INTRODUCTION
Cholesterol is an integral substance found in the human body. Globally, inflated serum cholesterol has developed into a significant lifestyle-oriented disorder that is raised with high morbidity and mortality (Singhal et al., 2019). Major concentration has been focused on drug therapy. Nonpharmacological activity includes dietary intervention, behavior modification, and regular exercise, which are commonly pursued to lower blood cholesterol levels and function as a tool to prevent and treat cardiovascular disease (Dunn-Emke et al., 2001). Although studies have reported many cholesterol-lowering agents, certain bacteria can diminish the cholesterol level present in the liquid system (Saavedra et al., 2003) and the blood serum (Hlivak et al., 2005). Currently, cholesterol oxidase (CHO) from bacterial species oxidizes cholesterol and participates in the initial step of cholesterol metabolism (Devi and Kanwar, 2017).
CHO is a flavoenzyme belonging to the oxidoreductase family that catalyzes the oxidation of 3β-hydroxysteroids, and the isomerization of cholest-5-en-3-one functions as an intermediate to form an end product 4-cholesten-3-one (4-CHN) (Kumari and Kanwar, 2012). Initially, CHO presence was detected in Rhodococcus erythropolis (Turfitt, 1946). Later, it was identified in numerous bacteria, including both Gram-positive and negative (Kumari and Kanwar, 2012). Two extracellular CHOs were characterized later, which reacted with cholesterol, and the obtained yield was diagnosed as 4-CHN (Rhee et al., 2002). According to Cruz et al. (2001), the bioconversion process superficially occurs due to the involvement of an aqueous and organic double-phasic system.
In the pharmaceutical industry, steroids can function as leading compounds among the most extensively used products available in the market. Chemical or microbial routes implicate the synthesis of steroid drugs that involve complex mechanisms. Initially, steroid precursors, a starting material engaged in the transformation of steroid to drug intermediates, were made for the production of the final material. Microbial transformation cleaves the steroid precursor convoluted side chain into an individual step which assimilates effective modification in the steroid nucleus (Rao et al., 2013). The manufacture of steroid drugs and hormones using microorganisms is potentially encouraged in large-scale industrial processes. The biotransformation related to chemical synthesis required sufficient tools for the massive production of natural or diversified steroid drugs. Later, with innumerable therapeutic advantages, such as increased potency, longer half-live in the bloodstream, easy deliverable method and lesser side effects, the natural bioconversion process has been focused on (Bortolini et al., 1997).
Commercially made steroid-based drugs exhibit a wide range of therapeutic applications and represent the maximum marketed products in the pharmaceutical industry after antibiotics. The production of steroidal compounds, such as estrogens, androgens, and progestogens, are commercially synthesized using a combination of microbiological and chemical processes (Fernández-Cabezón et al., 2018). In steroid transformations, 4-CHN is regarded as a precursor for the synthesis of other drug intermediates, such as androst-4-ene-3,17-dione and androsta-1,4-di-ene-3,17-dione, which are leading initial components for the synthesis of anabolic drugs and contraceptive hormones (Dogra and Qazi, 2001).
The synthesis of 4-CHN by chemical methods require many adverse solvents like chloroform, methanol, and benzene. At the same time, these methods take a long time with multistep processes to yield 4-CHN. In order to overcome these, a specific action for the manufacture of 4-CHN using microbes has been widely approached. For better bioconversion, the enzymes involved in the transformation of the steroid compounds have to be improved. Even though many methods are available, metal ions or surfactants are supplied to develop enzyme productivity, optimization of transformation system, and heterologous expression of the CHO gene, and are also applied to boost steroid bioconversion (Wu et al., 2015).
In modern drug discovery, a molecular docking method proves the action of small molecules in the binding site of a target protein. This protein–ligand or protein–protein docking plays a crucial role in predicting the orientation of the ligand where it binds to the receptor (Pagadala et al., 2017). ArgusLab is a new molecular modeling package used for molecular docking study. This produces crystallographic binding orientations. This functions as an effective teaching tool to illustrate molecular docking (Abdelouahab et al., 2008).
In this research, CHO from Bacillus cereus strain KAVK5 converts cholesterol into 4-CHN in a double-phasic (i.e., aqueous and organic) system. Optimization of the CHO productivity for the bioconversional process is assessed. Furthermore, produced 4-CHN has been investigated in silico molecular docking mode against obesity, diabetes, cancer, and bacterial proteins.
MATERIALS AND METHODS
Isolation and identification of B. cereus strain KAVK5
Bacillus cereus strain KAVK5 was isolated from a ghee sample collected in Chennai, Tamil Nadu, India. The ghee was continuously diluted with 9 ml sterile distilled water, further cultured in a cholesterol-incorporated nutrient medium, followed by incubation at 37°C for 24 hours. The strain KAVK5 was primarily resolved based on physical, morphological, and biochemical parameters (Bergey and Holt, 1994). Molecular identification of species was attained by partial amplification of the 16S rRNA gene segment. The amplified gene product was submitted to National Center for Biotechnology Information (NCBI) database, and the accession number was obtained.
Detection of CHO production from KAVK5 using qualitative and quantitative methods of analysis
Qualitative analysis of CHO
The production of CHO from KAVK5 was corroborated by the colony staining method. Progression of red color indication occurred due to quinoneimine dye formation, revealing the evidence of CHO-producing bacterial strains. Furthermore, substantial evidence was affirmed for the synthesis of CHO using the CHO indicator plate method. Strain KAVK5 was cultured on a CHO indicator plate and incubated at 30°C. As a result, the conversion of H2O2 turned the medium color into brown, thus substantiating CHO production (Lashkarian et al., 2010).
Quantitative analysis of CHO
CHO productivity from KAVK5 was calibrated according to the method defined by Yang and Zhang (2012). The enzyme produced in the medium was spectrophotometrically analyzed at 500 nm, and their action was calculated, as mentioned in Amutha and Kokila’s (2016) study.
Optimization of medium components for the production of CHO
Differently enriched medium components were modified for the production of CHO from KAVK5. In this study, five different media were initially optimized: media 1 (Ahmad et al., 1992); media 2, containing ammonium chloride (1.0 g/l), dipotassium phosphate (0.25 g/l), magnesium sulfate heptahydrate (0.25 g/l), ferrous sulfate heptahydrate (0.001 g/l), yeast extract (5.0 g/l), and steroid-Tween 80 (20 ml at pH 7.0), with the modification of Arima (1969); media 3 (Varma and Nene, 2003); media 4 (Wu et al., 2015); and media 5 (Lee et al., 1997) were designed. The enzymatic assay determined the volume of enzymes produced by the isolates at 12, 24, 32, 48, 64, and 80 hours. Among the five optimized media, medium 1 contains cholesterol (0.1 g/100 ml), glucose (2.0 g/100 ml), yeast extract (0.5 g/100 ml), NH4NO3 (0.2 g/100 ml), K2HPO4 (0.02 g/100 ml), and MgSO4.7H2O (0.03 g/100 ml) (pH 7.0) that produced maximum CHO, which is preferred for further course of action. Bacteria were cultivated at 37°C with shaking at 150 rev minute−1. Later, the production medium was adjusted with different carbon sources (sucrose, fructose, maltose, glucose, and xylose) at a concentration of 2.0%. Accordingly, nitrogen sources (yeast extract, beef extract, peptone, ammonium chloride, sodium nitrate, and ammonium nitrate) were added at a concentration of 0.5% and 0.2% by replacing yeast extract and ammonium nitrate in the basal medium, respectively. In the medium, CHO production was further enhanced with different metal ions at a concentration of 0.03% (zinc sulfate, manganese sulfate, ferrous sulfate, copper sulfate, and magnesium sulfate). Additionally, pH (6, 6.5, 7.0, 7.5, and 8.0) activity for CHO production was adjusted. Subsequently, the medium was scrutinized by different temperatures (30°C, 40°C, 50°C, 55°C, and 65°C). Finally, the optimized enriched media was explored using various times, such as 12, 24, 32, 48, 64, and 80 hours. The amount of CHO produced by the isolates was proposed by the enzymatic assay. Due to the effect of the components in the media, the fermentation media was decisively designed for the maximum production of CHO.
Purification and molecular weight determination of CHO from KAVK5
The strains of KAVK5 cultured in an optimized enriched medium were obtained and centrifuged at 4°C cooling condition for 15 minutes at 10,000× g. The pellet was discarded, and the remaining supernatant was preferred for the purification of the extracellular enzyme. The supernatant was exposed to ammonium sulfate (80% w/v) precipitation, followed by dialysis against 0.05 M sodium phosphate buffer maintained at pH 7.0 overnight. The protein precipitated within the dialyzed membrane and was immersed in the sodium phosphate buffer (0.05 M, pH 8.0). The protein substances were loaded on to the Diethylaminoethyl cellulose ion-exchange column pack and washed using a buffer. The collected precipitated protein was loaded, and the eluted sample was collected by adding 0.1–0.5 M NaCl in the sodium phosphate buffer. For each molarity, 10 fractions were collected; further CHO activity was explored at every fraction. At the end of the experiment, the fraction containing CHO was pooled out and the purified CHO molecular weight was resolved by the SDS-PAGE method (Laemmli, 1970). The purified fraction containing CHO was run on 12% gel along with the standard marker. The molecular weight was inspected by comparing the protein band with a standard marker.
Enzymatic bioconversion of cholesterol to 4-CHN using CHO
Biological modification of cholesterol to 4-CHN using CHO from KAVK5 was implemented in an aqueous/organic biphasic system. The conventional condition was maintained as follows: 1 g of cholesterol with 130 ml of aqueous and isoamyl alcohol organic mixture (10:3 v/v) in a 500 ml flask; the content was kept at 250× g, 30°C at 3 hours, and subsequently oxygen was provided (Liu et al., 1996).
Extraction and purification of 4-CHN from the bioconversion system
After the bioconversion process was completed, 4-CHN and other cholesterol derivatives were retrieved with an equal volume of ethyl acetate; subsequently, the content was treated with water to eliminate the enzyme. The organic layer was separated from the extract and condensed under a vacuum. The recovered product was loaded on silica gel (300–400 mesh) packed columns and washed with petroleum ether. The column was washed with solvents like ethyl acetate and petroleum ether in the ratio of 1:40 to 1:20 v/v to conduct gradient elution. The eluent in the column was pooled out in a 10 mL test tube and condensed in a rotary evaporator to obtain the final product of 4-CHN (Wu et al., 2015). Thin-layer chromatography was carried out to detect the presence of a derivative product from KAVK5. The silica gel plate was coated, and the eluent was spotted. These TLC plates were placed in a chromatography tank containing 1.5 ml of hexane with 0.5 mL of ethyl acetate. After drying, the plates were treated with iodine vapor until the appearance of spots in the plates.
Molecular docking studies of 4-CHN
In silico molecular docking studies were carried out to predict the mode of action of the ligand 4-CHN against proteins, such as pancreatic lipase (PL), alpha-ketoglutarate-dependent dioxygenase (FTO), sterol regulatory element-binding protein 1, fatty acid synthase (FAS), acetyl-CoA carboxylase (ACC) (obesity proteins), α-amylase (AMS), α-glucosidase (GLS), glycogen synthase kinase-3-alpha (GSK1), glycogen synthase kinase-3-beta (GSK2) (diabetes proteins); cyclin-dependent kinase-2 (CDK-2), human topoisomerase IIa (HT), vascular endothelial growth factor receptor-2 (VEGFR-2) (cancerous proteins), dihydropteroate synthase (2VEG), and DNA gyrase subunit B (3TTZ) (bacterial proteins).
Ligand preparation
4-CHN was selected as the ligand molecule. The structure was constructed using the ChemSketch tool and then converted into a 3D structure.
Receptor protein preparation
The Protein Data Bank (PDB), a comprehensive portal, was used to retrieve the 3D structure of large biological macromolecules, including proteins and nucleic acid. The structure of the proteins, such as PL (ID: 1GPL), FTO (ID: 4IE0), SREBP1 (ID: 1AM9), FAS (ID: 2JFD), ACC (ID: 2YL2), AMS (ID: 1C8Q), GLS (ID: 5KZW), GSK1 (ID: 2DFM), GSK2 (ID: 1GNG), CDK-2 (ID: 1AQ1), HT (ID: 4FM9), VEGFR-2 (ID: 1VR2), 2VEG (ID: 2VEG), and 3TTZ (ID: 3TTZ), were downloaded from PDBSum database. RasMol tool was used to visualize the 3D structure of proteins.
ArgusLab tool used for molecular docking studies and visualization of docked substances
ArgusLab is a molecular docking tool employed to generate a grid, calculate dock score, and assess the interaction between the inhibitors with the active site of targeted enzymes proteins. According to Tanguenyongwatana and Jongkon (2016), ArgusLab, a user-friendly program, affords quick calculation. It has an accessible user interface in molecular docking studies. ArgusLab 4.0.1 functions with grid resolution of 0.40 Å. Energy calculations appraised the stability of the docked structure and the number of hydrogen bonds formed (Thompson, 2004). The most satisfying binding configuration was developed based on the hydrogen bond interactions between 4-CHN and the binding site of proteins. PyMol software demonstrated molecular models and created publication-quality images. The final docked structures were then visualized using PyMol viewer, and the docking results were predicted.
Statistical analysis
In this report, the entire investigations were carried out in triplicate, and the results were revealed as mean ± standard deviation (SD). Using Duncan’s multiple range test (DMRT), significance differences were determined and P < 0.05 values were statistically significant. This data was provided using SPSS statistical version 16.0.
RESULTS AND DISCUSSION
Cholesterol is a fatty substance that abundantly grows in all the cells of the body. Accumulation of cholesterol deals with multiple disorders, includes stroke, coronary heart disease, peripheral vascular disease, high blood pressure, and diabetes (Pekkanen et al., 1990). Cholesterol-degrading bacteria can be discriminated from food samples such as yoghurt, whey, milk, cheese, and raw milk (Yehia et al., 2015). A few bacterial species not only absorb cholesterol as a carbon source (Kovalenko et al., 2004), but they also decompose using CHO enzyme and produce multiple intermediate products (Liu and Shan, 2006). However, in this report, B. cereus strain KAVK5 produced CHO and was extensively applied for biological modification into 4-CHN.
Identification of strain KAVK5
The cholesterol-lowering B. cereus strain KAVK5 was isolated from the collected ghee samples and determined according to morphological and biochemical characteristics. Bacillus subtilis SFF34 from the Korean traditional fermented flatfish degraded cholesterol at 0.2% in the medium (Kim et al., 2002). Accordingly, Pseudomonas sp., Bacillus sp., and Streptomyces sp. isolated from the soap and vegetable oil industrial wastes were potentially degraded cholesterol (Saranya et al., 2014). In this report, strain KAVK5 was Gram-positive and also exhibited a positive effect on catalase and oxidase test. Dash et al. (2015) identified B. subtilis BI19 molecularly by sequencing the 16S rDNA gene region. Molecular-level identification was crucial to confirm the organisms at the species level; consequently, the 16S rRNA gene region was targeted to perform amplification, followed by sequencing. The obtained amplified data were submitted to NCBI and the accession number obtained was KP792776, which stated the organism as B. cereus and named the strain KAVK5 (Fig. 1). The genus Bacillus was treated as beneficial bacteria for the production of extracellular enzymes. Majority (60%) of the commercially producing enzymes were obtained from Bacillus species (Burhan et al., 2003).
Qualitative and quantitative analyses of CHO from strain KAVK5
CHO harvested from KAVK5 was executed using qualitatively and quantitative methods. A red color pattern consummate around the bacterial colonies was detected in the colony staining test (Fig. 2A). Subsequently, the indicator plate method also strongly verified the production of CHO, which could transform the medium color into brown. The color formation revealed in the medium was due to azo components, thus leading to H2O2 generation (Fig. 2B). CHO production was scrutinized in Serratia marcescens W1 and Bacillus pumilus W8. The red and brown colors’ progression was well documented in both the colony staining method and the indicator plate assay (Wali et al., 2019). CHO is a commercially produced enzyme used to determine cholesterol found in the food, serum, and other clinical samples. Rhodococcus sp. from urban compost and dairy soil samples produced CHO. Maximum yield was enforced by optimizing the production medium using temperature, pH, carbon, and nitrogen sources (Kaur et al., 2015).
Furthermore, in this study, after the confirmation of CHO production in a qualitative method, the volume (U) of enzyme production was quantified using spectrophotometry. In this regard, five designed media components were acquired; one of the media exhibited maximal production of 0.86 U/mL. Later, this medium component could be embraced for further modification to enhance CHO production (Fig. 2C). In this research, the yield of CHO from KAVK5 had been expanded because of the large-scale formulation; accordingly, optimization of the fermentation medium [carbon, nitrogen (organic and inorganic), metal ion, pH, temperature, and incubation time] was majorly noted.
Optimization of CHO production and purification
In the microbial fermentation process, carbon sources function as a major component responsible for cellular substance synthesis to provide energy widely (Devi and Kanwar, 2017). Accordingly, Yazdi et al. (2001) revealed the beneficial effects of various steroids and sugars enacted as a carbon source on CHO production using Rhodococcus equi 2C. So, their findings proved that cholesterol was selected as the prime carbon source. Consequently, the carbon source in the medium was altered; in that matter, xylose displayed the greatest production of 1.19 U/mL (Fig. 3A), whereas, in the organic and inorganic nitrogen source of adjustment, peptone and sodium nitrate provide CHO (1.24 and 1.39 U/ml, respectively) (Fig. 3B). The utilization of nitrogen sources in the medium was an essential factor for the production of CHO. Many researchers had a high impact on organic and inorganic nitrogen sources. Of all the data, ammonium and yeast extract had more influence on CHO productivity (Devi and Kanwar, 2017). Modifying the medium using metal ion sources indicates that manganese sulfate yields 1.47 U/ml (Fig. 3C). pH has a great response to the productivity and synthesis of enzymes. CHO actively works best at a pH of 6.5–8.0 (Doukyu, 2009). Later, the yield was increased by about 1.68 U/ml, while adjusting the optimal pH at 7.0 (Fig. 3D) and, simultaneously, the production rate gradually increased up to 1.81 U/ml at 30°C (Fig. 3E). Different cultivation temperatures (34.2°C–39.8°C) were regulated for CHO production, and the result stated that the cell reached maximum growth for all the temperatures assessed (Devi and Kanwar, 2017). However, optimum growth was obtained at a lower temperature range of 35°C, with a maximum of 37°C. Moreover, cell growth was obtained for CHO production at 39°C (Lee et al., 1999). The enriched medium exhibited tremendous output, which was designed for further study. KAVK5 showed the highest productivity (1.86 U/ml) at 32 h of the incubation (Fig. 3F). The designed enriched medium used for the production of CHO from KAVK5 consisted of the following components: cholesterol (0.1 g.ml−1), xylose (2.0 g.ml−1), peptone (0.5 g.ml−1), sodium nitrate (0.2 g.ml−1), and manganese sulfate (0.03 g.ml−1). pH 7.0 was maintained at 30°C and incubated at 32 hours.
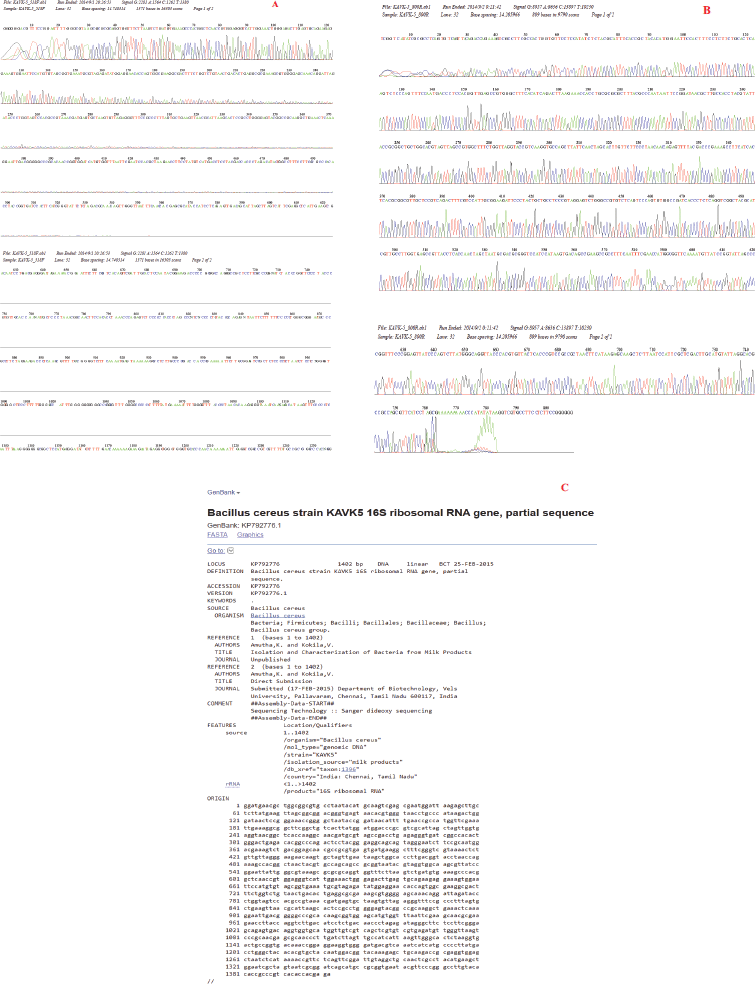 | Figure 1. 16S rRNA gene amplification of B. cereus strain KAVK5. (A) Forward chromatogram of B. cereus strain KAVK5. (B) Reverse chromatogram of B. cereus strain KAVK5. (C) NCBI data on the submitted gene sequence of B. cereus strain KAVK5. [Click here to view] |
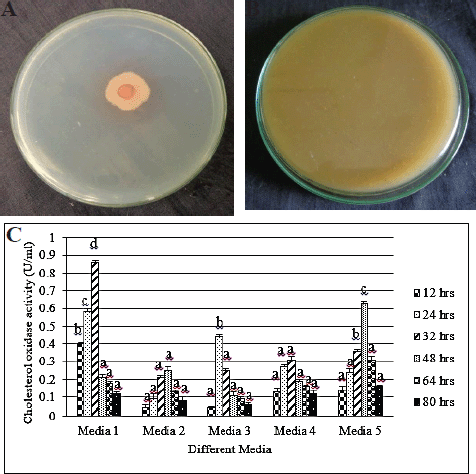 | Figure 2. (A) Colony staining method of B. cereus strain KAVK5. (B) Cholesterol oxidase indicator plate of B. cereus strain KAVK5. (C) Different media for the production of CHO from B. cereus strain KAVK5. Values are expressed as mean ± standard deviation for three samples (n = 3) in each group. Superscripts in the column show significant differences at the p < 0.05 level. Analysis of variance (ANOVA) was followed by DMRT. Comparison is made between media 1 and 5. [Click here to view] |
CHO was purified using differential ammonium sulfate precipitation, ion exchange, and gel filtration chromatography (Kumari and Kanwar, 2012). Bacillus subtilis from tiger excreta produced CHO, and purification was additionally processed by salting out the CHO using ammonium sulfate, followed by dialysis and riboflavin-affinity chromatography. This enzyme exhibited a molecular weight of about 105 kDa (Kumari and Kanwar, 2016). Bacillus subtilis YS01 produced CHO of approximately 56 kDa, which was widely filtered by ammonium sulfate precipitation, dialysis, DEAE cellulose ion-exchange chromatography, and Sephadex G-100 gel filtration (Aruna et al., 2017). In addition, CHO in the enriched medium was purified with the help of DEAE cellulose ion-exchange column chromatography. The purified CHO revealed its molecular weight at 32.8 kDa in the SDS-PAGE method (Fig. 4).
Production and purification of 4-CHN using enzymatic bioconversion method
The effect of the organic solvent, like isoamyl alcohol, used for the bioconversion study was examined in an organic/aqueous system. The conversion rate was higher in the aqueous/organic biphasic system, comparable to the simple aqueous phase (Wu et al., 2015).
Experimental condition
Different cholesterol concentrations were supplied to the bioconversion medium containing 100 mL of enzyme solution with 30 ml of organic solvent during the experimental period. Increasing the concentration of cholesterol decreased or delayed the bioconversion process, thus ultimately resulting in enzyme deactivation. Accordingly, 1 g of cholesterol was fixed up to achieve the conversional step, increasing the bioconversion rate. The biological conversion of cholesterol to 4-CHN produced H2O2, which functioned as a strong oxidant resulting in the denaturation of CHO protein (Wu et al., 2015).
In this study, the fermentation procedure was processed in a 0.5 l flask containing 30 ml of isoamyl alcohol, 100 ml of CHO from KAVK5 strain solution, and 1 g of cholesterol maintained at 30°C for 30 minutes at 250× g. After the bioconversion condition was completed, an equal amount of ethyl acetate was added to the component, and the organic layer was separated, followed by condensation. The residual substances were dropped onto a column chromatography (300–400 silica mesh), and 4-CHN was collected and subjected to thin-layer chromatography. The end product was collected and condensed under a vacuum evaporator, and the obtained yield was calculated, reaching a yield of about 1.23 g for every bioconversion step (Fig. 5). Researchers reported that the bioconversion process was achieved depending on the three main actions: mechanism of enzyme catalysis, metabolic pathway, and finally optimization of enzyme activity (Wu et al., 2015). According to them, petroleum ether used with enzyme solution increased its yield up to 4 g l−1 h−1, corresponding to the conversional rate at 90%. Shao et al. (2015) produced 4-CHN of about 0.61 g l−1 and 0.75 g l−1 with the conversional rates of 67% and 83% using genetically modified cells, respectively. The obtained product was spotted with standard 4-CHN on a silica gel plate and placed in a chromatography tank. Normal cholesterol’s retardation factor (Rf) exhibited a 0.5 value, and the standard 4-CHN revealed a 0.6 value. The purified compound in this study reported around 0.63 value.
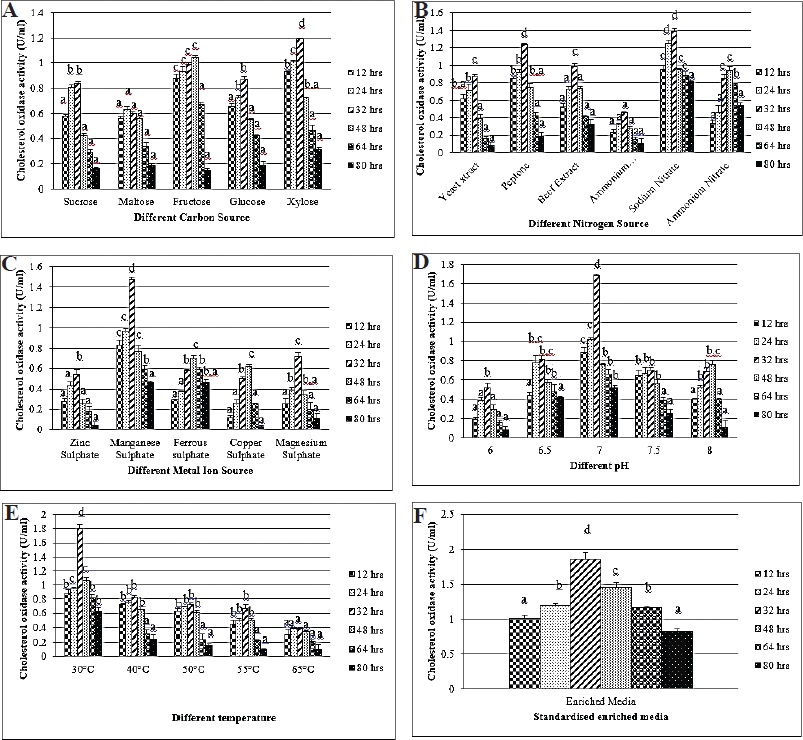 | Figure 3. (A) Different carbon sources for the production of CHO from B. cereus strain KAVK5. Values are expressed as mean ± standard deviation for three samples (n=3) in each group. Superscripts in the column show significant differences at the p < 0.05 level. ANOVA was followed by DMRT. Comparison is made between carbon sources. (B) Different nitrogen sources for the production of CHO from B. cereus strain KAVK5. Values are expressed as mean ± standard deviation for three samples (n = 3) in each group. Superscripts in the column show significant differences at the p < 0.05 level. ANOVA was followed by DMRT. Comparison is made between nitrogen sources. (C) Different metal ion sources for the production of CHO from B. cereus strain KAVK5. Values are expressed as mean ± standard deviation for three samples (n = 3) in each group. Superscripts in the column show significant differences at the p < 0.05 level. ANOVA was followed by DMRT. Comparison is made between metal ion sources. (D) Different pH for the production of CHO from B. cereus strain KAVK5. Values are expressed as mean ± standard deviation for three samples (n = 3) in each group. Superscripts in the column show significant differences at the p < 0.05 level. ANOVA was followed by DMRT. Comparison is made between pH. (E) Different temperatures for the production of CHO from B. cereus strain KAVK5. Values are expressed as mean ± standard deviation for three samples (n = 3) in each group. Superscripts in the column show significant differences at the p < 0.05 level. ANOVA was followed by DMRT. Comparison is made between temperatures. (F) Different incubation times for the production of CHO from B. cereus strain KAVK5. Values are expressed as mean ± standard deviation for three samples (n = 3) in each group. Superscripts in the column show significant differences at the p < 0.05 level. ANOVA was followed by DMRT. [Click here to view] |
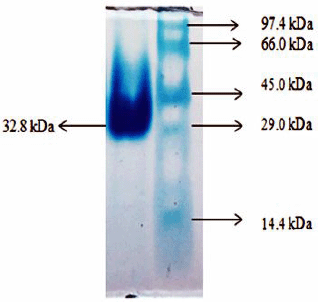 | Figure 4: Molecular weight determination of CHO from B. cereus strain KAVK5. [Click here to view] |
Molecular docking studies of 4-CHN against proteins
The molecular docking study is a fast-growing area to interpret and predict the interaction between a ligand and a receptor protein macromolecule. This report targeted the development of an active drug against obesity, diabetes, cancer, and bactericidal effect. We selected 4-CHN as a lead compound for the drug design. The molecular structure of 4-CHN used in this study was drawn using ACD ChemSketch (Fig. 6A), and the 3D structure of compounds is shown in Fig. 6B.
Interaction between 4-CHN with obesity-associated proteins
Obesity is a metabolic syndrome defined by extreme weight gain to height distribution primarily responsible for expanded fat deposition, associated with a larger calorie absorption related to the energy output (Mohapatra et al., 2015). Pancreatic lipase, a key enzyme, plays an active role in breaking dietary fats by hydrolysing the fat molecule into mono-acylglycerol, and fatty acids diminish the intake of triglycerides (Birari and Bhutani, 2007). In an anti-obesity study, this enzyme was widely focused on developing natural products (Sugiyama et al., 2007). According to Gurudeeban et al. (2012), the FTO gene interacted with obesity, which is considered a health risk factor around the world. The occurrence of obesity is due to the enlargement of fat mass, resulting from excess adipose cell size and number in the cytoplasm of adipocytes which participates in enzymes such as fatty acid synthase (Devlin et al., 2000). Inhibition of adipose tissue enlargement was considered as a target to control obesity disorder. This process can be regulated by transcriptional factors like sterol regulatory element-binding protein 1 (SREBP1) (Park et al., 2019). Acetyl-CoA carboxylase played a vital role in fatty acid metabolism and was designed as a potent target for developing a new therapeutic agent in controlling obesity (Gago et al., 2011). In the current study, PL, FTO, SREBP1, FAS, and ACC proteins codes for obesity disorder were docked against the ligand 4-CHN, and their proteins were downloaded from the PDB database, which is shown in Fig. 7(A–E).
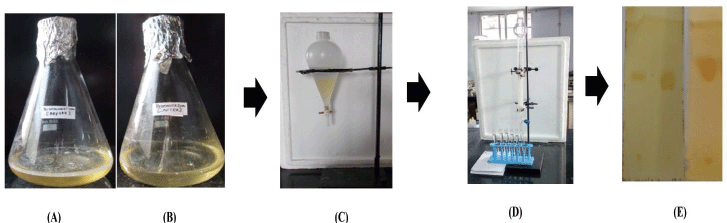 | Figure 5. Schematic representation of the bioconversion pathway. (A) Before the bioconversional process. (B) After the bioconversional process. (C) Separation of the organic layer. (D) Column chromatography for the separation of 4-CHN. (E) Thin-layer chromatography: (a) standard 4-CHN; (b) standard cholesterol; and (c) tested 4-CHN. [Click here to view] |
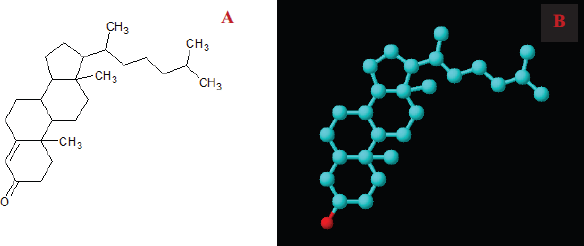 | Figure 6. Structure elucidation of 4-CHN. (A) 2-dimensional structure and (B) 3-dimensional structure. [Click here to view] |
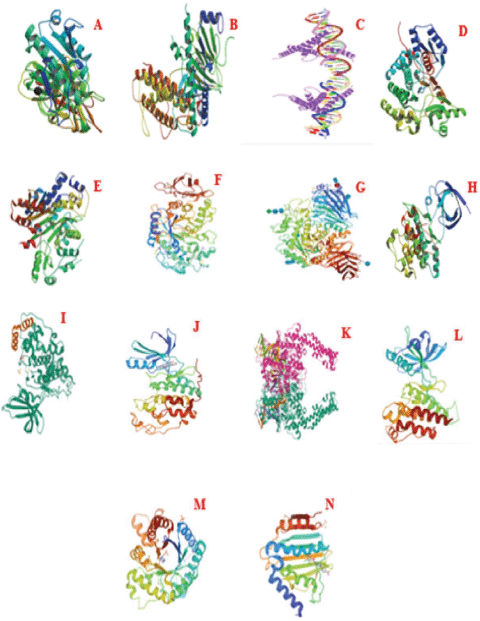 | Figure 7. Structure of proteins. (A) Pancreatic lipase. (B) Alpha-ketoglutarate-dependent dioxygenase. (C) Sterol regulatory element-binding protein 1. (D) Fatty acid synthase. (E) Acetyl-CoA carboxylase. (F) α-Amylase. (G) α-Glucosidase. (H) Glycogen synthase kinase-3-alpha. (I) Glycogen synthase kinase-3-beta. (J) Cyclin-dependent kinase-2. (K) Human topoisomerase IIa. (L) Vascular endothelial growth factor receptor-2. (M) Dihydropteroate synthase. (N) DNA gyrase subunit B. [Click here to view] |
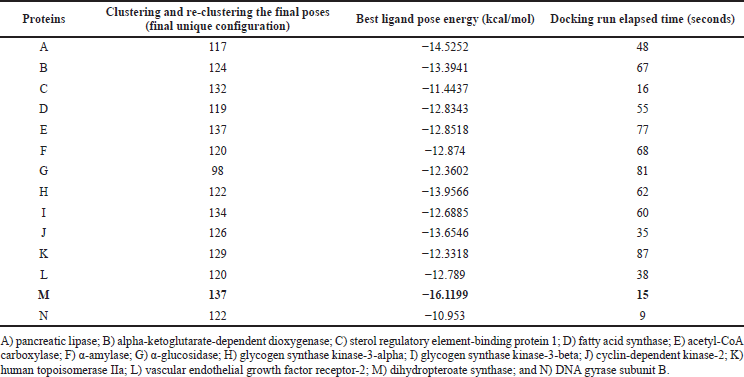 | Table 1. Docking energy of the docked complexes from ArgusLab software. [Click here to view] |
Interaction between 4-CHN with diabetes-associated proteins
α-Amylase, an enzyme also called endonuclease enzyme, is engaged in the digestion of polysaccharides, whereas α-glucosidase is situated in the intestinal part which participates in absorbing oligosaccharides (Telagari and Hullatti, 2015). These enzymes were treated as key enzymes that maintained blood glucose levels by slowing the digestive process and absorption of carbohydrates and incertins (Okechukwu et al., 2020). Type 2 diabetes mellitus appears more than that of the total diabetic population, which correlates with obesity-induced insulin resistance and subsequently secreted insulin from the β-cells of the pancreas. Glycogen metabolism plays an essential role in maintaining glucose homeostasis and is regulated by two enzymes like glycogen synthase and phosphorylase. Glycogen synthase kinase (GSK) is a negative regulator of the insulin signaling pathway composed of two similarly linked isoforms, namely GSK-3α and GSK-3β (Devi et al., 2018). Consequently, AMS, GLS, GSK1, and GSK2 were intended for docking against 4-CHN, and the respective proteins were downloaded from the PDB database (Fig. 7F–I).
Interaction between 4-CHN with cancer-associated proteins
Numerous types of enzymes could develop cancer. Among those, CDK-2, HT-Iiα, and VEGFR-2 were considered the most common enzymes actively involved in the development of cancerous cells (Sarkar et al., 2021). Accordingly, CDK-2, HT, and VEGFR-2 were targeted to achieve docking against the ligand, and the correspondent proteins were downloaded from the PDB database (Fig. 7J–L).
Interaction between 4-CHN with associated bacterial proteins
Recently, a difficulty faced by the researchers was the discovery of antimicrobial drugs and other therapies. Bacteria exhibited resistance to the drugs; this occurred due to the establishment of persister cells or biofilms (Puiu et al., 2017). On that account, the biological role of 4-CHN was ensured for antibacterial activity. Drugs potentially target the folate pathway by inhibiting the dihydropteroate synthase enzyme. Moreover, this activity was extensively required for bacterial and synthetic pathways, which also function as a target of interest for antibacterial agents (Potshangbam et al., 2020). DNA gyrase subunit B from bacteria was selected for the in silico docking study against ligand anthraquinones, proving antibacterial effect (Arevalo and Amorim, 2021). Based on this, 2VEG and 3TTZ were focused on for docking study against the ligand 4-CHN and the bacterial proteins were downloaded from the PDB database (Fig. 7M and N). This can be strongly proven as the best antibacterial agent.
In this docking study, a stable complex could be found upon the preferred orientation of one molecule (protein) bound to other molecules (ligand). This can be applied using ArgusLab software. The binding action plays a vital part in recognizing docking energy and stability of the receptor–ligand complex, which was authorized by hydrogen bonds. Interaction between the complex molecules produced an energy score, which is presented in Table 1, and the docked complex was visualized using PyMol software (Fig. 8). The interaction of docked complex produces binding residues, atoms, number of hydrogen bonds, and distances; the docking scores are listed in Table 2. The table also displays the amino acids in the active region of studied proteins interacting with 4-CHN.
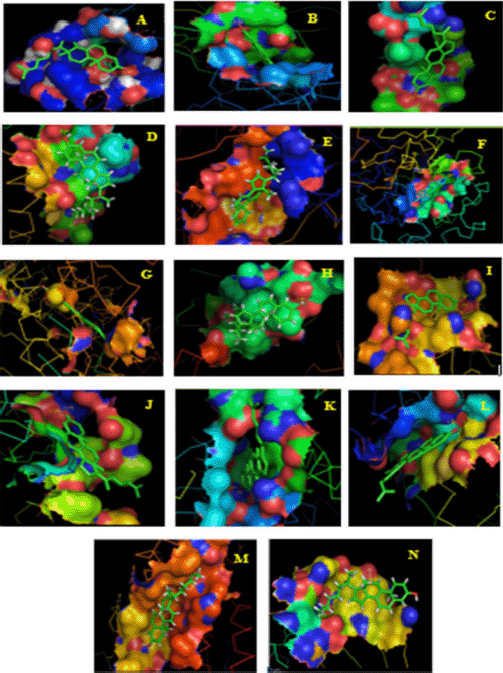 | Figure 8. Final docked conformations of 4-CHN with (A) pancreatic lipase; (B) alpha-ketoglutarate-dependent dioxygenase; (C) sterol regulatory element-binding protein 1; (D) fatty acid synthase; (E) acetyl-CoA carboxylase; (F) α-amylase; (G) α-glucosidase; (H) glycogen synthase kinase-3-alpha; (I) glycogen synthase kinase-3-beta; (J) cyclin-dependent kinase-2; (K) human topoisomerase IIa; (L) vascular endothelial growth factor receptor-2; (M) dihydropteroate synthase; and (N) DNA gyrase subunit B. [Click here to view] |
4-CHN interaction with proteins by the in silico method calculated the minimum binding energy (kcal/mol). In a molecular docking analysis, the parameters considered essential tools include hydrogen bond interactions, n–n interactions, binding energy, active site residues, and orientation of the phytochemicals with the reactors (Madeswaran et al., 2011). This result shows that there was a presence of a binding site between the protein and ligand. The docking was also valid by the development of a hydrogen bond between them. From the above docking result, 4-CHN docked well to these proteins responsible for obesity, diabetes, cancer, and bactericidal effect. The result of the Lipinski rule suggests that the analyzed compound acts as the best therapeutic pharmaceutical drug.
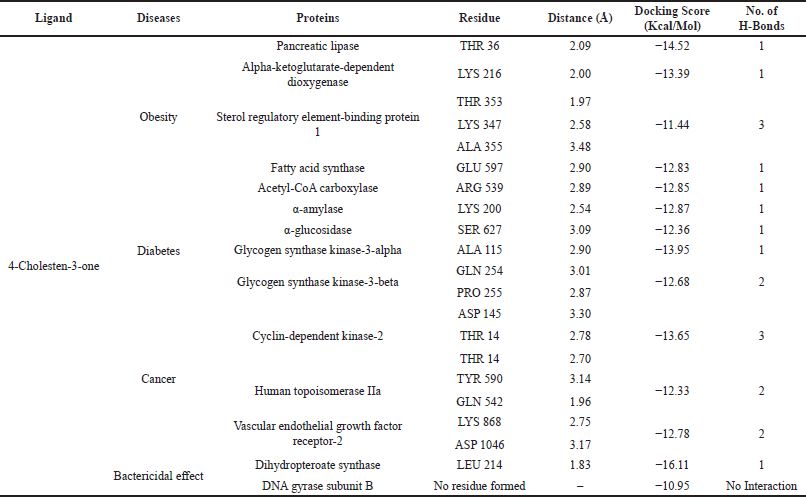 | Table 2. Docking score of 4-CHN with diseased proteins. [Click here to view] |
CONCLUSION
Bacillus cereus strain KAVK5 produced economically important CHO. The productivity was enhanced by the optimization of the fermentation medium, resulting in an improved bioconversion process. In a biotransformation system, this enzyme collaboratively provides a straightforward and reliable conversion of cholesterol into 4-CHN in an aqueous and organic double-phasic system. It was achievable to develop the superior quality of 4-CHN on an extensive range of processes and thus function as a pharmaceutical drug that could be predicted using an in silico docking model. 4-CHN is an active metabolite that acts as a ligand for various diseases. In this regard, this active molecule exhibits promising anti-obesity, antidiabetic, anticancer, and bactericidal disorder. However, based on our results, we can state that 4-CHN was likely bound to the protein and thus the interaction primarily recognized that this compound had a positive effect on the tested disorder. The research in this report concludes that the enzymatic production of 4-CHN provides a platform for developing steroid drugs in the pharmaceutical industry.
CONFLICT OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
FUNDING
This study was funded by VISTAS, Vels University Research Grant (No: UP12G9520010 dated 18.04.2013).
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Abdelouahab C, Abderrahmane B. Comparative study of the efficiency of three protein-ligand docking programs. J Proteomics Bioinform, 2008; 1:161–5. CrossRef
Ahmad S, Garg SK, Johri BN. Biotransformation of sterols: selective cleavage of the side chain. Biotechnol Adv, 1992; 10(1):1–67. CrossRef
Amutha K, Kokila V. Optimization of cholesterol oxidase production and 16S rRNA partial sequence of Bacillus cereus strain KAVK4 isolated from butter. J App Pharm Sci, 2016; 6(07):061–6. CrossRef
Arevalo JMC, Amorim JC. An in-silico analysis reveals 7,7′-bializarin as a promising DNA gyrase B inhibitor on gram-positive and gram-negative bacteria. Comput Biol Med, 2021; 135:104626; doi: 10.1016/j.compbiomed.2021.104626 CrossRef
Arima K, Nagasawa M, Bae M, Tamura G. Microbial transformation of sterols: Part I. Decomposition of cholesterol by microorganisms. Agric Biol Chem, 1969; 33(11):1636–43. CrossRef
Aruna K, Rao DM, Munawar TM. Purification and characterisation of cholesterol oxidase from novel native isolate Bacillus subtilis YS01 isolated from meat sample. Int J Biol Pharm Allied Sci, 2017; 6:1578–88.
Bergey DH, Holt JG. Bergey’s manual of determinative bacteriology. In: 9th edition, Williams and Wilkins, Baltimore, MD, 1994.
Birari RB, Bhutani KK. Pancreatic lipase inhibitors from natural sources: unexplored potential. Drug Discov Today, 2007; 12(19–20):879–89. CrossRef
Bortolini O, Medici A, Poli S. Biotransformations on steroid nucleus of bile acids. Steroids, 1997; 62(8–9):564–77. CrossRef
Burhan A, Nisa U, Gokhan C, Omer C, Ashabil A, Osman G. Enzymatic properties of a novel thermostable, thermophilic, alkaline and chelator resistant amylase from an alkaliphilic Bacillus sp. isolate ANT-6. Process Biochem, 2003; 38:1397–403. CrossRef
Cruz A, Fernandes P, Cabral JMS, Pinheiro HM. Whole-cell bioconversion of β-sitosterol in aqueous-organic two-phase systems. J Mol Catal B Enzym, 2001; 11:579–85. CrossRef
Dash BK, Rahman MM, Sarker PK. Molecular identification of a newly isolated Bacillus subtilis BI19 and optimisation of production conditions for enhanced production of extracellular amylase. Biomed Res Int, 2015; 859805:1–9. CrossRef
Devi S, Kanwar SS. Cholesterol oxidase: source, properties and applications. Insights Enzyme Res, 2017; 1:1; doi: 10.21767/2573-4466.100005 CrossRef
Devi VR, Sharmila C, Subramanian S. Molecular docking studies involving the inhibitory effect of gymnemic acid, trigonelline and ferulic acid, the phytochemicals with antidiabetic properties, on glycogen synthase kinase 3 (α and β). J App Pharm Sci, 2018; 8(04):150–60. CrossRef
Devlin MJ, Yanovski SZ, Wilson GT: Obesity: what mental health professionals need to know. Am J Psychiatry, 2000; 157:854–66. CrossRef
Dogra N, Qazi GN. Steroid biotransformation by different strains of Micrococcus sp. Folia Microbiol (Praha), 2001; 46(1):17–20. CrossRef
Doukyu N. Characteristics and biotechnological applications of microbial cholesterol oxidases. Appl Microbiol Biotechnol, 2009; 83(5):825–37. CrossRef
Dunn-Emke S, Weidner G, Ornish D. Benefits of a low-fat plant-based diet. Obes Res, 2001; 9(11):731; doi: 10.1038/oby.2001.100 CrossRef
Fernández-Cabezón L, Galán B, García JL. New insights on steroid biotechnology. Front. Microbiol, 2018; 9:958; doi: 10.3389/fmicb.2018.00958 CrossRef
Gago G, Diacovich L, Arabolaza A, Tsai SC, Gramajo C. Fatty acid biosynthesis in actinomycetes. FEMS Microbiol Rev, 2011; 35:475–97. CrossRef
Gurudeeban S, Satyavani K, Ramanathan T, Balasubramanian T. An in silico approach of alpha-ketoglutarate dependent dioxygenase FTO inhibitors derived From Rhizophora mucronata. Drug Invent Today, 2012; 4(11):594–8.
Hlivak P, Odraska J, Ferencik M, Ebringer L, Jahnova E, Mikes Z. One-year application of probiotic strain Enterococcus faecium M-74 decreases serum cholesterol levels. Bratisl Lek Listy, 2005; 106(2):67–72.
Kaur S, Kaur HP, Prasad B, Prasher M. Production and optimisation of cholesterol oxidase from Rhodococcus species. Int J Pharm Sci, 2015; 7(7):265–8.
Kim KP, Rhee CH, Park HD. Degradation of cholesterol by Bacillus subtilis SFF34 isolated from Korean traditional fermented flatfish. Lett Appl Microbiol, 2002; 35(6):468–72. CrossRef
Kovalenko NK, Kasumova SA, Muchnik FV. Screening of the strains of lactic acid bacteria possessing hypocholesterinemic activity and their practical use. J Microbiol, 2004; 66(3):33–42.
Kumari L, Kanwar SS. Cholesterol oxidase and its applications. Adv Microbiol, 2012; 2(2):49–65. CrossRef
Kumari L, Kanwar SS. Purification and characterisation of an extracellular cholesterol oxidase of Bacillus subtilis isolated from Tiger Excreta. Appl Biochem Biotechnol, 2016; 178:353–67. CrossRef
Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 1970; 227:680–5. CrossRef
Lashkarian H, Raheb J, Shahzamani K, Shahbani H, Shamsara M. Extracellular cholesterol oxidase from Rhodococcus sp: Isolation and molecular characterisation. Iran Biomed J, 2010; 14(1–2):49–57.
Lee MT, Chen WC, Chou CC. Medium improvement by orthogonal array designs for cholesterol oxidase production by Rhodococcus equi No. 23. Process Biochem, 1997; 32(8):697–703. CrossRef
Lee MT, Chen WC, Chou CC. Optimisation and kinetic analysis of cholesterol oxidase production by Rhodococcus equi no. 23 in submerged cultures. Enzyme Microb Technol, 1999; 25:598–604. CrossRef
Liu WH, Horng WC, Tsai MS. Bioconversion of cholesterol to cholest-4-en-3-one in aqueous/organic solvent two-phase reactors. Enzyme Microb Technol, 1996; 18:184–9. CrossRef
Liu ZQ, Shan HY. Cholesterol, not polyunsaturated fatty acids, is target molecule in oxidation induced by reactive oxygen species in membrane of human erythrocytes. Cell Biochem Biophys, 2006; 45(2):185–93. CrossRef
Madeswaran A, Umamaheswari M, Asokkumar K, Sivashanmugam T, Subhadradevi V, Jagannath P. Docking studies: In silico lipoxygenase inhibitory activity of some commercially available flavonoids. Bangladesh J Pharmacol, 2011; 6:133–8. CrossRef
Mohapatra S, Prasad A, Haque F, Ray S, De B, Ray SS. In silico investigation of black tea components on α-amylase, α-glucosidase and lipase. J Appl Pharm Sci, 2015; 42–7. CrossRef
Okechukwu P, Sharma M, Tan WH, Chan HK, Chirara K, Gaurav A, Al-Nema M. In-vitro antidiabetic activity and in-silico studies of binding energies of palmatine with alpha- amylase, alpha-glucosidase and DPP-IV enzymes. Pharmacia, 2020; 67(4):363–71. CrossRef
Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: a review. Biophys Rev, 2017; 9(2):91–102. CrossRef
Park YJ, Lee GS, Cheon SY, Cha YY, An HJ. The anti-obesity effects of Tongbi-san in a high-fat diet-induced obese mouse model, BMC Complement Altern Med, 2019; 19:1; doi: 10.1186/s12906-018-2420-5 CrossRef
Pekkanen J, Linn S, Heiss G, Suchindran CM, Leon A, Rifkind BM, Tyroler HA. Ten-year mortality from cardiovascular disease in relation to cholesterol level among men with and without preexisting cardiovascular disease. N Engl J Med, 1990; 322(24):1700–7. CrossRef
Potshangbam AM, Rathore RS, Nongdam P. Discovery of sulfone-resistant dihydropteroate synthase (DHPS) as a target enzyme for kaempferol, a natural flavonoid. Heliyon, 2020; 6:e033; doi: 10.1016/j.heliyon.2020.e03378 CrossRef
Puiu RA, Dolete G, Ene AM, Nicoar? B, Vl?sceanu GM, Holban AM, Grumezescu AM, Bolocan A. 2- properties of biofilms developed on medical devices. Biofilms and Implantable Med Devices: Infect and Control, 2017; 25–46. CrossRef
Rao SM, Thakkar K, Pawar K. Microbial transformation of steroids: current trends in cortical side chain cleavage. Quest, 2013; 1(2):16–20.
Rhee CH, Kim KP, Park HD. Two novel extracellular cholesterol oxidases of Bacillus sp. isolated from fermented flatfish. Biotechnol Lett, 2002; 24(17):1385–9. CrossRef
Saavedra L, Taranto MP, Sesma F, De Valdez GF. Homemade traditional cheeses for the isolation of probiotic Enterococcus faecium strains. Int J Food Microbiol, 2003; 88(2-3):241–5. CrossRef
Saranya S, Shekinah S, Rajagopal T, Vijayakumar J, Ponmanickam P. Isolation and characterisation of cholesterol degrading bacteria from soap and vegetable oil industrial waste. Indian J Biotechnol, 2014; 13(4):508–13.
Sarkar B, Ullah MA, Islam SS, Rahman MH, Araf Y. Analysis of plant derived phytochemicals as anti-cancer agents targeting cyclin dependent kinase-2, human topoisomerase IIa and vascular endothelial growth factor receptor-2. J Recept Signal Transduct Res, 2021; 41(3):217–33. CrossRef
Shao M, Rao Z, Zhang X, Xu M, Yang T, Li H, Xu Z, Yang S. Bioconversion of cholesterol to 4-cholesten-3-one by recombinant Bacillus subtilis expressing choM gene encoding cholesterol oxidase from Mycobacterium neoaurum JC-12. J Chem Technol Biotechnol, 2015; 90:1811–20. CrossRef
Singhal N, Maurya AK, Mohanty S, Kumar M, Virdi JS. Evaluation of bile salt hydrolases, cholesterol-lowering capabilities, and probiotic potential of Enterococcus faecium isolated from rhizosphere. Front Microbiol, 2019; 10:1567; doi: 10.3389/fmicb.2019.01567 CrossRef
Sugiyama H, Akazome Y, Shoji T, Yamaguchi A, Yasue M, Kanda T, Ohtake Y. Oligomeric procyanidins in apple polyphenol are main active components for inhibition of pancreatic lipase and triglyceride absorption. J Agric Food Chem, 2007; 55:4604–9. CrossRef
Tanguenyongwatana P, Jongkon N. Molecular docking study of tyrosinase inhibitors using ArgusLab 4.0.1: A comparative study. Thai J Pharm Sci, 2016; 40(1):21–5.
Telagari M, Hullatti K. In-vitro α-amylase and α-glucosidase inhibitory activity of Adiantum caudatum Linn. and Celosia argentea Linn. extracts and fractions. Indian J pharmacol, 2015; 47(4):425–9. CrossRef
Thompson M. ArgusLab 4.0.1. Planaria Software LLC, Seattle, Washington, 2004.
Turfitt GE. The microbiological degradation of steroids; oxidation of hydroxy-steroids to keto-derivatives by Proactinomyces spp. Biochem J, 1946; 40(1):79–81. CrossRef
Varma R, Nene S. Biosynthesis of cholesterol oxidase by Streptomyces lavendulae NCIM 2421. Enzyme Microb Technol, 2003; 33:286–91. CrossRef
Wali H, Rehman FU, Umar A, Ahmed S. Cholesterol degradation and production of extracellular cholesterol oxidase from Bacillus pumilus W1 and Serratia marcescens W8. Biomed Res Int, 2019; 1359528:1–9. CrossRef
Wu K, Li W, Song J, Li T. Production, purification, and identification of cholest-4-en-3-one produced by cholesterol oxidase from Rhodococcus sp. in aqueous/organic biphasic system. Biochem Insights, 2015; 8(1):1–8. CrossRef
Yang S, Zhang H. Optimization of cholesterol oxidase production by Brevibacterium sp. employing response surface methodology. Afr J Biotechnol, 2012; 11(33):8316–22. CrossRef
Yazdi MT, Malekzadeh F, Zarrini GH, Faramarzi MA, Kamranpour N, Khaleghparast SH. Production of cholesterol oxidase by a newly isolated Rhodococcus sp. World J Microbiol Biotechnol, 2001; 17(7):731–7. CrossRef
Yehia HM, Hassanein WA, Ibraheim SM. Purification and characterisation of the extracellular cholesterol oxidase enzyme from Enterococcus hirae. BMC Microbiol, 2015; 15:178. doi: 10.1186/s12866-015-0517-2. CrossRef