, which fits the previous theoretical simulations (Supplementary Fig. S1).
Peptide synthesis and purification
The HEA-9 peptide was synthetically produced using the solid-phase approach and Fmoc chemistry. Reverse-phase HPLC was used to check the HEA-9’s pureness, and the chromatogram revealed its synthesis with high purity >95.34% (Fig. 1), which was in agreement with the standard purity requirements for peptide synthesis that was necessary for in vitro research. The HEA-9 ESI-MS analysis result (Fig. 1) shows significant peaks in the +3, +4, +5, and +6 charge states of 930.8, 698.1, 558.7, and 465.8 Da, respectively, supporting the peptide’s identification.
Bacterial susceptibility assay
The HEA-9 peptide showed similar potency against two Gram-positive bacteria strains, sensitive (ATCC29213) and MDR (BAA-41) S. aureus, and two Gram-negative bacteria strains, susceptible (25922) and MDR (BAA-2452) E. coli. The MIC value was 12.5 μM (Table 3). The MBC value for the HEA-9 peptide against all examined strains of bacteria was consistent with the MIC values, indicating that the peptide possesses bactericidal activity. The MIC and MBC values of HEA-9 against all bacterial strains are listed in Table 3.
Checkerboard assay results
The checkerboard microdilution technique was used to determine the MIC of ciprofloxacin, doxycycline, ampicillin, and rifampicin with the HEA-9 peptide against control and MDR strains of Gram-positive and Gram-negative bacteria. We investigated 16 antimicrobial combinations, as shown in Table 4. In the ampicillin–HEA-9 and rifampicin–HEA-9 combinations against sensitive and MDR E. coli bacteria, the MIC value of the HEA-9 peptide was dramatically decreased (99.22% reduction). In addition, we notice a significant drop in the MIC values of conventional antibiotics. For example, the combination of doxycycline with HEA-9 leads to a significant drop in HEA-9’s MIC (99.22% reduction) against the sensitive strain of S. aureus. Furthermore, in the case of MDR S. aureus, the four medication combinations have the same significant influence on the HEA-9 MIC value (87.52% reduction).
Determination of FIC
After calculating the FIC indices for each combination, researchers found that 93.75% of the groups using the combinations showed synergistic activity (FIC 0.5) against the target microorganisms. With a FIC value of 0.039, the combination of HEA-9-ampicillin against E. coli (25922) had the highest synergistic activity of all combinations. Only the ciprofloxacin-HEA-9 combination against S. aureus (29213) showed an additive effect with a FIC value of 0.75 out of the 16 combinations studied. In Table 4, a summary of all of the FIC index results against all of the bacteria examined can be found.
Antibiofilm activity of HEA-9
The HEA-9 antibiofilm activity was examined against MDR S. aureus (BAA-41) and MDR E. coli (ATCC 2452). After 4 hours of exposure, the MBEC was set as the peptide concentration necessary to inhibit biofilm regrowth. The MBEC values for HEA-9 against MDR S. aureus (BAA-41) and MDR E. coli (ATCC 2452) were 25 and 100 μM, respectively, which was significantly higher than the MIC value against planktonic cells. The MBCbs values for HEA-9 against MDR S. aureus (BAA-41) and MDR E. coli (ATCC 2452) were 25 and 100 μM, respectively, matching the corresponding MBECs (Table 5).
MTT cell proliferation assay
The MTT assay was performed to measure Vero cells proliferation as an indication of the toxicity of HEA-9 and its selectivity after they were exposed to eight concentrations of HEA-9 peptide (400, 200, 100, 50, 25, 12.5, 3.125, and, 6.25 μM). At a half-maximal inhibitory concentration (IC50) value, the concentration was 54 μM (Fig. 2). This number is more than the effective MIC of HEA-9 against all planktonic strains. Therefore, based on this, we may conclude that HEA-9 is relatively not toxic to Vero cells.
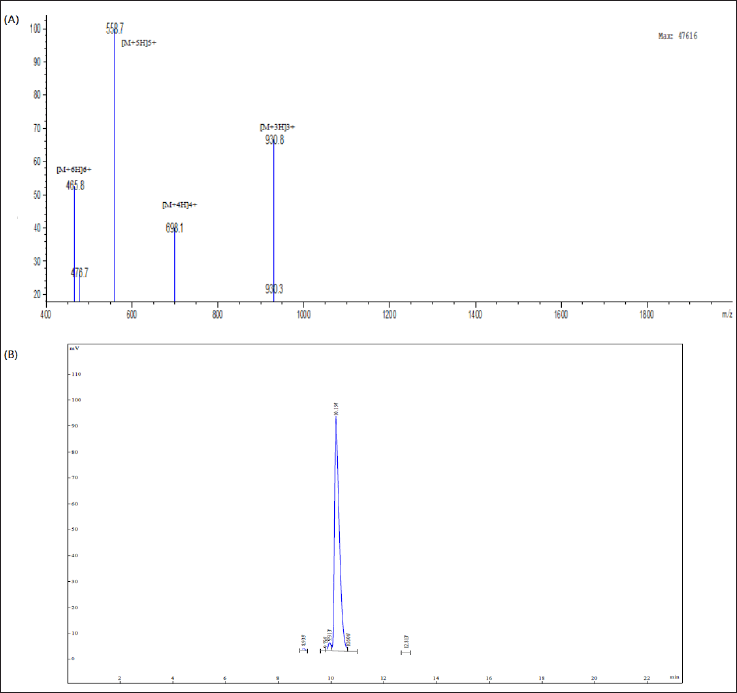 | Figure 1. (A) The analytical RP-HPLC chromatogram of the synthetic modified hybrid peptide HEA-9. (B) The HEA-9 peptide ESI-MS analysis report shows significant peaks in the +3, +4, +5, and +6 charge states of 930.8, 698.1, 558.7, and 465.8 Da, respectively. [Click here to view] |
Hemolytic assay
The hemolysis assay determined the HEA-9’s toxicity to red blood cells. In this study, eight concentrations of HEA-9 (200, 100, 50, 25, 12.5, 6.25, 3.125, and 1.56 μM) were incubated with a 4% blood suspension for 1 hour to assess the hemolysis percentage (Fig. 3). The maximum reported hemolysis percentage at the 200 μM concentration was 26.10% ± 4.42%. On the other hand, the HEA-9 peptide showed only 1.15% ± 0.13% hemolysis at the MIC/MBC concentration (12.5 μM), indicating its safety.
DISCUSSION
The high prevalence of bacterial infection with MDRs poses a challenge to the healthcare system because these bacteria are resistant to most conventional antibiotics on the market and may form biofilms, which function as massive barriers for medications to pass through (Fernandes, 2006; Ventola, 2015). Eight of the 12 antibiotics that have been launched since 2000 have widespread resistance to clinical isolates. Moreover, within 1 year of its market introduction, resistant isolates to the most current antibiotic combination, ceftazidime/avibactam, were reported (Shields et al., 2017). As a result, creating new classes of antibiotics is urgently needed to combat the rise in bacterial resistance to currently existing antibiotics (Simões et al., 2010).
AMPs have been proposed as possible antibacterial substitutes for standard antibiotics. However, the antibacterial mechanism of AMPs is significantly different when compared to traditional antibiotics (Wang et al., 2016b). Antibiotics disrupt the inner biosynthesis of RNA, DNA, peptidoglycan, proteins, and folic acid (Neu and Gootz, 1996), while AMPs are less susceptible to drug resistance since their processes are mainly connected to interactions with the bacterial cell membrane (Andersson et al., 2016; Moravej et al., 2018).
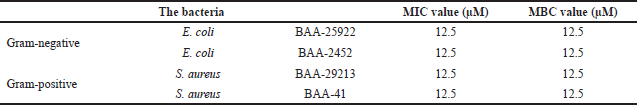 | Table 3. MIC and MBC values of HEA-9 against all investigated bacterial strains (the results represent triplicates). [Click here to view] |
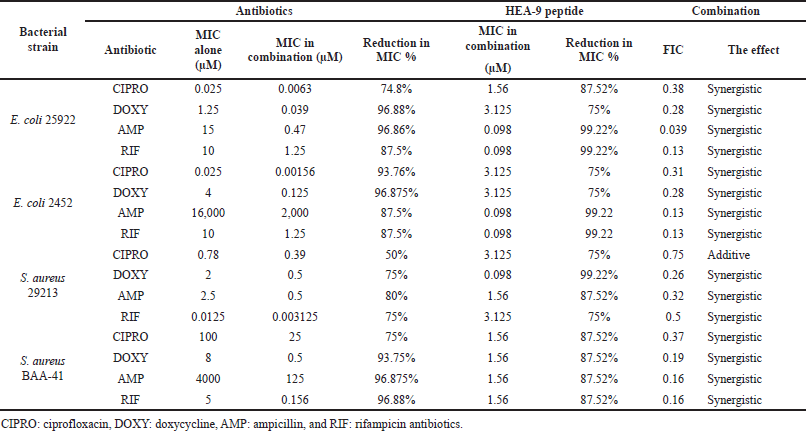 | Table 4. MIC of conventional antibiotics–HEA-9 peptide combination against Gram-negative and Gram-positive bacteria. The percentage of reduction in MIC value for the drug in combination compared to the MIC for the drug alone. [Click here to view] |
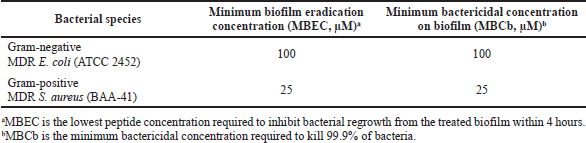 | Table 5, Antibiofilm activity of HEA-9 toward different bacterial species. [Click here to view] |
Several efforts have been undertaken over the years to improve the efficacy of AMPs against pathogens and minimize their unwanted cytotoxicity to eukaryotic cells (Eckert, 2011). One successful method for producing novel AMPs with enhanced antibacterial activity, but with reduced cytotoxicity, is hybridizing and modifying various AMP sequences (Klubthawee et al., 2020; Wei et al., 2016). The present study applied both the hybridization and sequence modification techniques to design the HEA-9 peptide. Thereafter, the physicochemical parameters of the AMP were evaluated using several online tools.
Two unique peptides were chosen for the design: BMAP-27 and cecropin A. Each of these peptides has its own set of issues that prevent it from further development and use in clinical practice. Cecropin A, for example, is thought to be safe, yet it is ineffective against S. aureus species (Moore et al., 1996). The N-terminal amphipathic alpha-helix domain, which corresponds to the first 7–8 residues, has widespread usage to develop a significant number of new peptides, including cecropin A (1–8)-melittin (4–12) (Wu et al., 2014). Therefore, a third peptide was attached to the ninth amino acid residue, the hybrid peptide N-terminal domain. On the other hand, BMAP-27 is a very effective peptide but has substantial toxicity toward human erythrocytes. This toxicity is thought to be caused by the hydrophobic C-terminal residues (Gennaro and Zanetti, 2000; Lee et al., 2011). Consequently, a sequence from the third to the 14th amino acids for the C-terminal domain was chosen for the novel peptide design of HEA-9. After that, the first lysine residue was replaced with glutamic acid to improve the physicochemical properties. Finally, HEA-9 consisted of 21 amino acids with improved helicity of 90.48% (Table 1). It had two negatively charged glutamic acid residues, two positively charged arginine residues, and eight positively charged lysine amino acid residues, which contributed to the (+8) net charge. Thus, the peptide was expected to interact with the negatively charged components of bacterial cell membranes, including the lipoteichoic acid of Gram-positive bacteria and the lipopolysaccharides (LPS) groups in Gram-negative bacteria. Additionally, HEA-9 contains nine hydrophobic residues [five phenylalanine (F), one valine (V), one isoleucine (I), and two leucine (L)] with a total hydrophobic ratio of 43%. The hydrophobic residues were expected to interact with the membrane’s hydrophobic core, anchoring the peptide to the membrane and enabling it to penetrate more into the hydrophobic core, therefore facilitating its antimicrobial activity (Malanovic and Lohner, 2016). Moreover, the GRAVY score of HEA-9 suggests moderate hydrophilicity, while the stability and aliphatic indices predicted through the APD software demonstrated exceptional thermostability and stability (Table 2).
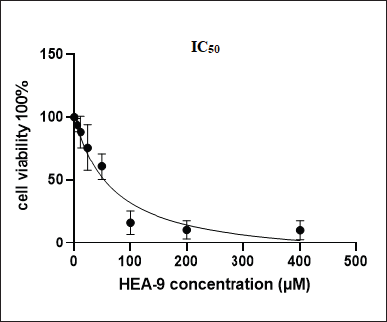 | Figure 2. VERO cells viability after exposure to different concentrations of the HEA-9 peptide. [Click here to view] |
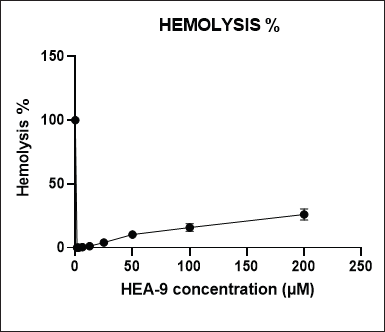 | Figure 3. Hemolytic effect of HEA-9 peptide on human erythrocytes after 1 hour of incubation. The results were measured at λ = 450 (results represent triplicates). [Click here to view] |
When the HEA-9 peptide was tested for antimicrobial vulnerability, the new rationally designed modified hybrid peptide HEA-9 exhibited a wide antimicrobial activity against all tested bacterial strains. Furthermore, the MIC/MBC values of HEA-9 against the control and MDR Gram-positive and Gram-negative strains were both 12.5 μM, showing that the new peptide has bactericidal properties (Table 3). This also may indicate that the peptide targets a common component in both Gram-positive and Gram-negative bacteria and is not affected by the differences in the cell wall components. Moreover, AMPs are known to attack different targets, structures, or types of lipids including core structures such as charged phospholipids that collectively end up causing cell death (Halder and Karmakar, 2022; Ko et al., 2020). On the contrary, parent peptide MIC values of cecropin A against both S. aureus and E. coli were reported to be 64 μM and 0.5 μM, respectively (Lee et al., 2013). Nevertheless, the MIC values of BMAP-27 were within 2–4 μM against both S. aureus and E. coli (Yang et al., 2019).
Combination treatment with conventional antibiotics, often known as synergistic studies, is commonly used to evaluate the antibacterial efficacy of the conventional antibiotics–peptides combinations. This method is particularly effective in reducing the chance of resistance, increasing combined medication effectiveness, and, most significantly, lowering the effective dose of both the peptide and the antibiotics, thus lowering its toxic effects and production expenditure (Gill et al., 2015; Zharkova et al., 2019). The synergistic experiments of HEA-9 with four conventional antibiotics (ciprofloxacin, doxycycline, ampicillin, and rifampicin) against all tested bacterial strains (both control and resistant strains of S. aureus and E. coli) resulted in significantly lower effective MICs for both HEA-9 and the antibiotics. Fifteen out of the 16 combinations exhibited synergistic activity, and ampicillin–HEA-9 had the lowest FIC index against E. coli (ATCC25922), with a FIC of 0.039 μM. The remaining combination (ciprofloxacin–HEA-9 combination against the sensitive strain of S. aureus ATCC 29213) exhibited additive behavior (Table 4). Moreover, the lowest MIC value for HEA-9 was 0.098 μM, which was reported in the combination of rifampicin–HEA-9 and ampicillin–HEA-9 against sensitive and MDR strains of E. coli. Furthermore, rifampicin and ampicillin MICs in the combinations were reduced by 93.75% and 96.86%, respectively, against E. coli (ATCC25922) and by 93.75% and 87.5%, respectively, against MDR E. coli (ATCC 2452). Furthermore, the combination of doxycycline and HEA-9 against S. aureus (ATCC29213) demonstrated synergistic action, with the MIC of HEA-9 being reduced by 99.22% (0.098 μM) (Table 4).
The results of HEA-9 and conventional antibiotic combinations revealed that the combined drugs increase each other’s activities, suggesting the combined agents have a different mode of action. Except for ampicillin, which inhibits bacterial growth by inhibiting cell wall synthesis, all antibiotics examined in this study had intracellular targets, including protein and inhibition of nucleic acid synthesis (Walsh, 2003). One proposed explanation for the synergistic effect is that AMPs might degrade the peptidoglycan layer, increasing membrane permeability and, therefore, facilitating antibiotics’ entrance, increasing their intracellular concentration, and promoting their efficacy (Mahlapuu et al., 2016; Zhang et al., 2014).
The antibiofilm activities of HEA-9 were also investigated, and it displayed significant antibiofilm activity against MDR E. coli (ATCC 2452) and MDR S. aureus (ATCC BAA-41). Both bacterial strains showed the same MBEC and MBCb values for HEA-9, but these values were eight and two times greater than the corresponding MIC values against the planktonic forms of MDR E. coli (ATCC 2452) and MDR S. aureus (ATCC BAA-41), respectively. As mentioned previously, this difference may be due to peptide interaction with a different component of the EPS for each strain (Donlan and Costerton, 2002). Overall, those findings indicate that HEA-9 is a potentially effective antibiofilm agent, particularly against MDR S. aureus (ATCC BAA-41). It has been shown that MDR S. aureus is a significant cause of health-related and community-associated infections due to its ability to form biofilms on tissues and medical devices (Tong et al., 2015).
Finally, HEA-9’s peptide toxicity against mammalian cells was also investigated using the Vero cell line. Antimicrobial activity is maximized at concentrations that have no detectable effect on mammalian cell viability. The IC50 value of HEA-9 was reported to be 54 μM, which is more than four times higher than the MIC/MBC against the planktonic cells of both Gram-positive and Gram-negative bacteria (12.5 μM). Furthermore, it was found that cell viability at the indicated dose to suppress MDR S. aureus (25 μM) biofilm is about 76%.
In addition, the hemolytic impact of HEA-9 was evaluated, and the highest hemolysis was 26.10% ± 4.42% at a concentration of 200 μM, which was 16 times higher than the concentration required to kill the planktonic cells of both the sensitive and resistant strains of E. coli and S. aureus. On the other hand, it caused only 1.15% ± 0.13% of hemolysis at the MIC value (12.5 μM) of the peptide. It was also shown that, at the dose required to suppress MDR S. aureus biofilm formation, HEA-9 only induced 4.08% ± 0.83% hemolysis which renders HEA-9 safer to use in the treatment of bacterial infections in relation to the parent peptide BMAP-27 that demonstrated hemolytic activity at 6.2 μM (Lange, 2011; Skerlavaj et al., 1996).
CONCLUSION
Designing an AMP utilizing computer-aided technologies is regarded as one of the fastest medication development procedures. This study used cecropin A and BMAP-27 sequences to design a novel modified hybrid peptide, HEA-9, demonstrating improved antibacterial efficacy against planktonic Gram-positive and Gram-negative bacterial cells and a low toxicity profile against normal cells. Moreover, HEA-9 demonstrated selectivity, reduced cytotoxicity, and a reduced hemolytic effect on mammalian erythrocytes. In addition, when combined with four different antibacterial agents, the antimicrobial activity and toxicity profiles were significantly enhanced. Furthermore, HEA-9 displayed safe and significant antibiofilm properties, especially against MDR S. aureus strains.
FUNDING
This study was supported by the Deanship of Research at Jordan University of Science and Technology (Grant No. 2022/20).
CONFLICTS OF INTEREST
The authors report no conflicts of interest in this work.
ETHICAL APPROVAL
The institutional review board approved the Jordan University of Science and Technology has approved the study protocol.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Albsoul-Younes A, Wazaify M, Yousef AM, Tahaineh L. Abuse and misuse of prescription and nonprescription drugs sold in community pharmacies in Jordan. Subst Use Misuse, 2010; 45(9):1319–29. CrossRef
Almaaytah A, Zhou M, Wang L, Chen T, Walker B, Shaw C. Antimicrobial/cytolytic peptides from the venom of the North African scorpion, Androctonus amoreuxi: biochemical and functional characterization of natural peptides and a single site-substituted analog. Peptides, 2012; 35(2):291–9. CrossRef
Andersson DI, Hughes D, Kubicek-Sutherland JZ. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist Updates, 2016; 26:43–57. CrossRef
Azmi F, Skwarczynski M, Toth I. Towards the development of synthetic antibiotics: designs inspired by natural antimicrobial peptides. Curr Med Chem, 2016; 23(41):4610–24. CrossRef
Baeumlisberger D, Arrey TN, Rietschel B, Rohmer M, Papasotiriou DG, Mueller B, Beckhaus T, Karas M. Labeling elastase digests with TMT: informational gain by identification of poorly detectable peptides with MALDI-TOF/TOF mass spectrometry. Proteomics, 2010; 10(21):3905–9. CrossRef
Beaufays J, Lins L, Thomas A, Brasseur R. In silico predictions of 3D structures of linear and cyclic peptides with natural and non-proteinogenic residues. J Pept Sci, 2012; 18(1):17–24. CrossRef
Cama J, Leszczynski R, Tang P, Khalid A, Lok V, Dowson C, Ebata A. To push or to pull? In a post-COVID world, supporting and incentivizing antimicrobial drug development must become a governmental priority. ACS Infect Dis, 2021; 7:2029–42. CrossRef
Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob Agents Chemother, 2007; 51(4):1398–406. CrossRef
Ceri H, Olson M, Morck D, Storey D, Read R, Buret A, Olson B. The MBEC assay system: multiple equivalent biofilms for antibiotic and biocide susceptibility testing. Methods Enzymol, 2001; 337:377–85. CrossRef
CLSI. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI, Wayne, PA, 2014.
Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci, 2000; 25(3):147–50. CrossRef
Ciumac D, Gong H, Hu X, Lu JR. Membrane targeting cationic antimicrobial peptides. J Colloid Interface Sci, 2019; 537:163–85. CrossRef
Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev, 2002; 15(2):167–93. CrossRef
Eckert R. Road to clinical efficacy: challenges and novel strategies for antimicrobial peptide development. Future Microbiol, 2011; 6(6):635–51. CrossRef
Epand RM, Epand RF. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim Biophys Acta, 2009; 1788(1):289–94. CrossRef
Gupta UK, Mahanta S, Paul S. In silico design of small peptide-based Hsp90 inhibitor: a novel anticancer agent. Med Hypotheses, 2013; 81(5):853–61. CrossRef
Fernandes P. Antibacterial discovery and development—the failure of success? Nat Biotechnol, 2006; 24(12):1497–503. CrossRef
Fjell CD, Hiss JA, Hancock RE, Schneider G. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov, 2012; 11(1):37–51. CrossRef
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM (Ed.). The proteomics protocols handbook, Humana Press, Totowa, NJ, pp 571–607, 2005. CrossRef
Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics, 2008; 24(18):2101–2. CrossRef
Gelband H, Molly Miller P, Pant S, Gandra S, Levinson J, Barter D, White A, Laxminarayan R. The state of the world’s antibiotics 2015. Wound Healing South Afr, 2015; 8(2):30–4.
Gennaro R, Zanetti M. Structural features and biological activities of the cathelicidin–derived antimicrobial peptides. Peptide Sci, 2000; 55(1):31–49. CrossRef
Giangaspero A, Sandri L, Tossi A. Amphipathic α helical antimicrobial peptides. A systematic study of the effects of structural and physical properties on biological activity. Eur J Biochem, 2001; 268(21):5589–600. CrossRef
Gill EE, Franco OL, Hancock RE. Antibiotic adjuvants: diverse strategies for controlling drug–resistant pathogens. Chem Biol Drug Des, 2015; 85(1):56–78. CrossRef
Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol, 2018; 16(2):91–102. CrossRef
Huang Y, Huang J, Chen Y. Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cell, 2010; 1(2):143–52. CrossRef
Halder A, Karmakar S. An evidence of pores in phospholipid membrane induced by an antimicrobial peptide NK-2. Biophy Chem, 2022; 282:106759. CrossRef
Huan Y, Kong Q, Mou H, Yi H. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front Microbiol, 2020; 11:582779. CrossRef
Jiang Y, Chen Y, Song Z, Tan Z, Cheng J. Recent advances in design of antimicrobial peptides and polypeptides toward clinical translation. Adv Drug Deliv Rev, 2021; 170:261–80. CrossRef
Jureti? D, Sonavane Y, Ili? N, Gajski G, Goi?-Bariši? I, Tonki? M, Kozic M, Maravi? A, Pellay FX, Zorani? L. Designed peptide with a flexible central motif from ranatuerins adapts its conformation to bacterial membranes. Biochim Biophys Acta Biomembr, 2018; 1860(12):2655–68. CrossRef
Kardani K, Bolhassani A. Exploring novel and potent cell penetrating peptides in the proteome of SARS-COV-2 using bioinformatics approaches. PLoS One, 2021; 16(2):e0247396. CrossRef
Klubthawee N, Adisakwattana P, Hanpithakpong W, Somsri S, Aunpad R. A novel, rationally designed, hybrid antimicrobial peptide, inspired by cathelicidin and aurein, exhibits membrane-active mechanisms against Pseudomonas aeruginosa. Sci Rep, 2020; 10(1):1–17. CrossRef
Ko SJ, Park E, Asandei A, Choi JY, Lee SC, Seo CH, Luchian T, Park Y. Bee venom-derived antimicrobial peptide melectin has broad-spectrum potency, cell selectivity, and salt-resistant properties. Sci Rep, 2020; 10(1):10145. CrossRef
Kourkouta L, Koukourikos K, Iliadis C, Plati P, Dimitriadou A. History of antibiotics. Sumerian J Med Healthcare, 2018; 1:51–5. CrossRef
Kumar P, Kizhakkedathu JN, Straus SK. Antimicrobial peptides: diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules, 2018; 8(1).
Lange AB. Crustacean cardioactive peptide in the Chagas’ disease vector, Rhodnius prolixus: presence, distribution and physiological effects. Gen Comp Endocrinol, 2011; 174(1):36–43. CrossRef
Lee E, Jeong KW, Lee J, Shin A, Kim JK, Lee J, Lee DG, Kim Y. Structure-activity relationships of cecropin-like peptides and their interactions with phospholipid membrane. BMB Rep, 2013; 46(5):282. CrossRef
Lee EK, Kim YC, Nan YH, Shin SY. Cell selectivity, mechanism of action and LPS-neutralizing activity of bovine myeloid antimicrobial peptide-18 (BMAP-18) and its analogs. Peptides, 2011; 32(6):1123–30. CrossRef
Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov, 2013; 12(5):371–87. CrossRef
Liévin-Le Moal V, Servin AL. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev, 2006; 19(2):315–37. CrossRef
Lundstedt E, Kahne D, Ruiz N. Assembly and maintenance of lipids at the bacterial outer membrane. Chem Rev, 2021; 121(9):5098–123. CrossRef
Mahlapuu M, Håkansson J, Ringstad L, Björn C. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol, 2016; 6:194. CrossRef
Malanovic N, Lohner K. Antimicrobial peptides targeting Gram-positive bacteria. Pharmaceuticals, 2016; 9(3):59. CrossRef
Masadeh M, Ayyad A, Haddad R, Alsaggar M, Alzoubi K, Alrabadi N. Functional and toxicological evaluation of the MAA-41: a novel rationally designed antimicrobial peptide using hybridization and modification methods from LL-37 and BMAP-28. Curr Pharm Des, 2022; 28:2177–88. CrossRef
Masadeh M, Ayyad A, Haddad R, Alsaggar M, Alzoubi K, Alrabadi N; Functional and toxicological evaluation of MAA-41: a novel rationally designed antimicrobial peptide using hybridization and modification methods from LL-37 and BMAP-28. Curr Pharm Des, 2022; 28(26):2177–88. CrossRef
McDermott P, Zhao S, Wagner D, Simjee S, Walker R, White D. The food safety perspective of antibiotic resistance. Anim Biotechnol, 2002; 13(1):71–84. CrossRef
Mohr KI. History of antibiotics research. How to overcome the antibiotic crisis. Curr Top Microbiol Immunol, 2016; 398:237–72. CrossRef
Moore AJ, Beazley WD, Bibby MC, Devine DA. Antimicrobial activity of cecropins. J Antimicrob Chemother, 1996; 37(6):1077–89. CrossRef
Moravej H, Moravej Z, Yazdanparast M, Heiat M, Mirhosseini A, Moosazadeh Moghaddam M, Mirnejad R. Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb Drug Resist, 2018; 24(6):747–67. CrossRef
Neu HC, Gootz TD. Antimicrobial chemotherapy. Medical microbiology. 4th edition, University of Texas Medical Branch, Galveston, TX, 1996.
Ron?evi? T, Puizina J, Tossi A. Antimicrobial peptides as anti-infective agents in pre-post-antibiotic era? Int J Mol Sci, 2019; 20(22). CrossRef
Roy S, Maheshwari N, Chauhan R, Sen NK, Sharma A. Structure prediction and functional characterization of secondary metabolite proteins of Ocimum. Bioinformation, 2011; 6(8):315–9. CrossRef
Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen M, Clancy CJ. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother, 2017; 61(3):e02097–16. CrossRef
Simões M, Simões LC, Vieira MJ. A review of current and emergent biofilm control strategies. LWT Food Sci Technol, 2010; 43(4):573–83. CrossRef
Skerlavaj B, Gennaro R, Bagella L, Merluzzi L, Risso A, Zanetti M. Biological characterization of two novel cathelicidin-derived peptides and identification of structural requirements for their antimicrobial and cell lytic activities. J Biol Chem, 1996; 271(45):28375–81. CrossRef
Toke O. Antimicrobial peptides: new candidates in the fight against bacterial infections. Peptide Sci, 2005; 80(6):717–35. CrossRef
Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev, 2015; 28:603–61. CrossRef
Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther, 2015; 40(4):277.
Walsh C. Antibiotics: actions, origins, resistance. American Society for Microbiology (ASM), Washington, DC, 2003. CrossRef
Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res, 2016a; 44(D1):D1087–93. CrossRef
Wang S, Zeng X, Yang Q, Qiao S. Antimicrobial peptides as potential alternatives to antibiotics in food animal industry. Int J Mol Sci, 2016b; 17(5):603. CrossRef
Wei XB, Wu RJ, Si DY, Liao XD, Zhang LL, Zhang RJ. Novel hybrid peptide cecropin A (1–8)-LL37 (17–30) with potential antibacterial activity. Int J Mol Sci, 2016; 17(7):983. CrossRef
Wu R, Wang Q, Zheng Z, Zhao L, Shang Y, Wei X, Liao X, Zhang R. Design, characterization and expression of a novel hybrid peptides melittin (1–13)-LL37 (17–30). Mol Biol Rep, 2014; 41(7):4163–9. CrossRef
Yang S, Lee CW, Kim HJ, Jung HH, Kim JI, Shin SY, Shin SH. Structural analysis and mode of action of BMAP-27, a cathelicidin-derived antimicrobial peptide. Peptides, 2019; 118:170106. CrossRef
Zhang Y, Liu Y, Sun Y, Liu Q, Wang X, Li Z, Hao J. In vitro synergistic activities of antimicrobial peptide brevinin-2CE with five kinds of antibiotics against multidrug-resistant clinical isolates. Curr Microbiol, 2014; 68(6):685–92. CrossRef
Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics, 2008; 9(1):40. CrossRef
Zharkova MS, Orlov DS, Golubeva OY, Chakchir OB, Eliseev IE, Grinchuk TM, Shamova OV. Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics—a novel way to combat antibiotic resistance? Front Cell Infect Microbiol, 2019; 9:128. CrossRef
SUPPLEMENTARY MATERIAL
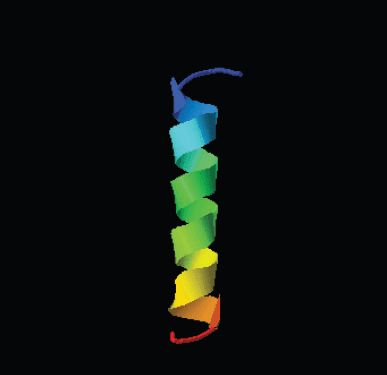 | Supplementary Figure S1. Structure of the HEA-9 peptide as predicted by I-TASSER. [Click here to view] |