INTRODUCTION
The Chikungunya virus (CHIKV) belonging to the Alphavirus genus and the Togaviridae family has caused significant epidemics because it can spread locally throughout nonendemic areas. The virus is transmitted to humans via mosquito bites from infected Aedes aegypti or Aedes albopictus. Chikungunya presents with symptoms similar to dengue fever, such as fever, skin rash, and acute arthralgia (Enserink, 2006), so it is frequently misdiagnosed. Although CHIKV has a low mortality rate, this chronic disease causes significant symptoms that cannot be ignored (Ganesan et al., 2017).
Chikungunya outbreak occurred in 24 locations across Indonesia between 2001 and 2003 (Laras et al., 2005), impacting Western and Central Indonesia in 2009 and 2010, with cases rising from approximately 3,000 per year to 83,000 and 52,000 cases per year, respectively (Harapan et al., 2019). After 2010, the number of cases detected per year dropped to 3,000.
The CHIKV viral genome is composed of a single-stranded RNA (~11.8 kb in length) that encodes four nonstructural proteins (nsP1, nsP2, nsP3, and nsP4) as well as three key structural proteins (e.g., nucleocapsid) and two envelope glycoproteins, E1 and E2, which form spikes on the virus surface. The E1 and E2 glycoproteins are CHIKV virulence factors because they bind to cell receptors in the clathrin region and stimulate endocytosis (Solignat et al., 2009; Zhang et al., 2006). Internalization of the virus occurs when E2 interacts with these receptors, whereas E1 facilitates virus fusion with the host cell, allowing the virus to complete its life cycle (Fields and Kielian, 2013; Weaver and Lecuit, 2015). In addition, viral envelope glycoproteins also serve as a target for neutralizing antibodies as host immune responses (Cho et al., 2008). It has been shown that 15 monoclonal antibodies (MAbs) can neutralize the trimeric E1E2 structure, with epitope sites dispersed throughout the E1E2 glycoprotein region (Fong et al., 2014).****
Chikungunya is detected by serological (hemagglutination inhibition), immunoassay [enzyme-linked immunosorbent assay (ELISA) and immunofluorescence assay],, molecular [polymerase chain reaction (PCR)], and immunochromatography assays (Johnson et al., 2016). Typically, serological assays and IgM detection by ELISA can be performed on day 5 after symptoms start. The PCR approach is the most accurate for CHIKV identification, but it requires technical expertise as well as costly materials and equipment (Prat et al., 2014). Immunochromatographic detection of CHIKV is significantly faster and does not require technical expertise. It is a paper-based diagnostic tool that is portable, practical, and accurate for diagnosing CHIKV, making it particularly useful for health professionals. Commercially available immunochromatographic kits for the identification of CHIKV have recently become available but have limited sensitivity to Indonesian CHIKV strains (Burdino et al., 2016; Kosasih et al., 2013).
MAbs are antigen-recognition proteins commonly used in immunoassays and manufactured using hybridoma technology, which is a time-consuming and costly process (Okabayashi et al., 2015). The single-chain variable fragment (scFv) in the antigen-binding fragment (Fab) is composed of a variable region of the heavy (VH) and light (VL) chains connected by a short linker peptide (Bach et al., 2001). Since the scFv is a smaller protein than a MAb, it can be produced in recombinant form by Escherichia coli host cells (Maynard and Georgiou, 2000). Escherichia coli was chosen as a host cell for recombinant protein expression because of its high expression level, low cost of growth media, and ease of scaling up (Rosano and Ceccarelli, 2014).
This study aimed to develop an anti-E1E2 CHIKV scFv that could sensitively identify the Indonesian Chikungunya (CHIKV) strain via an in silico approach (Norman et al., 2020; Yamashita, 2018) involving the modelling of Indonesian E1E2 CHIKV (hereafter referred to as E1E2-Ind) and the scFv in a fusion form with the biotin acceptor domain (BAD), which allows site-specific conjugation of the scFv to streptavidin on nitrocellulose membrane. The scFv was derived from the three-dimensional (3D) Fab structure from African anti-E1E2 CHIKV. Coarse-grained molecular dynamic (CGMD) and all-atom MD simulations were applied to evaluate the binding of the scFv to E1E2-Ind. The scFv was successfully expressed in E. coli; then, the biotinylated form was immobilized to nitrocellulose membranes for the detection of E2 CHIKV and NS1 DENV. The promising results demonstrate the potential for further development of an immunochromatography kit to detect CHIKV.
MATERIALS AND METHODS
Homology modelling of E1E2 CHIKV Ind
The E1E2-Ind sequence was obtained from GenBank (AHA87256.1), and the template search for homology modelling was performed using protein BLAST/blastp (https://blast.ncbi.nlm.nih.gov/). Subsequently, the 3D structure of E1E2-Ind was modelled using Modeller9.18 (http://salilab.org/modeller/), and the best 3D structure was selected based on the lowest discrete optimized protein energy score, as a lower value reflects the associated model is the more native-like (Hardianto et al., 2017). The model was further evaluated by the Ramachandran plot generated by the PROCHECK webserver (http://www.ebi.ac.uk/) and then subjected to iterative loop refinement using Modeller9.18 until more than 90% of residues occupied the most favourable regions in the Ramachandran plot.
Design of anti-E1E2 scFv and its complex with E1E2-Ind
The anti-E1E2 scFv was designed and infused with a BAD to form anti-E1E2-scFv-BAD (hereafter referred to as scFv-BAD), the structure of which originated from the African anti-E1E2 CHIKV Fab (Fab CHK152) (PDB ID: 3J30) (Voss et al., 2010). The design of scFv began with the search for conserved domains in 3J30 using the NCBI-conserved domain website (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to obtain the conserved VL and VH sequences. Then, a linker (GGGGS)4 was added between VL and VH, before the attachment of BAD (PDB ID: 2EBV) to the C-terminal to form scFv-BAD. All steps in the design of the scFv-BAD structure were conducted using BIOVIA Discovery Studio Visualizer 2019, and the model was further iteratively refined using Modeller9.18.
The docking of scFv-BAD to E1E2-Ind was adopted from the 3D structure coordinates of African anti-E1E2 CHIKV Fab (PDB ID: 3J30) and CHIKV E1E2 (PDB ID: 3J2W). The scFv-BAD and E1E2-Ind models were superimposed to 3J30 and 3J2W in BIOVIA Discovery Studio Visualizer 2019 to obtain the scFv-BAD and E1E2-Ind complex.
CGMDs simulations
The previously described protocol using the pmemd.cuda programme as implemented in Amber16 was used to conduct the CGMD simulations. The pdb4amber programme in AmberTools16 was employed to handle the disulphide bonds of Cys residues and various charge states of His side chains in the pdb files (Machado et al., 2019). Subsequently, SIRAH Tools was used to map the all-atom structures to CG models. Each resulting CG model was solvated in an octahedral box using WT4 water models, where the minimum distance of the box from the solute was 2.0 nm. The radii of the solvent-solute interaction were scaled by 0.7 to improve the initial hydration of the CG model. WT4 water models were randomly replaced by CG models of Na+ and Cl− ions to yield a 0.15 M salt concentration. A 1.2 nm cut-off was used for nonbond interactions, and particle mesh Ewald was used for long-range interactions. The Langevin thermostat and Berendsen barostat were implemented for all simulations. In the initial step of the CGMD simulations, solvent and side chains were relaxed by 5,000-step energy minimization, while the backbone beads were restrained by 1,000 kJ mol−1 nm−2. In the next stage, 5,000 ns energy minimization was performed without restraint; then, while a 1,000 kJ mol−1 nm−2 restraint was applied to the CG protein model, the system was equilibrated in NVT conditions at 300 K for 5 ns. The system was further equilibrated for 25 ns of NVT conditions at 300 K, where a 100 kJ mol−1 nm−2 restraint was imposed on the backbone beads. The production stage was performed in the NPT condition at 1 bar and 300 K, resulting in 2 μs CGMD trajectories. The snapshot frames in CGMD trajectories were written every 1 ns and analyzed using SIRAH Tools.
All-atom MD simulations
All-atom MD simulations were performed with the graphical processing unit-based pmemd programme of Amber16, and the inputs were prepared with the LEaP programme. The cysteine residues that form disulphide bonds were renamed from CYS to CYX, and the neutral histidine residues protonated in the delta and epsilon sections were renamed from HIS to HID and HIE. Neutralizing ions, as well as the model water molecule TIP3P, were added to the system with a minimum radius of 10 Å from the protein surface. It was then minimized, followed by heating to 27°C, and equilibrated for 1,000 ps with a cut-off value of 9 Å. The structure minimization was performed through the steepest descent and conjugate gradient methods. Subsequently, it was gradually heated until it reached temperature stability of 27°C (323°K) for 60 ps before the system density and pressure were equilibrated for 1,000 ps. The restraint was released gradually during equilibrium; then, the production stage proceeded for 10 ns. The calculation for energy interaction was performed using the MMGBSA.py programme, whereas other analyses such as hydrogen bond (H-bond) were performed using cpptraj in AmberTools16.
Construction of recombinant scFv in E. coli Origami B (DE3)
The scFv-BAD amino acid sequence was reverse-translated into a nucleotide sequence using Emboss on Expasy; then, the scFv-BAD codon was optimized based on the E. coli codon preferences using the GCUA software (http://gcua.schoedl.de/), and the guanosine-cytidine (GC) percentage was calculated by an online tool (https://jamiemcgowan.ie/bioinf/gc.html). scFv-BAD was designed with the addition of NdeI and BamH1 restriction sites for insertion into the pET21b (+) vector and synthesized by GenScript (Hong Kong). The pET21b-[scFv-BAD] plasmid was used to transform E. coli Origami B (DE3) by the heat shock method (Froger and Hall, 2007). The positive transformants were grown on a liquid Luria Bertani medium containing 100 μg/ml ampicillin and 40 μg/ml kanamycin and incubated overnight at 37°C. Subsequently, 50 μl of the culture was transferred to a flask containing 450 ml of fresh super broth medium, 100 μg/ml ampicillin, 0.05% glucose (Nacalai, Japan), and 4 M Biotin (Nacalai, Japan) and incubated at 37°Cand 200 rpm until the OD600 reached ~0.5. Then, 1 mM isopropyl-L-D-thiogalactoside was added to induce scFv-BAD expression and incubated overnight at 20°C before the cells were collected by centrifugation (8,000 rpm, 20 minutes, 4°C). Cell pellets were resuspended in binding buffer (20 mM phosphate buffer pH 7.4 and 0.5 M NaCl) and sonicated intermittently (40 W, 20 kHz) for 2 minutes on and 2 minutes off, five times. The supernatant containing dissolved protein was separated, while the pellets containing insoluble protein were resuspended in binding buffer and characterized by SDS-PAGE.
Purification of biotinylated scFv-BAD
The soluble protein was purified using the TALON metal affinity resin, washed in varying concentrations of imidazole (100, 250, and 500 mM), and characterized by SDS-PAGE. All supernatants containing biotinylated scFv-BAD were collected and concentrated using ultra cut-off 3K Amicon (Millipore, USA). The protein concentration was determined using the A280 nanodrop (Thermo Scientific, USA) before protein purification using the AKTA explorer (GE Healthcare, USA) via anion exchange and reevaluated by SDS-PAGE.
SDS-PAGE and western blotting
Initially, SDS-PAGE was performed following Laemmli (1970) at 300 V for 95 minutes; then, western blot analysis was conducted according to Kim (2017). An equal concentration of protein sample was separated by 12% SDS-PAGE gel, and then the protein was transferred to a polyvinylidene fluoride membrane (Millipore, USA). The membrane was soaked overnight in blocking buffer (super blocking in tris buffer saline) (Thermo Scientific, USA) at 4°C and then washed three times with TBST (Sigma Aldrich, USA) for 5 minutes. Subsequently, the streptavidin horse radish peroxidase (-HRP) antibody (1:500,000; Sigma Aldrich) was added and then washed six times using TBST for 5 minutes, before incubation at room temperature for 5 minutes with the detection reagent (Sigma Aldrich). The membrane was analyzed with ChemiDoc XRS+ Imager chemiluminescence (BioRad, USA) in a dark room.
Biotinylated scFv ELISA
The sensitivity assay of biotinylated scFv against NS1 DENV and bovine serum albumin (BSA) was analyzed by the ELISA method. All procedures were conducted at room temperature. The E2 CHIKV protein (ProSpec, Japan) was immobilized on a 96-well microtiter plate, rinsed once with phosphate buffer saline-Tween (PBST),, soaked in a blocking buffer (Abcam, USA), and shaken for 1 hour. The blocking buffer was removed by washing three times with PBST. Subsequently, 300 μl of biotinylated scFv, NS1, and BSA in varying concentrations ranging from 0.2 to 5.3 μM was added to each well and incubated for 1 hour before washing three times with PBST. Next, the streptavidin-HRP conjugate was added (Sigma Aldrich) and incubated for an hour, followed by washing four times with PBST. The o-phenylenediamine dihydrochloride substrate (Sigma Aldrich) was kept in a dark room for 5–10 minutes before 100 μl was added to the wells, and the enzyme activity was stopped by the addition of 100 μl of 2 N H2SO4. The absorbance was measured using a spectrophotometer (Benchmark Plus, BioRad) at 490 nm.
The attachment of biotinylated scFv to nitrocellulose membranes via the streptavidin-biotin system
Briefly, 0.5 μl streptavidin (1 mg/ml) was dripped onto a nitrocellulose membrane in the test line position and incubated overnight before 0.5 μl of biotinylated scFv (0.8 mg/ml) was dripped onto the test line and incubated overnight. The membrane was placed in a 96-well plate containing 100 μl PBST pH 7.4 [Tween-20 0.05% in phosphate buffer saline (PBS)] and incubated for 20 minutes, then immersed in a staining solution containing 0.025% Coomassie brilliant blue, destained, and dried. The appearance of the test line indicates the presence of biotinylated scFv bound to streptavidin, with streptavidin or biotinylated scFv only used as a negative control.
Conjugation of ScFv-link C with gold nanoparticles
The scFv-Link C used in this study belongs to the Research Centre of Molecular Biology and Bioinformatics, Universitas Padjadjaran. AuNPs were synthesized based on the thermal citrate reduction method with modifications (Kimling et al., 2006). Briefly, 10 ml of hydrogen chloroaurate (HAuCl4) was heated with constant stirring until the temperature reached 80°C–90°C; then, 1% sodium citrate was added rapidly. The colour of the solution changed from clear, dark purple to dark red, indicating the formation of the AuNP colloid. The AuNP colloid was then cooled at room temperature, and the pH was adjusted to 7.4. The gold nanoparticles were characterized by their absorption using a visible light spectrophotometer (Beckman Coulter, USA).
Fifty microlitre of scFv-Link C (100 μg/ml) was added to 450 μl of AuNP and incubated for 60 minutes before centrifugation to separate the pellets and supernatant. The pellets which contained AuNP-scFv-Link C were resuspended in a wash buffer (1% BSA in Tris pH 7.4), and the AuNP-scFv-Link C conjugate was resuspended in a conjugate diluent before analysis using a visible light spectrophotometer (Keshvari et al., 2016).
The immunochromatographic assay
The nitrocellulose membrane was cut into 2.5 × 0.4 cm sections; then, the control line was marked at the top and the test line at the bottom. Briefly, 0.5 μl (1 mg/ml) of the L protein was dripped on the control line, with 0.5 μl of streptavidin (1 mg/ml) dripped on the test line and incubated overnight. Subsequently, 0.5 μl of biotinylated scFv (0.8 mg/ml) was dripped on the test line and incubated overnight. A conjugate pad (Whatman standard 17; 0.7 × 0.4 cm) was prepared and treated with 10 μl of AuNP-scFv-Link C and dried at room temperature overnight. The sample pad (2 × 0.4 cm) was treated with 0.5 μl (0.9 mg/ml) of E2 CHIKV and then incubated in a desiccator at room temperature overnight. The wick for the absorption pad (1.8 × 0.4 cm) was prepared, and each membrane was fixed to a 6 cm backing card with 1–2 mm gap overlaps in each layer (Fig. 1). The assay was initiated by adding 100 μl of 10 mM PBS buffer in pH 7.4 containing 0.05% Tween-20 (v/v), and then this was incubated until the signal was observed. The specificity of the developed immunochromatographic assay was validated by checking the negative sample, which was NS1 DENV (Abcam, UK) (Sebastian and William, 2016).
RESULTS
In silico design of an E1E2-Ind model
The electron cryomicroscopy (cryo-EM) structure of African E1E2-CHIKV (PDB ID: 3J2W, hereafter referred to as E1E2-African) had a high sequence homology of 94% to E1E2-Ind (GenBank AHA87256.1); therefore, it was considered a suitable template for E1E2-Ind modelling. Homology modelling of E1E2-Ind was performed using 3J2W as the template, with the best model having 92.4% of its amino acid residues in the most favoured regions in the Ramachandran plot (Fig. S1), 85.4% for E2-Ind (Fig. S2). Since an acceptable model should possess more than 90% residues in the most favoured regions of the Ramachandran plot, the E2-Ind model was subjected to loop refinements using Modeller9.18, increasing the percentage of residues in the most favoured regions to 90.4%. Iterative loop refinements were also applied to the E1-Ind model to improve the number of residues in the generously allowed regions (Fig. S1).
The sequence alignment of E1E2-African and E1E2-Ind (Fig. 2) identified some missense mutations but did not provide information on which mutations affect the binding of specific antibodies. Fortunately, the 3J2W comes with the cryo-EM structure of a neutralizing antibody Fab CHK152 (3J30), so a structural alignment was performed of E1E2-African (3J2W) and the E1E2-Ind model. The Fab has a high neutralizing activity against CHIKV, with an EC50 of 0.013 μg/ml (Sun et al., 2013), and thus can be used to develop an immunochromatographic assay for the detection of CHIKV.
The structural alignment of the E1E2-Ind model and 3J2W identified a mutation of G694S at the epitope of E1E2-Ind binding to neutralizing antibody Fab CHK152 (Fig. 2). The mutation may affect the binding of Fab CHK152 and cause a lower sensitivity in the immunochromatographic assay; thus, the binding of Fab-CHK152-derived scFv (E1E2-scFv) to E1E2-Ind should be evaluated through an in silico approach before conducting wet-lab experiments.
In silico design of the E1E2-scFv complex
The E1E2-scFv complex was designed based on the Fab CHK152 (PDB ID: 3J30) structure. Variable domains in 3J30 were identified using the NCBI-conserved domain website. The VL domain consists of 109 amino acid residues (IVLTQSPASL AVSLGQRATI SCRASESVDS YGNSFMNWYQ QKPGQPPKLL IYRASNLESG IPARFSGSGS RTDFTLTINP VEADDVATYY CQQSNEDPFT FGSGTKLEI), whereas the VH contains 113 amino acid residues (LQQSGPELVK PGVSVKISCK ASGYSFTSFY IYWVKQRPGQ GLEWIGWIFP GSTNTKYNEK FKGKATLTAD TSSSTASMQL SSLTSEDSAV YFCARVDGYA MDYWGQGTSV TVS). Then, the C-terminus of VL was joined to the N-terminus of VH through an oligopeptide linker (GGGGS)4, resulting in E1E2-scFv. The streptavidin-biotin complex was used to attach E1E2-scFv to the nitrocellulose membrane, requiring the biotinylating of E1E2-scFv at the exposed lysine residues (Fig. S4), which are assisted by BirA. Biotinylating may lead to the blocking of the scFv paratope; however, to reduce the potency of biotinylating on the scFv paratope, we appended the BAD at the C-terminus, yielding scFv-BAD. Lysine residues in the coil structure of BAD (Fig. S4) are highly specific to biotinylating by BirA (Fairhead and Howarth, 2015). The use of BAD and BirA yielded site-specific protein biotinylating, including antibody-avidin conjugation (Asai et al., 2005; Chattopadhaya et al., 2006).
The quality of scFv-BAD was assessed using the Ramachandran plot. The initial model possessed less than the desired 90% of residues in the most favoured regions (Fig. S4) and therefore was subjected to an iterative loop refinement step to increase the number of residues to 90.8% (Fig. S3).
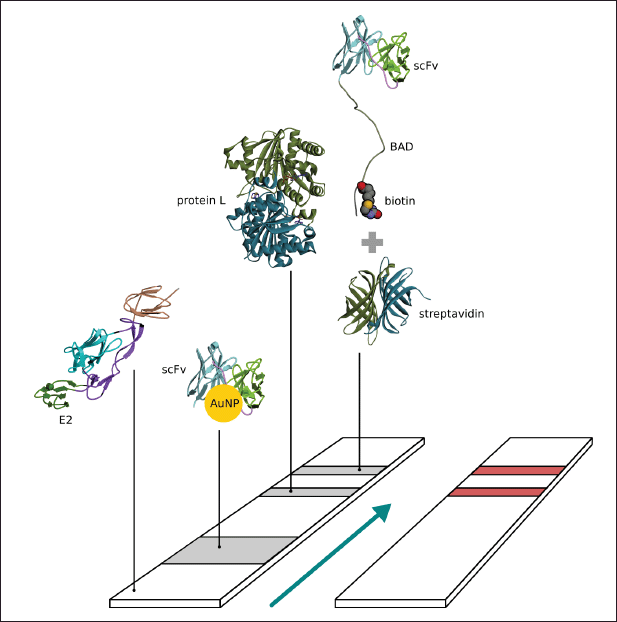 | Figure 1. Schema of paper based immunochromatography for CHIKV detection. [Click here to view] |
MD simulations
The E1E2-African crystal structure (3J2W) with Fab CHK152 (3J30) binds to the epitope at the B and arch2 domains of E2 (Fig. 2). Since only one amino acid mutation occurs on the epitope, these structures were adopted for the initial docking position of E1E2-Ind to scFv-BAD by applying a superimposition step. Subsequently, the complex of E1E2-Ind and scFv-BAD was subjected to CGMD simulation to yield a 2 μs trajectory as CGMD simulation can enhance the sampling of the protein conformational space (Machado et al., 2019), thus allowing observation of the dynamic behaviour of E1E2-Ind docked on scFv-BAD. Additionally, the simulation may give information regarding the effect of the linker and BAD introduction. A CGMD simulation of the complex of E1E2-African and scFv-BAD was also performed for comparison.
The analyses of the root mean square fluctuation (RMSF) plots (Fig. 3) revealed the dynamic behaviour of E1E2-Ind docked on scFv-BAD and E1E2-African docked on scFv-BAD. E1-Ind shows greater conformational changes than E1-African within residues 180 and 215 in domain II (Fig. 3A). Nevertheless, the E1-African also exhibits higher fluctuations than the E1-Ind spanning from residues 50–130 in domain II and FL. For E2, the Indonesian predominantly experiences higher fluctuations than the African (Fig. 3A). As shown in Figure 3B and C, E1E2-Ind displays a larger tilt than E1E2-African. Similarly, higher fluctuations are primarily observed on the scFv-BAD interacting with E1E2-Ind more than the E1E2-African (Fig. 3A). These higher fluctuations occur on the light chain and the linker of the scFv-BAD. Meanwhile, in some parts of the heavy chain, scFv-BAD binding to E1E2-African shows higher fluctuations than the binding to E1E2-Ind (Fig. 3A), which are also portrayed on their structure visualizations (Fig. 3B and C).
In the next step, we refined both complexes of E1E2-African and E1E2-Ind binding to scFv-BAD from the last frame of each CGMD simulation through all-atom MD simulations. The intermolecular interactions were analyzed in both complexes showing that the noncovalent bonds are contributed by residues from the E2 chain and the complementarity determining region of scFv-BAD. Also, the tilt of E1E2-Ind on scFv-BAD intensifies the intermolecular interactions, as summarized in Table S1. Salt bridge interaction increased by threefold, twofold for H-bonds. New interactions between E1E2-Ind and scFv-BAD also appear after CGMD and all-atom MD simulations which involve π-cation, π-anion, π–π, and π-alkyl (Table S1). Similarly, the intermolecular interaction profile of E1E2-African also improves after CGMD and all-atom MD simulations. Detailed analysis of H-bonds (Table S2) from 10 ns all-atom MD-trajectory revealed that scFv-BAD forms more H-bonds with E1E2-Ind than E1E2-African. Nonetheless, H-bonds formed by scFv-BAD and E1E2-African are more conserved than those of scFv-BAD and E1E2-Ind (Table S3). All interactions formed by scFv-BAD with E1E2-Ind and E1E2-African are through the E2 chain.
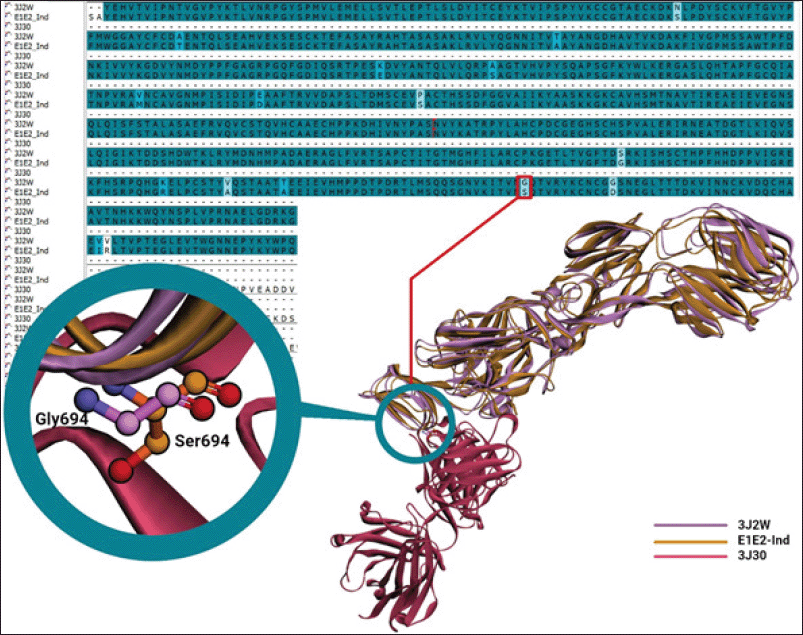 | Figure 2. Sequence and structural alignments of E1E2-African (3J2W), binding to Fab CHK152 (3J30), and E1E2-Ind. Purple and orange denote E1E2-African and E1E2-Ind, whereas magenta is Fab CHK152. [Click here to view] |
The molecular mechanics generalized-born surface area (MMGBSA) binding energy (ΔGMMGBSA) from a single 10 ns trajectory from each complex was calculated (Table S4), showing that the E1E2-Ind complex exhibits slightly stronger binding to scFv-BAD (ΔGMMGBSA = −46.9123 kcal/mol) than E1E2-African (−42.8413 kcal/mol). Nonbonded interaction energy terms, including van der Waals (EvdW) and electrostatic (Eelectrostatic), are also stronger for E1E2-Ind than E1E2-African (Table S4).
Optimization of scFv-BAD codon and construction of the expression vector
The scFv-BAD codon was optimized using the E. coli K12 codon table available in GCUA, resulting in 90% of optimum codons and 53.4% GC content. Subsequently, scFv-BAD was constructed in the pET21b(+) vector with the order: VL (109 aa) - (GGGGS)4 (20 aa) - VH (113 aa) - BAD (45 aa) and 6× His-Tag at the C-terminal (Fig. 4).
Expression of scFv-BAD protein
The characterization of the expressed protein using SDS-PAGE showed that scFv-BAD with a molecular weight of 30 kDa was expressed in E. coli Origami DE3, but scFv-BAD was more expressed in the insoluble fraction (Fig. 5). Overexpression of exogenous proteins in E. coli may lead to protein aggregation that causes the formation of insoluble inclusion bodies (Gupta et al., 2020). For the convenience of the purification process, only protein in the soluble fraction was collected for further purification.
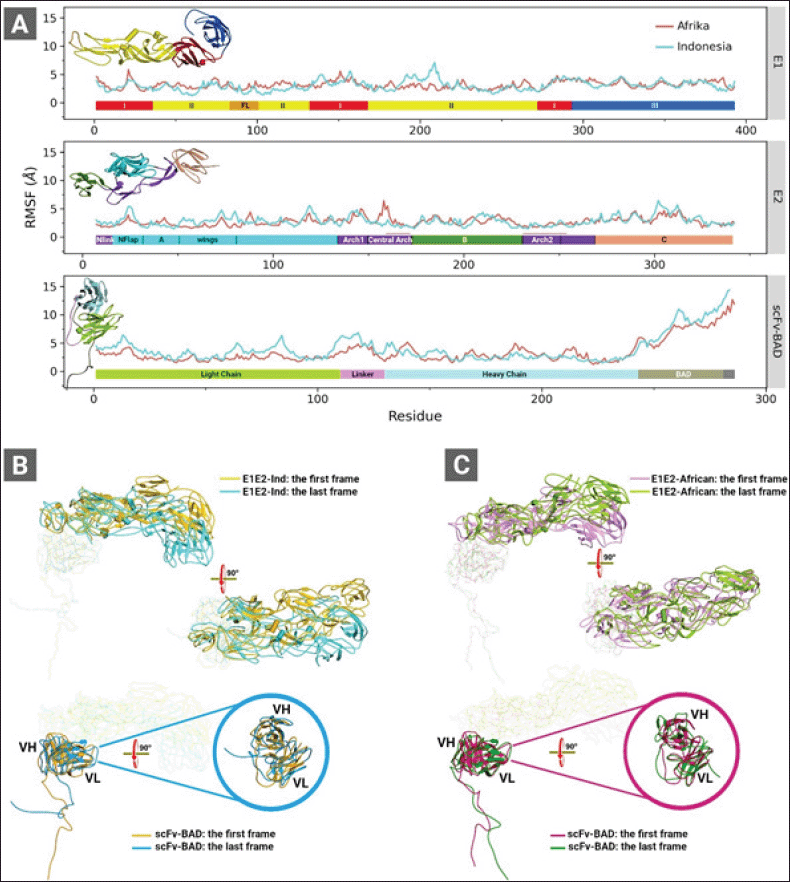 | Figure 3. (A) RMSF graphs of the complexes of E1E2-Ind and E1E2-African binding to scFv-BAD. (B) The first and the last CGMD trajectory snapshots of the complex of E1E2-Ind (top panel) and scFv-BAD (lower panel). (C) The first and the last CGMD trajectory snapshots of the complex of E1E2-African (top panel) and scFv-BAD (lower panel). These analyses were obtained from 2 μs CGMD simulation trajectories. Colour bars in (A) represent domains of E1 (top panel), E2 (middle panel), and scFv (lower panel). [Click here to view] |
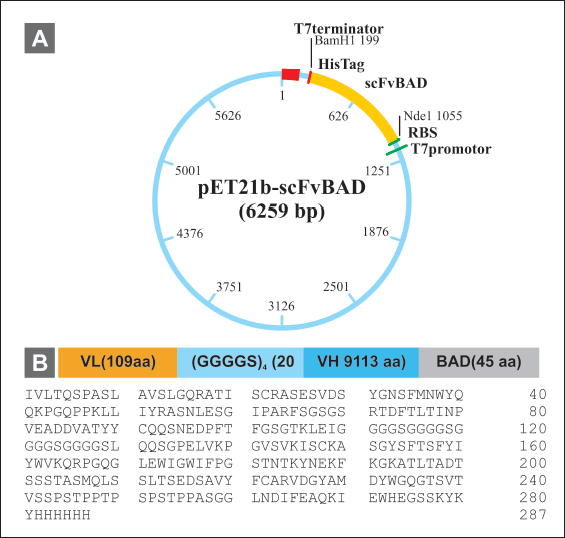 | Figure 4. Construction of scFv-BAD expression vector. (A) Map of pET 21b(+)-scFv-BAD. (B) Map of scFv-BAD and the amino acid sequences. [Click here to view] |
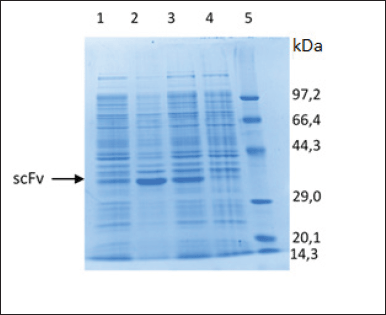 | Figure 5. Characterization of scFv-BAD fusion with SDS-PAGE. (1) Soluble fraction after induction and sonication. (2) Insoluble fraction after induction and sonication. (3) Insoluble fraction after induction without sonication. (4) Insoluble fraction before induction. (5) Protein marker. [Click here to view] |
Purification and characterization of scFv-BAD
Firstly, scFv-BAD was purified based on the ability of His-Tag to interact with the cobalt column; however, there were still many other protein bands; therefore, the purification was continued using the anion exchange method to yield a single band, as shown in Figure 6. Characterization of biotinylated scFv by western blotting using streptavidin-HRP showed that biotinylated scFv with a molecular weight of ~30 kDa was expressed (Fig. 6).
Sensitivity of biotinylated scFv
The interaction of biotinylated scFv with 10 μM of the commercial E2 CHIKV, NS1 DENV, and BSA was assessed by ELISA. The results in Figure 7 and Table S5 indicate that scFv-BAD was more sensitive to E2 CHIKV when the scFv concentration was above 0.6 μM, indicating that the sensitivity of biotinylated scFv to E2 CHIKV is not too high compared to NS1 and BSA.
Specificity of the immunochromatographic assay
The specificity of the interaction of scFv to E2 CHIKV using the NS1 DENV antigen for comparison (Fig. 8) revealed sensitivity to E2 CHIKV until 0.2 μl of 0.9 mg/ml. However, if the concentration of NS1 DENV was higher (0.6 μl of 1 mg/ml), it gave a positive result; thus, false positive results will be obtained if the concentration of NS1 is sufficiently high. Given that the NS1 protein is structurally distinct from CHIKV E2, this finding requires additional investigation and is a limitation of the qualitative immunochromatographic approach.
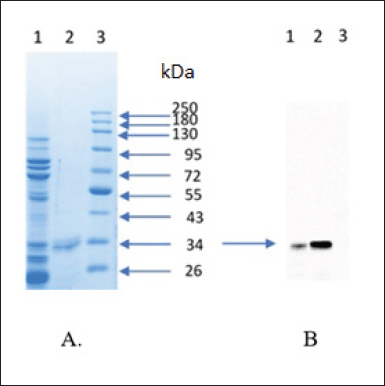 | Figure 6. Purification and characterization of scFv-BAD; (A) SDS-PAGE result of purification scFv-BAD; (1) with His-tag affinity chromatography; (2) with anion exchange chromatography; (3) protein molecular weight marker. (B) Characterization of scFv-BAD with western blot; (1) the result of His-tag affinity chromatography; (2) the result of anion exchange chromatography; (3) protein molecular weight marker. [Click here to view] |
DISCUSSION
The commercially available immunochromatographic kits for Chikungunya have low sensitivity in detecting Indonesian CHIKV strains (Burdino et al., 2016; Kosasih et al., 2013), possibly due to mutations occurring on E1E2-Ind. It is of note that epitope sites for antibody recognition are spread over the surface of the E1E2 glycoprotein (Fong et al., 2014). Fortunately, the specific Fab against E1E2-African (Fab CHK152) with high neutralizing activity is already deposited in the Protein Data Bank (3J2W), and this 3D structure can be used to design the scFv to detect E1E2-Ind. Only one mutation with a weak similarity (G694S) occurs at the epitope of E1E2-Ind interacting with Fab CHK152, but it may affect the binding of E1E2-Ind to scFv-BAD.
According to the trajectory analyses of CGMD and all-atom MD simulations, the G694S mutation significantly tilts E1E2-Ind from its initial docking on scFv-BAD, with a slight tilt also observed on the docking of E1E2-African on scFv-BAD. Thus, such movement on E1E2-Ind may be due to the introduction of the linker (GGGGS)4. E1E2-Ind and scFv-BAD are likely to adjust their binding by changing intermolecular interactions to respond to the G694S mutation and linker introduction. Interestingly, despite the G694S mutation, E1E2-Ind more strongly binds to scFv-BAD than E1E2-African. Nonetheless, Welch’s t-test calculation suggests that the ΔGMMGBSA values of E1E2-Ind and E1E2-African are not significantly different; therefore, scFv-BAD can sensitively detect E1E2-Ind.
The scFv-Link C scFv was conjugated to AuNP, resulting in an indicator dye for the immunochromatographic test system. The biotinylated scFv was placed at the test line, with the difference between these two scFv’s being the linker used to connect each protein fragment, whereas the linker used for scFv-Link C contained cysteine binds AuNP covalently. Meanwhile, the linker for scFv-BAD contained no cysteine but had additional amino acids in the BAD sequence at the end of the C-terminal. BAD functions as an anchor via the biotin-streptavidin interaction to keep the small scFv in place and make the antigen-binding site point up to facilitate interaction if there is a CHIKV antigen in the sample. The attachment of biotin-BAD onto the nitrocellulose membrane via the streptavidin-biotin bond is a strong noncovalent bond, resistant to organic solvents, detergents, and proteolytic enzymes, and can withstand extreme changes in temperature and pH (Green, 1975; Hermanson, 2013). Streptavidin was initially immobilized onto the nitrocellulose membrane, thus allowing a dipole-dipole interaction between the peptide bond and the nitrate ester from the nitrocellulose membrane. Hence, the electrostatic force would stabilize streptavidin immobilization onto the membrane (Bahadir and Sezgintürk, 2016).
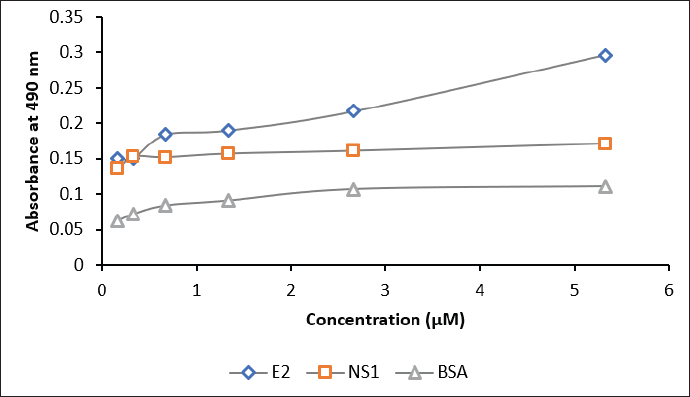 | Figure 7. The sensitivity assay of biotinylated scFv against E2 antigen (blue), NS1 (red), and BSA (grey). Concentrations of scFv: 0.2, 0.3, 0.6, 1.3, 2.6, and 5.3 μM. [Click here to view] |
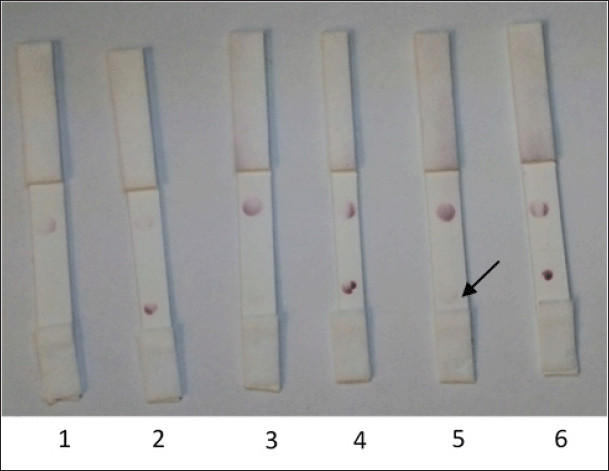 | Figure 8. The antigen volume optimization for the attachment onto the nitrocellulose membranes. (1) NS1 DENV (0.2 μl); (2) E2 CHIKV (0.2 μl); (3) NS1 DENV (0.4 μl); (4) E2 CHIKV (0.4 μl); (5) NS1 DENV (0.6 μl); (6) E2 CHIKV (0.6 μl). [Click here to view] |
The E. coli Origami B (DE3) expression system was used since it supports the biotinylation process (Santala and Lamminmaki, 2004) and can perform efficient folding for the production of scFv and Fab antibody fragments (Levy et al., 2001; Venturi et al., 2002). The anti-E1E2 scFv-BAD expressed in this study was found in the insoluble fraction rather than the soluble fraction, possibly due to the newly formed inclusion bodies containing partially folded protein. However, it might still have some advantages, such as the ease of extraction from bacterial cells by cell lysis because the inclusion bodies have different sizes and densities compared to other cellular components. Nevertheless, isolating pure active protein from inclusion bodies is challenging and involves several steps, such as dissolving inclusion bodies and repairing denatured proteins (Gupta et al., 2020).
In preparing the anti-EIE2 CHIKV immunochromatographic rapid test, the AuNP- scFv-Link C conjugate was coated onto a conjugate pad made from glass fibres because this material has no affinity for protein. The developed CHIKV immunochromatographic rapid test was constructed using the spot test method with a control line as the initial prototype for system optimization. The control line was used to validate the system using immobilized antibodies that recognize the Fc region of anti-CHIKV antibodies, such as the L protein. The results indicated that scFv could be used as a substitute for IgM in immunochromatographic and ELISA methods, but the sensitivity requires further improvement, as well as the use of whole viruses, both living and dead.
CONCLUSION
Biotinylated scFv was designed and expressed in E. coli Origami B (DE3) to produce a sufficient amount of biotinylated scFv for ELISA and immunochromatography; however, the protein purification requires optimization to obtain pure anti-E1E2 CHIKV scFv-BAD. Overall, the purified anti-E1E2 CHIKV scFv-BAD protein produced in this study is specific for the target antigen if an appropriate concentration and volume of purified protein are used in the immunochromatography-based rapid test.
ACKNOWLEDGMENTS
The author would like to thank Universitas Bakti Tunas Husada, the BUDI-DN, and the PKPI scholarship program for their extraordinary support. Thanks are due to Prof. Ikuo Fujii and the Biology Lab of Osaka Prefecture University, which have helped smooth the research process.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest regarding this study.
ETHICAL APPROVAL
This article does not contain any studies with human participants or animals performed by any of the authors. This study did not require informed consent.
AUTHORS’ CONTRIBUTIONS
KN, BA, and TA made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data. KN, SG, AH, and NAH contributed to drafting the article or revising it critically for important intellectual content. ARRF, AH, BA, and MY conceived and designed the experiments, analyzed and interpreted the data, and contributed to the analysis of the data. SG and KN provided final approval of the version to be published.
FUNDING
This research was supported by the Internal Grant of Universitas Padjadjaran to Shabarni Gaffar (Contract No. 1959/ UN6.3.1/PT.00/2021).
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Asai T, Trinh R, Ng PP, Penichet ML, Wims LA, Morrison SLA. Human biotin acceptor domain allows site-specific conjugation of an enzyme to an antibody-avidin fusion protein for targeted drug delivery. Biomol Eng, 2005; 21:145–55; doi:10.1016/j.bioeng.2004.10.001 CrossRef
Bach H, Mazor Y, Shaky S, Shoham-Lev A, Berdichevsky Y, Gutnick DL, Benhar I. Escherichia coli maltose-binding protein as a molecular chaperone for recombinant intracellular cytoplasmic single chain antibodies. J Mol Biol, 2001; 312:79–93; doi:10.1006/jmbi.2001.4914 CrossRef
Bahadir EB, Sezgintürk MK. Lateral flow assays: principles, designs and labels. TrAC Trends Anal Chem, 2016; 82:286–306; doi:10.1016/j.trac.2016.06.006 CrossRef
Burdino E, Calleri G, Caramello P, Ghisetti V. Unmet needs for a rapid diagnosis of Chikungunya virus infection. Emerg Infect Dis, 2016; 22:1837–9; doi:10.3201/eid2210.151784 CrossRef
Chattopadhaya S, Tan L, Yao S. Strategies for site-specific protein biotinylation using in vitro, in vivo and cell-free systems: toward functional protein arrays. Nat Protoc, 2006; 1:2386–98; doi:10.1038/nprot.2006.338 CrossRef
Cho B, Jeon BY, Kim J, Noh J, Kim J, Park M, Park S. Expression and evaluation of Chikungunya virus E1 and E2 envelope proteins for serodiagnosis of Chikungunya virus infection. Yonsei Med J, 2008; 49:828–35; doi:10.3349/ymj.2008.49.5.828 CrossRef
Enserink M. Infectious diseases. Massive outbreak draws fresh attention to little-known virus. Science, 2006; 311:1085; doi:10.1126/science.311.5764.1085a CrossRef
Fairhead M, Howarth M. Site-specific biotinylation of purified proteins using BirA. Methods Mol Biol, 2015; 1266:171–84; doi:10.1007/978-1-4939-2272-7_12 CrossRef
Fields W, Kielian M. A key interaction between the alphavirus envelope proteins responsible for initial dimer dissociation during fusion. J Virol, 2013; 87:3774–81; doi:10.1128/JVI.03310-12 CrossRef
Fong RH, Banik SS, Mattia K, Barnes T, Tucker D, Liss N, Lu K, Selvarajah S, Srinivasan S, Mabila M, Miller A, Muench MO, Michault A, Rucker JB, Paes C, Simmons G, Kahle KM, Doranz BJ. Exposure of epitope residues on the outer face of the chikungunya virus envelope trimer determines antibody neutralizing efficacy. J Virol, 2014; 88:14364–79; doi:10.1128/JVI.01943-14 CrossRef
Froger A, Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp, 2007; 6:253; doi:10.3791/253 CrossRef
Ganesan VK, Duan B, Reid SP. Chikungunya virus: pathophysiology, mechanism, and modeling. Viruses, 2017; 9:368; doi:10.3390/v9120368 CrossRef
Green NM. Avidin. Adv Prot Chem, 1975; 29:85–133; doi:10.1016/S0065-3233(08)60411-8 CrossRef
Gupta V, Sudhakaran IP, Islam Z, Vaikath NN, Hmila I, Lukacsovich T, Kolatkar PR, El-Agnaf OMA. Expression, purification and characterization of α-synuclein fibrillar specific scFv from inclusion bodies. PLoS One, 2020; 15:1–17; doi:10.1371/journal.pone.0241773 CrossRef
Harapan H, Michie A, Mudatsir M, Nusa R, Yohan B, Wagner AL, Sasmono RT, Imrie A. Chikungunya virus infection in Indonesia: a systematic review and evolutionary analysis. BMC Infect Dis, 2019; 19:243; doi:10.1186/s12879-019-3857-y CrossRef
Hardianto A, Yusuf M, Liu F, Ranganathan S. Exploration of charge states of balanol analogues acting as ATP-competitive inhibitors in kinases. BMC Bioinform, 2017; 18:572; doi:10.1186/s12859-017-1955-7 CrossRef
Hermanson G. Bioconjugate techniques. 3rd edition, Academic Press, London, UK, 2013.
Johnson BW, Russell BJ, Goodman CH. Laboratory diagnosis of Chikungunya virus infections and commercial sources for diagnostic assays. J Infect Dis, 2016; 214:S471–4; doi:10.1093/infdis/jiw274 CrossRef
Keshvari F, Bahram M, Farhadi K. Sensitive and selective colorimetric sensing of acetone based on gold nanoparticles capped with l?cysteine. JICS, 2016; 13:1–6; doi:10.1007/s13738-016-0856-4 CrossRef
Kim B. Western blot techniques. Methods Mol Biol, 2017; 1606:133–9; doi:10.1007/978-1-4939-6990-6_9 CrossRef
Kimling J, Maier M, Okenve B, Kotaidis V, Ballot H, Plech A. Turkevich method for gold nanoparticle synthesis revisited. J Phys Chem B, 2006; 110:15700––7; doi:10.1021/jp061667w CrossRef
Kosasih H, de Mast Q, Widjaja S, Sudjana P, Antonjaya U, Ma’roef C, Riswari SF, Porter KR, Burgess TH, Alisjahbana B, van der Ven A, Williams M. Evidence for endemic chikungunya virus infections in Bandung, Indonesia. PLoS Negl Trop Dis, 2013; 7:e2483; doi:10.1371/journal.pntd.0002483 CrossRef
Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 1970; 227:680–5; doi:10.1038/227680a0 CrossRef
Laras K, Sukri NC, Larasati RP, Bangs MJ, Kosim R, Djauzi, Wandra T, Master J, Kosasih H, Hartati S, Beckett C, Sedyaningsih ER, Beecham HJ 3rd, Corwin AL. Tracking the re-emergence of epidemic chikungunya virus in Indonesia. Trans R Soc Trop Med Hyg, 2005; 99:128–41; doi:10.1016/j.trstmh.2004.03.013 CrossRef
Levy R, Weiss R, Chen G, Iverson BL, Georgiou G. Production of correctly folded Fab antibody fragment in the cytoplasm of Escherichia coli trxBgor mutants via the coexpression of molecular chaperones. Protein Expr Purif, 2001; 23:338–47; doi:10.1006/persiapan.2001.1520 CrossRef
Machado MR, Barrera EE, Klein F, Sónora M, Silva S, Pantano S. The SIRAH 2.0 force field: Altius, Fortius, Citius. J Chem Theory Comput, 2019; 15:2719–33; doi:10.1021/acs.jctc.9b00006 CrossRef
Maynard J, Georgiou G. Antibody engineering. Annu Rev Biomed Eng, 2000; 2:339–76; doi:10.1146/annurev.bioeng.2.1.339 CrossRef
Norman RA, Ambrosetti F, Bonvin AMJJ, Colwell LJ, Kelm S, Kumar S, Krawczyk K. Computational approaches to therapeutic antibody design: established methods and emerging trends. Brief Bioinform, 2020; 21:1549–67; doi:10.1093/bib/bbz095 CrossRef
Okabayashi T, Sasaki T, Masrinoul P, Chantawat N, Yoksan S, Nitatpattana N, Chusri S, Morales Vargas RE, Grandadam M, Brey PT, Soegijanto S, Mulyantno KC, Churrotin S, Kotaki T, Faye O, Faye O, Sow A, Sall AA, Puiprom O, Chaichana P, Kurosu T, Kato S, Kosaka M, Ramasoota P, Ikuta K. Detection of chikungunya virus antigen by a novel rapid immunochromatographic test. J Clin Microbiol, 2015; 53:382–8; doi:10.1128/JCM.02033-14 CrossRef
Prat CM, Flusin O, Panella A, Tenebray B, Lanciotti R, Leparc-Goffart I. Evaluation of commercially available serologic diagnostic tests for chikungunya virus. Emerg Infect Dis, 2014; 20:2129–32; doi:10.3201/eid2012.141269 CrossRef
Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol, 2014; 5:172; doi:10.3389/fmicb.2014.00172 CrossRef
Santala V, Lamminmaki U. Production of a biotinylated single-chain antibody fragment in the cytoplasm of Escherichia coli. J Immunol Methods, 2004; 284:165–75; doi:10.1016/j.jim.2003.10.008 CrossRef
Sebastian C, William H. Immunochromatografi: format and aplications. Indo Am J Pharm Res, 2016; 6(07):1–10.
Solignat M, Gay B, Higgs S, Briant L, Devaux C. Replication cycle of chikungunya: a re-emerging arbovirus. Virology, 2009; 393:183–97; doi:10.1016/j.virol.2009.07.024 CrossRef
Sun S, Xiang Y, Akahata W, Holdaway H, Pal P, Zhang X, Diamond MS, Nabel GJ, Rossmann MG. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife, 2013; 2:e00435; doi:10.7554/eLife.00435.001 CrossRef
Venturi M, Seifert C, Hunte C. High level production of functional antibody Fab fragments in an oxidizing bacterial cytoplasm. J Mol Biol, 2002; 315:1–8; doi:10.1006/jmbi.2001.5221 CrossRef
Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Blanc CG, Crublet E, Thompson A, Bricogne G, Rey FA. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature, 2010; 468:709–14; doi:10.1038/nature09555 CrossRef
Weaver SC, Lecuit M. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med, 2015; 372:1231–9; doi:10.1056/NEJMra1406035 CrossRef
Yamashita T. Toward rational antibody design: recent advancements in molecular dynamics simulations. Int Immunol, 2018; 30:133–40; doi:10.1093/intimm/dxx077 CrossRef
Zhang JL, Gou JJ, Zhang ZY, Jing YX, Zhang L, Guo R, Yan P, Cheng NL, Niu B, Xie J. Screening and evaluation of human single-chain fragment variable antibody against hepatitis B virus surface antigen. Hepatobiliary Pancreat Dis Int, 2006; 5:237–41.
SUPPLEMENTARY MATERIAL
Supplementary material available in the journals website link: https://japsonline.com/admin/php/uploadss/3772_pdf.pdf