INTRODUCTION
With the considerable developments made in the field of bioinformatics, in silico approaches of drug repurposing have become a more attractive approach recently (DeOliveira and Lang, 2018). Various antibiotics including N-thiolated β lactams and derivatives (Frezza et al., 2008; Kuhn et al., 2004), erythromycin (Chlebda-Sieragowska et al., 2007, 2013), clarithromycin (Van Nuffel et al., 2015), fluoroquinolones (Yadav and Talwar, 2019), etc. have already been proven to be anticancer agents. The potential of tetracyclines in cancer therapy was first proposed in the 1980’s (Kroon et al., 1984), and molecules such as doxycycline, minocycline, and COL-3 (Rudek et al., 2001) have proven their role in cancer treatment, exerting the effects through different mechanisms. Matrix metalloproteinases (MMPs) are a reliable drug target in pathological conditions such as autoimmune disorders, central nervous system-related disorders, and cancer. But the development of specific MMPs inhibitors is laborious due to the shared structural similarity and overlapping substrate specificity between members of MMPs family which results in broad-spectrum actions on multiple MMPs causing undesirable side effects (Gooljarsingh et al., 2008; Vandenbroucke and Libert, 2014; Verma, 2012).
Recent development of high throughput screening has paved the way to develop specific MMPs with high affinity (Arkadash et al., 2017). Several natural products are also known to be inhibitors of collagenases and gelatinases (Leyon et al., 2005) and most MMPs inhibitors are chelating agents and target the zinc ion located in the catalytic site thereby blocking its activity (Jacobsen et al., 2010). Another class of small molecule-based inhibitors are designed in a way that they could fit to the S’ pocket of MMPs located close to the catalytic site; this site is crucial for substrate recognition (Cathcart et al., 2015; Overall and Kleifeld, 2006). Novel MMP-13 pyrimidine derivative inhibitors utilize this mechanism of inhibition (Nara et al., 2017). Small molecules that make use of exosites for inhibition are also developed with the help of computational studies. This allosteric mechanism usually targets PEX domains of MMPs (Remacle et al., 2012; Udi et al., 2013).
The current study investigated the repurposing potentials of four tetracycline molecules, viz, demeclocycline (C21H21ClN2O8), eravacycline (C27H31FN4O8), lymecycline (C29H38N4O10), and omadacycline (C29H40N4O7) of which two are halogenated (Fig. 6), by analyzing their binding affinity with promising drug targets such as collagenase MMPs and gelatinase MMPs.
MATERIALS AND METHODS
Protein and ligand preparation for docking
High resolution protein 3D structures derived from crystallographic techniques were downloaded from Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB). The downloaded protein PDB structures were prepared for docking as given in the Supplementary Methodology (SM1). Careful examination was done to make sure there are no missing residues in the binding region. Similarly, the drugs (also referred to as ligands) were downloaded from the PubChem database and prepared for docking using Autodock Tools 1.5.6 (ADT) as detailed in SM1. Details of the PDB and Pubchem structures are summarized in Table 1.
Docking studies
Docking studies were conducted using ADT and Vina (Trott and Olson 2010), where the receptor is kept rigid and the ligand is flexible. Algorithm searches are done to look through different ‘poses’ in the prescribed site in the receptor (Supplementary Methodology SM2) and to arrive at the preferred orientation. Scoring functions are employed to finally predict the strength of association or binding affinity between the receptor and ligand. At the end of docking, results are depicted as binding energy/affinity for each pose in kcal/mol with root mean square deviation (RMSD) values.
Molecular dynamic (MD) simulation studies
MD simulation for the protein molecule and protein–ligand complexes were conducted using GROMACS 5.7.4 (2021.4) package (Berendsen et al., 1995; Lemkul 2019). Both ligand and protein were prepared for simulation using the CHARMM force field. Pdb2gmx was used to generate receptor topology and the CGenFF server (Vanommeslaeghe et al., 2010) was used to create the ligand topology. The protein/ligand complexes were solvated in a cubic box with TIP3P water molecules maintaining the edge of the box at least 1.0 nm from all atoms. The system was neutralized by adding Na/Cl to replace solvent molecules. The neutralized system further subjected to energy minimization helps in removing the irregular torsions. The energy minimization was carried out using the steepest descent minimization algorithm for 50,000 steps using 10 kJ/mol/nm as the maximum force. Further to reduce unrestrained dynamics, the system was well equilibrated before stimulation. The equilibration was carried out in two steps, amount of substance (N), volume (V) and temperature (T) and amount of substance (N), pressure (V) and temperature (T). The temperature was maintained constant at 300 K using the Berendsen thermostat (Berendsen et al., 1984). The MD simulation was carried out for 10 ns and the trajectory files obtained were analyzed using xmgrace and visualization using PyMOL (DeLano Number 40 March 2002).
 | Table 1. RCSB-PDB IDs of proteins and pubchem compound identification number (CIDs) of ligand molecules used in the study. [Click here to view] |
RESULTS
Binding affinity of the tetracyclines to 30S complex
The binding affinity of the selected ligand molecules was initially analyzed against their primary target, 30S ribosomal subunit (1FJG) in our in silico systems and the results are in Table 2. As expected, all four tetracyclines (Demeclocycline −10.3 Kcal/mol, Eravacycline −10.3 Kcal/mol, Lymecycline −10.2 Kcal/mol, Omadacycline −9.7 Kcal/mol) showed very strong binding affinity in the ligand binding sockets of 30S complex. On the other hand, Ceftobiprole, a known Penicillin Binding Protein (PBP) binding ligand, gave lesser binding affinity toward 30S complex (8.7 Kcal/Mol) but resulted in strong binding affinity (−9.4 Kcal/Mol) with its known target PBP1b (3VMA). 2D images showing the molecular interactions of tetracyclines with 30S ribosomal subunit are detailed in Figure S1.
Binding affinity of the tetracyclines to collagenases and gelatinases
Out of the four tetracyclines tested, eravacycline showed the strongest molecular interactions with all the collagenase enzymes. The highest binding affinity of −9.7 Kcal/mol with MMP-9 was obtained for eravacycline (Table 3). The details of hydrogen bonds and hydrophobic interactions between eravacycline and MMPs are detailed in Table 4. All the other ligand molecules showed relatively lesser binding affinity ranging between −6.0 and −8.6 Kcal/mol (Table 3). The broad spectrum MMP inhibitor doxycycline, (Jung et al., 2016) that acts as a reference for our study, returned lesser binding scores compared to the experimental tetracyclines. Collagenase selective inhibitors cipemastat (MMP1) (Hemmings et al., 2001), MMP-8i (MMP8) (Kumar et al., 2018), and WAY170523 (MMP13) (Akhtar et al., 2017) gave good docking scores for their respective target protein molecules (Table 3). It was interesting to note that, eravacycline exhibited higher binding affinity to these targets than their selective inhibitors except in the case of WAY170523 which was −10.1 Kcal/mol. The co-crystallized ligand of 3O2X also resulted in strong binding affinity (−10.8 Kcal/mol) and we used this for our validations as per the Supplementary Methodology (SM3) and results are shown in Figure 1a. The binding score of eravacycline to gelatinases was also mostly higher than their selective inhibitors ARP101 (−8.6 Kcal/mol) and SB3CT (−8.1 Kcal/mol) that are known to inhibit MMP2 (Jo et al., 2011) and MMP9 (Qin et al., 2015) respectively. 2D images showing the molecular interactions of eravacycline with collagenases and gelatinases are detailed in Figures S2 and S3, respectively. However, (2~{S}) -2-[2- [4-(4-methoxyphenyl)phenyl] sulfanylphenyl] pentanedioic acid (B9Z), the co-crystallized ligand found in the MMP9 structure had a strong binding affinity in our docking simulations and we also used this interaction for validating our protocol (Fig. 1b). The other tertracyclin ligands exhibited a lower value for binding affinity than that of these selective inhibitors.
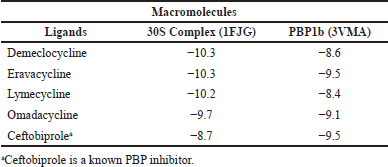 | Table 2. Binding affinity (Kcal/mol) of the selected tetracycline antibiotic molecules and ceftobiprole to their normal targets. [Click here to view] |
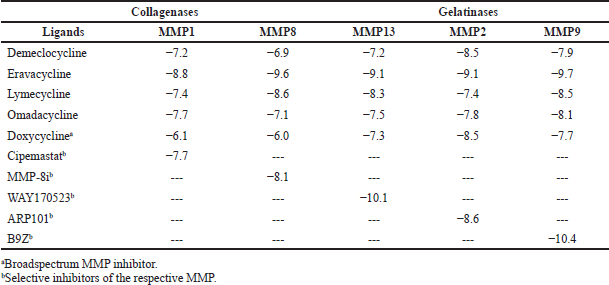 | Table 3. Binding affinity (Kcal/mol) of the selected antibiotic molecules to different matrix metalloproteinases. [Click here to view] |
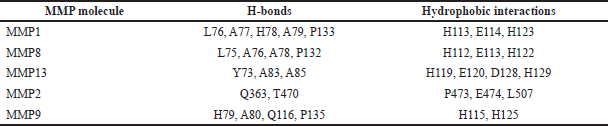 | Table 4. The amount of hydrogen bonds and hydrophobic interactions of the results obtained from the docking studies performed between eravacycline and MMPs. [Click here to view] |
MD trajectory analysis
Considering the highest binding affinity of eravacycline to MMP9, we conducted the MD simulation of this interaction. Three separate runs were made using protein alone, protein complexed with its co-crystallized ligand (B9Z), and protein complexed with eravacycline. The best ‘pose’ (with the lowest binding energy) obtained from the docking studies was used to make protein-ligand complexes for subsequent simulations. The RMSD, the root mean square fluctuation (RMSF) of residues and the radius of gyration of protein or protein-ligand complex were analyzed after the completion of 10 ns MD simulation. The intermolecular hydrogen bonding between the ligand and protein were also analyzed using appropriate gromacs commands. The RMSD graph predicts the stability of molecules in the solvated, charge neutralized system (Fig. 2). The structure of protein-ligands complexes was found to be within 0.15 nm from the backbone indicating a better stability as compared to the protein alone, where it varied slightly higher at 0.2 nm. The RMSF data showed a decrease in the residue fluctuation and thereby increasing the stability of the amino acid residues after binding with the ligands (Fig. 3). Even the residues from 175 to 180 (that forms a loop) which peaked at 0.2 nm in the protein ligand complex, were having a higher fluctuation (0.41 nm) in the protein alone simulation. Further, the compactness of protein or protein-ligand complex was extracted from the MD trajectory by executing g_gyrate (Fig. 4). Here again, the protein-ligand complex was consistently showing a radius of 1.50 nm, whereas in the protein alone simulation, it was at 1.53 nm in the initial phase later coming down to 1.50 nm. Finally, the hydrogen bonds formed between the protein and ligand were analyzed by the g_hbond tool in gromacs. It was found to have two to three hydrogen bonds formed by B9Z and one to two bonds with eravacycline but the number was more consistent in eravacycline throughout the simulation than in B9Z (Fig. 5).
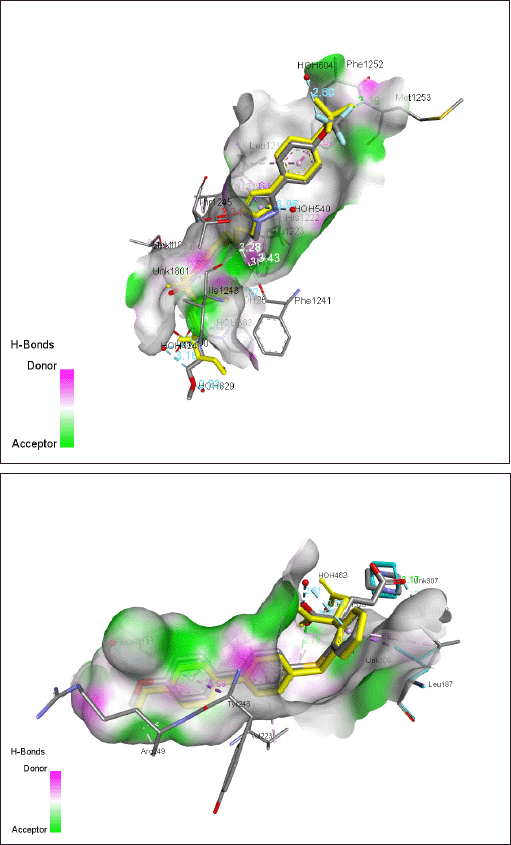 | Figure 1. Superimposed 3D images showing the docking ‘pose’ and interactions of the ligand with respect to that of the co-crystallised ligand. (a) the 3O2 ligand in 3O2X (MMP13) in gray color. ‘Yellow’ 3O2 ligand represents the superimposed ‘pose’ of the docking result. [Click here to view] |
DISCUSSION
Tetracyclines are protein synthesis inhibitors that act by blocking the stable binding of aminoacyl tRNA to the A site located in the 30S subunit of ribosomes during translation. Six binding sites of tetracyclines on the 30S subunits have been identified so far among which the most occupied primary binding site is located at the base of the head of the 30S subunit (Pioletti et al., 2001). Tetracycline utilizes the hydrophilic surface of the molecule to interact with the irregular minor groove of helix 34 (h34) and the loop of h31 of the 16S rRNA (Goldman et al., 1980). In all the six binding sites, residues of 16S rRNA contribute much of the binding activity, but the role of protein subunits such as S4, S9, S17, and S7 cannot be neglected (Goldman et al., 1983, Oehler et al., 1997). Our results are consistent with these previous observations and all the four molecules exhibited very strong binding affinity at or above −9.7 Kcal/mol and the binding interactions involved several of the nucleotide residues of 16S rRNA in common. On the other hand, ceftobiprole, a fifth generation cephalosporin that is known to exert its effect by binding to PBP1b (Kumar et al., 2014), showed stronger affinity with 3VMA than 1FJG. It was also interesting to note that the ligand nucleotide interactions, that were prevalent with tetracycline molecules, are absent with ceftobiprole (Fig. S1).
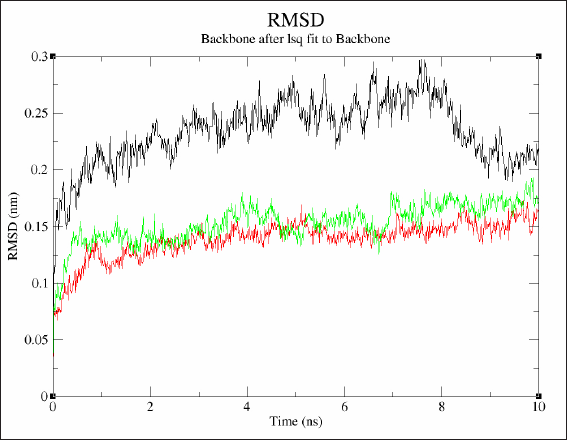 | Figure 2. Superimposed images showing the RMSD of protein or protein ligand complexes from three different MD simulation analysis. Black - Protein alone, Green - Protein complexed with eravacyline, Red - Protein complexed with B9Z [Click here to view] |
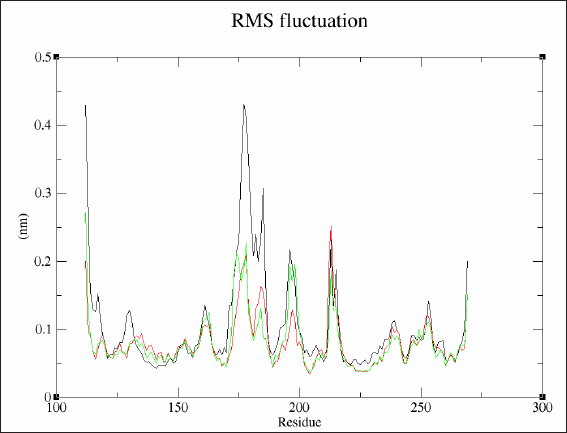 | Figure 3. Superimposed images showing the RMSF of protein or protein ligand complexes from three different MD simulation analysis. Black - Protein alone, Green - Protein complexed with eravacyline, Red - Protein complexed with B9Z. [Click here to view] |
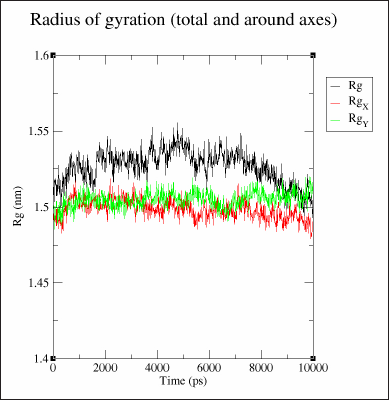 | Figure 4. Superimposed images showing the radius of gyration of protein or protein ligand complexes from three different MD simulation analysis. Black - Protein alone, Green - Protein complexed with eravacyline, Red - Protein complexed with B9Z [Click here to view] |
 | Figure 5. Graphical representation of the number of hydrogen bonds between protein and ligand during the molecular dynamic simulation. (a) Number of hydrogen bonds between MMP9 and B9Z. (b) Number of hydrogen bonds between MMP9 and eravacycline. [Click here to view] |
Considering the importance of MMPs in many neoplastic conditions, their inhibitors are of intense research to use in therapeutics. The candidate ligand molecules under study were therefore docked against two groups of MMPs (Collagenases and Gelatinases) in order to investigate their binding affinity, simultaneously running known specific inhibitors for each MMP. It was observed that all the ligand molecules were able to interact with all the target proteins as the ?G values were all in the negative range, which is indicative of their spontaneous occurrence, but the degree of interaction strength varies greatly. In general, non-covalent interactions like hydrogen bonding, hydrophobic interactions and electrostatic interactions are taken into account while studying protein-ligand interactions. Eravacycline, which showed highest binding affinity with different MMPs exhibited several of the above types of interactions on top of the fluorine interaction with different residues. Eravacycline (XeravaTM) is a novel fully synthetic flurocycline antibacterial within tetracycline class of antimicrobial drugs (Zhanel et al., 2016) that got approved by Food and Drug Administration (FDA) in August 2018 following the IGNITE 1 through IGNITE 4 trials (Alosaimy et al., 2020; Solomkin et al., 2017; Zhanel et al., 2016). It consists of a general tetracyclic core scaffold with modification at C-7 (fluorine) and C-9 (pyrrolidineacetamide group). We further our study to analyze the stability of eravacycline-MMP9 interaction in MD simulation which showed a stable interaction between them. The number of hydrogen bonds were consistent throughout the simulation and the RMSD of the protein was further minimized after the ligand binding. The individual amino acid residue fluctuation was also decreased after the ligand binding. All these results are indicative of a strong inhibitory binding of eravacycline to the catalytic domain of MMP9.
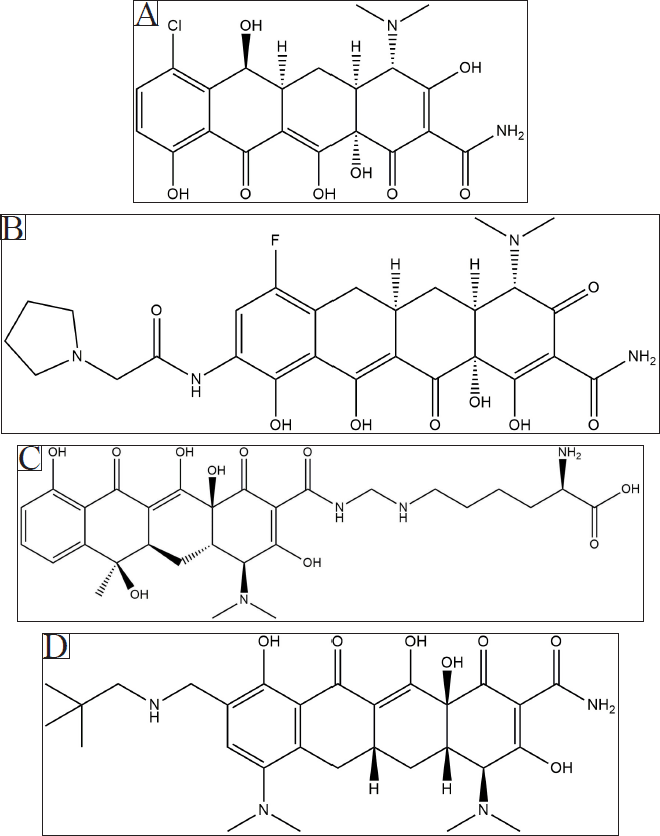 | Figure 6. Chemical structure depiction of the tetracycline molecules, viz, demeclocycline (A), eravacycline (B), lymecycline (C), and omadacycline (D) used for the study. [Click here to view] |
CONCLUSION
Based on our results, eravacycline was found to be a strong inhibitor of MMPs. It is noteworthy that, among the four tetracyclines studied, only eravacycline had a fluorine atom and the computational experiments revealed, eravacycline has a strong interaction between its fluorine atom and target amino acid residues in MMP2 and MMP13. We did MD simulation of the eravacycline-MMP9 complex, but we expect similar results with other eravacycline-MMP interactions since the docking score is very high even in the absence of a halogen interaction. Eravacycline is therefore strongly recommended for further in vitro studies such as gelatin zymography in order to promote it for repurposing as an MMP inhibitor to be studied in pre-clinics. It is especially important because MMPs are a reliable drug target in many pathologies including cancer. If the Eravacycline-MMP interaction is proven to be stable in the in vitro systems, repositioning of this clinical antibiotic to an anticancer drug is worthwhile to be studied.
ABBREVIATIONS
B9Z - (2~{S}) -2-[2- [4-(4-methoxyphenyl)phenyl] sulfanylphenyl] pentanedioic acid
CID - Compound identification number
FDA - Food and Drug Administration
MD - Molecular dynamic
MMP - Matrix metalloProteinase
PDB - Protein data bank
PBP - Penicillin binding protein
PEX - A protein domain (eg hemopexin)
RCSB - Research Collaboratory for Structural Bioinformatics
RMSD - Root mean square deviation
RMSF - Root mean square fluctuation
FUNDING
There is no funding to report.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Akhtar N, Khan NM, Ashruf OS, Haqqia TM. Inhibition of cartilage degradation and suppression of PGE2 and MMPs expression by pomegranate fruit extract in a model of posttraumatic osteoarthritis. Nutrition, 2017; 33(January):1–13. CrossRef
Alosaimy S, Abdul-Mutakabbir JC, Kebriaei R, Jorgensen SCJ, Rybak MJ. Evaluation of eravacycline: a novel fluorocycline. Pharmacotherapy, 2020; 40(3):221–38. CrossRef
Arkadash V, Yosef G, Shirian J, Cohen I, Horev Y, Grossman M, Sagi I, Radisky ES, Shifman JM, Papo N. Development of high affinity and high specificity inhibitors ofmMatrix metalloproteinase 14 through computational design and directed evolution. J Biol Chem, 2017; 292(8):3481–95. CrossRef
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys, 1984; doi:10.1063/1.448118 CrossRef
Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun, 1995; 91:43–56; doi:10.1016/0010-4655(95)00042-e. CrossRef
Cathcart J, Pulkoski-Gross A, Cao J. Targeting matrix metalloproteinases in cancer: bringing new life to old ideas. Genes Dis, 2015; 2(1):26–34. CrossRef
Chlebda-Sieragowska E, Merwid-L?d A, Szumny D, Trocha M, Fereniec-Go??biewska L, G?barowska E, Szel?g A. Action of orally administered macrolides–erythromycin and roxithromycin–on B16 melanoma-transplanted mice. Pharmacol Rep, 2013; doi:10.1016/s1734-1140(13)71310-0 CrossRef
Chlebda-Sieragowska E, Merwid-Ld A, Szumny D, Trocha M, Fereniec-Goêbiewska L, Gêbarowska E, Kowalski P, Szelg A. Antitumor effect of macrolides - erythromycin and roxithromycin in B16 melanoma-transplanted mice. Pharmacol Rep PR, 2007; 59(1):269–74.
DeLano WL. PyMol: an open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography, Number 40, March 2002. Available via http://legacy.ccp4.ac.uk/newsletters/newsletter40.pdf
DeOliveira, EAM, Lang KL. Drug repositioning: concept, classification, methodology, and importance in rare/orphans and neglected diseases. J Basic Appl Pharm Sci, 2018; 8(08):157–65. CrossRef
Frezza M, Garay J, Chen D, Cui C, Turos E, Dou QP. Induction of tumor cell apoptosis by a novel class of N-Thiolated beta-lactam antibiotics with structural modifications at N1 and C3 of the lactam ring. Int J Mol Med, 2008; 21(6):689–95. CrossRef
Goldman RA, Cooperman BS, Strycharz WA, Williams BA, Tritton TR. Photoincorporation of tetracycline into escherichia coli ribosomes. FEBS Lett, 1980; 118(1):113–18. CrossRef
Goldman RA, Hasan T, Strycharz HWA, Cooperman BS. Photoincorporation of tetracycline into escherichia coli ribosomes. Identification of the major proteins photolabeled by native tetracycline and tetracycline photoproducts and implications for the inhibitory action of tetracycline on protein synthesis. Biochemistry, 1983; 22(2):359–68. CrossRef
Gooljarsingh LT, Lakdawala A, Coppo F, Luo L, Fields GB, Tummino PJ, Gontarek RR. Characterization of an exosite binding inhibitor of matrix metalloproteinase 13. Protein Sci Pub Protein Soc, 2008; 17(1):66–71. CrossRef
Hemmings FJ, Farhan M, Rowland J, Banken L, Jain R. Tolerability and pharmacokinetics of the collagenase-selective inhibitor trocade in patients with rheumatoid arthritis. Rheumatology, 2001; 40(5):537–43. CrossRef
Jacobsen JA, Jourden JLM, Miller MT, Cohen SM. To bind zinc or not to bind zinc: an examination of innovative approaches to improved metalloproteinase inhibition. Biochim Biophys Acta, 2010; 1803(1):72–94. CrossRef
Jo YK, Park SJ, Shin JH, Kim Y, Hwang JJ, Cho DH, Kim JC. ARP101, a selective MMP-2 inhibitor, induces autophagy-associated cell death in cancer cells. Biochem Biophys Res Commun, 2011; 404(4):1039–43. CrossRef
Jung JJ, Razavian M, Kim HY, Ye Y, Golestani R, Toczek J, Zhang J, Sadeghi MM. Matrix metalloproteinase inhibitor, doxycycline and progression of calcific aortic valve disease in hyperlipidemic mice. Sci Rep, 2016; 6(1):1–7. CrossRef
Kroon AM, Dontje BHJ, Holtrop M, Van Den Bogert C. The mitochondrial genetic system as a target for chemotherapy: tetracyclines as cytostatics. Cancer Lett, 1984; 25:33–40; doi:10.1016/s0304-3835(84)80023-3 CrossRef
Kuhn D, Coates C, Daniel K, Chen D, Bhuiyan M, Kazi A, Turos E, Dou QP. Beta-lactams and their potential use as novel anticancer chemotherapeutics drugs. Front Biosci J V Lib, 2004; 9(September):2605–17. CrossRef
Kumar H, Jo MJ, Choi H, Muttigi MS, Shon S, Kim BJ, Lee SH, Han IB. Matrix metalloproteinase-8 inhibition prevents disruption of blood-spinal cord barrier and attenuates inflammation in rat model of spinal Ccrd injury. Mol Neurobiol, 2018; 55(3):2577–90. CrossRef
Kumar KM, Anitha P, Sivasakthi V, Bag S, Lavanya P, Anbarasu A, Ramaiah S. In silico study on penicillin derivatives and cephalosporins for upper respiratory tract bacterial pathogens. 3 Biotech, 2014; 4:241–51; doi:10.1007/s13205-013-0147-z CrossRef
Lemkul JA. From proteins to perturbed hamiltonians: a suite of tutorials for the GROMACS-2018 molecular simulation package [article v1.0]. Living J Comput Mol Sci, 2019; 1(1):5068. CrossRef
Leyon PV, Lini CC, Kuttan G. Inhibitory effect of Boerhaavia Diffusa on experimental metastasis by B16F10 melanoma in C57BL/6 mice. Life Sci, 2005; 76(12):1339–49. CrossRef
Nara H, Kaieda A, Sato K, Naito T, Mototani H, Oki H, Yamamoto Y, Kuno H, Santou T, Kanzaki N, Terauchi J, Uchikawa O, Kori M. Discovery of novel, highly potent, and selective matrix metalloproteinase (MMP)-13 inhibitors with a 1,2,4-Triazol-3-Yl moiety as a zinc binding group using a structure-based design approach. J Med Chem, 2017; 60(2):608–26. CrossRef
Oehler R. Polacek N, Steiner G, Barta A. Interaction of tetracycline with RNA: photoincorporation into ribosomal RNA of Escherichia Coli. Nucleic Acids Res, 1997; 25:1219–24; doi:10.1093/nar/25.6.1219 CrossRef
Overall CM, Kleifeld O. Towards third generation matrix metalloproteinase inhibitors for cancer therapy. Br J Cancer, 2006; 94:941–46; doi:10.1038/sj.bjc.6603043 CrossRef
Pioletti M, Schlünzen F, Harms J, Zarivach R, Glühmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J, 2001; 20(8):1829–39. CrossRef
Qin LH, Huang W, Mo XA, Chen YL, Wu XH. LPS induces occludin dysregulation in cerebral microvascular endothelial cells via MAPK signaling and augmenting MMP-2 levels. Oxid Med Cell Longev, 2015; (July):120641. CrossRef
Remacle AG, Golubkov VS, Shiryaev SA, Dahl R, Stebbins JL, Chernov AV, Cheltsov AV, Pellecchia M, Strongin AY. Novel MT1-MMP small-molecule inhibitors based on insights into hemopexin domain function in tumor growth. Cancer Res, 2012; 72(9):2339–49. CrossRef
Rudek MA, Figg WD, Dyer V, Dahut W, Turner ML, Steinberg SM, Liewehr DJ, Kohler DR, Pluda JM, Reed E. Phase I clinical trial of oral COL-3, a matrix metalloproteinase inhibitor, in patients with refractory metastatic cancer. J Clin Oncol Official J Am Soc Clin Oncol, 2001; 19(2):584–92. CrossRef
Solomkin J, Evans D, Slepavicius A, Lee P, Marsh A, Tsai L, Sutcliffe JA, Horn P. Assessing the efficacy and safety of eravacycline vs ertapenem in complicated intra-abdominal infections in the investigating gram-negative infections treated with eravacycline (IGNITE 1) trial: a randomized clinical trial. JAMA Surg, 2017; 152(3):224–32. CrossRef
Trott O, Olson AJ. AutoDock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem, 2010; 31(2):455–61. CrossRef
Udi Y, Fragai M, Grossman M, Mitternacht S, Arad-Yellin R, Calderone V, Melikian M, Toccafondi M, Berezovsky IN, Luchinat C, Sagi I. Unraveling hidden regulatory sites in structurally homologous metalloproteases. J Mol Biol, 2013; 425(13):2330–46. CrossRef
Vandenbroucke RE, Libert C. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat Rev Drug Discov, 2014; 13(12):904–27. CrossRef
Van Nuffel AMT, Sukhatme V, Pantziarka P, Meheus L, Sukhatme VP, Bouche G. Repurposing drugs in oncology (ReDO)-clarithromycin as an anti-cancer agent. Ecancermedicalscience, 2015; 9(February):513. CrossRef
Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Lopes I, Mackerell AD. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem, 2010; 31(4):671–90. CrossRef
Verma RP. Hydroxamic acids as matrix metalloproteinase inhibitors. Matrix Metalloproteinase Inhibitors, 2012; doi:10.1007/978-3-0348-0364-9_5 CrossRef
Yadav V, Talwar P. Repositioning of fluoroquinolones from antibiotic to anti-cancer agents: an underestimated truth. Biomed Pharmacother, 2019; 111(March):934–46. CrossRef
Zhanel GG, Cheung D, Adam H, Zelenitsky S, Golden A, Schweizer F, Gorityala B, Lagacé-Wiens PRS, Walkty A, Gin AS, Hoban DJ, Karlowsky JA. Review of eravacycline, a novel fluorocycline antibacterial agent. Drugs, 2016; 76(5):567–88. CrossRef