INTRODUCTION
Today several serious threats afflict humanity worldwide. One of the most severe threats is the rise in antibiotic resistance, where bacterial mutations decrease the efficacy of antibiotics (Ciabuschi et al., 2020). Many bacteria have developed means of resistance to antimicrobials, which can spread to other bacteria reducing the activeness of subsequent antimicrobial treatments. Antibiotic resistance is a growing threat that already causes more than 50,000 deaths per year in the EU and US alone, and several hundreds of thousands of casualties are estimated in the rest of the world. Subsequently, infectious disease experts have called for strategies to minimize the risk of spreading antibiotic resistance. Although the mechanisms which lead to antimicrobial resistance are biological, the motivations behind current levels and methods are determined by a wide range of factors, including individual, psychological, social, cultural, political, and economic forces (Chambers et al., 2020).
The Mine, Model, Manipulation, Measure, and Manufacture (5Ms) strategy is used to overproduce the desired product and gain a deeper knowledge of the intelligence system of microorganisms. “Mine” is the initial step toward understanding the internal connections in large datasets, such as omics. The procedure “Model” is used to construct hypotheses from “Mine.” “Manipulation” is required to test the aforementioned hypotheses and to point out a viable path to real applications. “Measure” refers to the process of revealing the phenotype of a genome-modified microorganism, while “Manufacture” refers to the process of scaling it up in industrial fermenters. Subsequently, the desired products can be manufactured and scaled up to the industrial level using this 5Ms plan (Gao et al., 2017).
Psychology is the study of the mind and its functions, which considers all possible conditions of the organism and involves the analyses of the acquired information from the surroundings, followed by information processing and finally responding (Baker, 2012). Moreover, behavioral psychology is a branch of psychology that focuses on the connection between the organism’s behavior and the mind, which has been shifted through the years between the perspective of nurture and nature. Currently, it is believed that the behavior of any organism has a genetic basis, which could also get influenced somehow by environmental factors. In recent years, a lot of attention has been given to investigating the genetic basis of behavior and intelligence in different higher organisms (Brennan, 2004; Flint, 1999; Plomin and Spinath, 2004), including humans (Hettema et al., 2003).
In this review, the genetic basis of classical conditioning and the probability of the connection with the noncoding RNAs were discussed, and whether that concept could be applied to enhance antibiotic sensitivity was also discussed.
MOLECULAR PSYCHOLOGY
As a part of behavioral researches in higher and lower organisms, several studies on the genetic background of learning and memory are progressing rapidly in invertebrates (Carew and Sahley, 1986; Giurfa and Sandoz, 2012; Menzel and Benjamin, 2013), especially Drosophila (Iliadi, 2009; Malik and Hodge, 2014; Tully and Quinn, 1985). The question was related to the single-celled organism if it has such evidence of biological intelligence. There was no direct relation between behavioral psychology and microbes. In order to conceptionally connect behavioral psychology to the microbial level, the connection between biology and psychology based on the molecular level should be introduced and a brief understanding of the term “molecular psychology” is required.
Psychology in term is rooted in biology, since not so long ago the field was greatly transformed by the development and breakthroughs of molecular biology tools. Those tools of molecular biology and genetics were applied successfully in uncountable studies to investigate the organism’s behavior and the brain system, which raised the term “molecular psychology” as the study of behavior and its underlying brain systems using the tools of molecular biology (Craddock and Owen, 1996; Demkow and Wola?czyk, 2017; Plomin, 1995).
Jennings (1906) argued that one of the relevant mechanisms underlying protozoan behavior was learning. He observed that repeated aversive stimulation of the ciliate Stentor roeselii resulted in a characteristic sequence of distinct adaptive behaviors, which may be interpreted as an elementary form of learning which is considered a key feature of learning in “higher” organisms. Single cells have the ability to carry out a form of information processing that neuroscientists have traditionally attributed to networks of cells. For example, Table 1 illustrates molecules/pathways suggested to be involved in learning and memory, with homologs in ciliates (Gershman et al., 2021).
The question remains, to what extent do molecular biology and genomics affect the psychological concepts in higher organisms? Five breakthrough discoveries made during the past two decades were suggested to be the main pillars of genomic psychology (Canli, 2007). The previous findings linked genetic variations to personality and to brain function and also suggested such interaction with environmental factors affecting mental health, leading to identifying the neural and molecular correlates of these gene–environment interactions. However, none of the findings showed evidence in regards to the behavioral psychology of the single cell organisms. Lack of information in such scope was the main authors’ motivation in the current review to align the evidences and previous art in context to hint something related to a perspective about the philosophical term “microbial psychology” or better known as “bacterial intelligence”, with special regard to the classical conditioning phenomena and its connection with the non-coded RNAs.
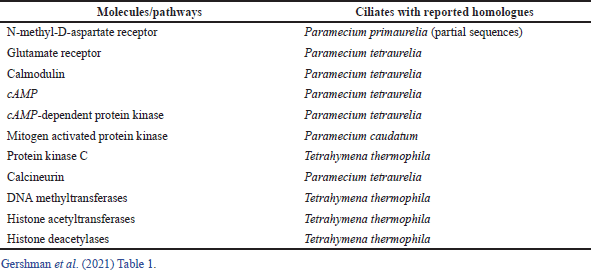 | Table 1. Molecules/pathways suggested involved in learning and memory, with homologs in ciliates. [Click here to view] |
CLASSICAL CONDITIONING THEORY
There is a theory for the learning process in higher organisms which has been well studied through the years in psychology called “classical conditioning theory,” which involves learning a new behavior via the process of association. In simple terms, two stimuli are linked together to produce a newly learned response in a person or animal (Andersson, 2016; Bitterman, 2006; Lorenzetti et al., 2011). Classical conditioning is the process where the combination of stimuli produces a specific response in an organism. The response of the conditioned organism can occur positively or negatively according to the connotation of the stimuli. The interest in studying this association process helped largely in deciphering critical problems in clinical psychiatry (Follette and Dalto, 2015). Classical conditioning was and still is under extensive studies to clarify the genetic basis of the association. However, a direct connection between stress and classical conditioning has been reported (Schreurs and Burhans, 2015).
Very limited studies provide a possible connection between classical conditioning and microbial level. However, there are plenty of researches reporting varied bacterial behavior such as symbiosis, mutualism, commensalism, and parasitism, including the mechanisms of biofilm formation, swimming, swarming, cell-to-cell communication, and stress response (Niu and Wang, 2012). All those studies came together to point out that bacteria are adaptive microorganisms. Just like other living organisms, the latter have the genetic material that enables them to receive the surrounding signals (sensing), process them, and then respond. Since bacteria already have different abilities of sensing the environmental signals, processing them, and responding, obviously that is the reason why some bacterial species are considered more adaptable and on an industrial scale valuable than others. The size of the genetic material in bacteria is also an advantage over higher organisms in regard to behavioral studies. Several reports showed that the bacterial ability to adapt to the environment is a result of multiple and complex genetic regulation networks (Brooks et al., 2011; Lozada-Chávez et al., 2006; McAdams et al., 2004). Gandhi et al. (2007) suggested that the presence of polycistronic RNA in bacteria and its involvement in the complex regulation process of biochemical networks are already evidence of a direct relation between genomic adaptation and applied classical conditioning. However, the complexity of those mechanisms in bacteria will never reach the same level as gene regulatory networks in higher organisms, as simply genetic complexity can be measured by genome size and the number of the Open Reading Frames (ORFs) included (Davidson and Peter, 2015; Huang and Kauffman, 2013; Somogyi and Sniegoski, 1996; Walhout, 2011).
If microorganisms could be considered as a useful candidate for behavioral studies, then why should not psychological concepts like classical conditioning be considered to explain bacteria and microorganisms’ behavior and therefore applied to the microbial level?
NONCODING RNAS (lncRNAs)
Long noncoding RNAs (lncRNAs) are a group of nonprotein-coding RNAs with a length exceeding 200 nucleotides. They will not be translated into proteins but affect the binding of DNA, mRNA, microRNA, and proteins and regulate gene expression at the transcriptional, posttranscriptional, translational, and posttranslational levels (Karakas and Ozpolat, 2021). lncRNAs, potential RNA polymerase II transcription byproducts, have been reported to regulate cellular processes, such as chromosome and genome modification, transcription activation and interference, and nuclear transport. lncRNAs functions are dependent on their position, so they are categorized in terms of position in the genome to protein-coding genes: they can be classified as sense, antisense, bidirectional, intronic, intergenic, and enhancer lncRNAs. They also may be classified according to length, function, location, and targeting mechanism. An unveiled value of lncRNAs was reported which encodes small peptides to fine-tune general biological processes in a tissue-specific manner (Chen et al., 2021).
The recent two decades have revealed that lncRNAs pervasively exist in the eukaryotic system. lncRNA (i) might indirectly affect controlling the expression or subcellular localization of the key protein factors, (ii) might act as a molecular platform, bringing diverse proteins together into an ribonucleoprotein (RNP) complex, or a decoy by sponging and prevent them from associating with their targets, and (iii) might be organizers of nuclear architecture. More efforts are required to provide insights into how these special transcripts are controlled and function, especially in the nucleus (Song et al., 2021).
BACTERIAL GENETIC COMPOSITION
Most bacteria have single, covalent closed, circular chromosomes, as opposed to the linear chromosomes found in eukaryotic cells. Many have circular chromosomes and linear plasmids, while some have linear chromosomes and linear plasmids. In several cases, multiple chromosomes have been discovered, including Brucella, Leptospira interrogans, Burkholderia, and Vibrio cholera (The Desk Encyclopedia of Microbiology, 2004). Borrelia and Streptomyces have linear chromosomes, and most strains contain both linear and circular plasmids (Miller et al., 2013). The Escherichia coli chromosome is around 1.35 mm long, which is longer than the bacterial cell a hundred times, but the circular DNA is looped and supercoiled to fit the chromosome into the limited space inside the cell (Guentzel, 1996). The bacterial genome is considered as the first level of the genome composition. However, the second level is considered as the transcribed genes, which are categorized into two main groups: protein-coding and noncoding RNA. The third level in the bacterial genome composition is the translated proteins, which involve structure and functional proteins.
An example to give about dynamic interaction in bacterial genetic levels is that of transmembrane proteins and sensors, which are involved in the reception of transduction signals emitted by other cells in the surrounding environment. It has been thought for a long time that the acquisition model of these signals is a single-component one before admitting that the complexity and simultaneity of the induced intracellular phenomena are far from responding to this hypothesis. The double-component model was then introduced with the implication of different levels of the genome, from gene to protein-coding RNA (Hellingwerf, 2005). This said, the question arises more about the role of noncoding RNAs in bacterial signal acquisition and social intelligence. Nucleotide sequence differences are in the form of single nucleotide polymorphism, insertion–deletion mutations, and simple sequence repeats. These individual single and/or multi nucleotide differences can lead to different observable or hidden traits, in addition to other ones that could possibly be silent on the genotypic level (Ismail and Essawi, 2012).
BACTERIAL INTELLIGENCE CONCEPT
The definition of “bacterial intelligence” is likely to have emerged from observations of the rudimentary cooperation of single cells (Ben-Jacob, 2009; Ben Jacob et al., 2004; Ben-Jacob and Shapira, 2005) and then was further developed by other authors (Ford, 2009, 2006; Hellingwerf, 2005). The study of biology focused on a generic vision of the large biological systems, and it gradually progressed towards the study of minutiae. This therefore led to a loss of the real contextual scale, and the unit cell was no longer considered as a whole organism. Ford (2009, 2006) and others (Hellingwerf, 2005; Ben Jacob et al., 2004) questioned this by initiating the first observations of a rudimentary form of cooperation and “social intelligence” in bacteria. British Psychologist Richardson (2012) concluded in his study that unicellular intelligence might provide the key to understanding intelligence in complex vertebrates, including humans (Lyon, 2015).
Due to its existence, bacteria have been able to convert the complex matter into easy matter to metabolize molecules to ensure its survival. This ability is not only at the service of the individual cell, but also for a structured hierarchical organization in the colony and communication as with coordination (Ben Jacob et al., 2004). Beyond a basic signal transfer in the form of physical interactions and biochemical exchange, the bacterial cell communicates its internal physiological state in a global context of cooperation and decision-making. The involvement of the dynamic and self-conscious genome in the biochemical regulation of this synchronized process between cells is a form of “pragmatic intelligence,” in which the experience of the bacterial community is exchanged and the core’s “wisdom” is the only determinant of the response. This “smart” process transforms the colony into a super brain and deserves a categorization of bacterial species according to their intelligence quotient (Pinto and Mascher, 2016). It is important to note that the understanding of these forms of communication and intelligence in bacteria was an inspiration for other studies, especially those on the cooperation and resistance mechanism in cancer cells (Ben-Jacob et al., 2012).
BACTERIAL NONCODING RNAs AND STRESS RESPONSE
Several reports studied small regulatory RNAs in bacteria (Dutta and Srivastava, 2018; Wagner and Romby, 2015; Waters and Storz, 2009). It all started with the genomewide studies of bacterial gene expression, which has shifted from microarray technology to second-generation sequencing platforms. Transcriptome analysis via RNA-seq has a number of advantages, such as annotation-independent detection of transcription, improved sensitivity, and increased dynamic range. Early studies have uncovered the novelty of several coding sequences and noncoding RNA in microorganisms. That reveals the similarity of a transcriptional landscape with a eukaryote. The basic RNA-seq protocols have been improved to fit the rapid progress of the studies of RNA biology, with special regard to the noncoding RNAs. Our understanding of gene expression and genome content depends mainly on the further refinements and improvement of the current techniques (Croucher and Thomson, 2010).
Expression of the small RNA, which is tightly regulated at the level of transcription, can help the cell cope with environmental stress by redirecting cellular metabolism (Table 2,) (Azhikina et al., 2015; De Bruijn, 2016; De La Fuente and Martínez-Guitarte, 2016; Fröhlich et al., 2013; Gottesman et al., 2006; Michaux et al., 2014; Sonnleitner et al., 2011). A large number of these small RNAs act by pairing to their target mRNAs. The outcome of pairing can be either stimulation or inhibition of translation. Many of the well-studied stress response regulons have now been found to include regulatory RNAs (Gottesman et al., 2006; Plomin and Spinath, 2004; Wagner and Romby, 2015).
STRESS RESPONSE AND CLASSICAL CONDITIONING
Classical conditioning (or respondent conditioning) refers to acquiring a new behavior via the process of association. In that regard, stresses are considered as a valuable classical conditioning tool (Li, 2012; Moreira and Volpato, 2004; Shors et al., 1992; Wood and Shors, 1998). It has been studied well enough in higher animal models though there is still limited available literature on the nonneural organisms (Clark et al., 2002; Dayan et al., 2000; Hesslow and Yeo, 2002).
This curiosity about the presence of a direct link between stress and classical conditioning is in fact induced by the abovementioned “social” behavior of bacteria. Under limiting conditions, the adaptive mutagenesis induced in the cell is not necessarily in favor of its own survival but rather of the general framework of the entire colony. Morphotypes observed during stress are characteristic and involve social resistance (Crespi, 2001; Ben Jacob et al., 2004; Tarnita, 2017; West et al., 2007). These facts have shifted the standard definition of the bacterial genome from a static unit of storage of genetic information to a cybernetic, self-conscious, and dynamic agent. Figure 1 illustrates a diagram of the model for general stress response in E. coli that shows that under stress conditions microorganisms may develop signal transduction systems to sense environmental stresses and to control the coordinated expression of genes involved in cellular defense mechanisms. These evolved protective or adaptive networks assist microorganisms in modifying their environments and/or surviving the stress condition (Chung et al., 2006).
CLASSICAL CONDITIONING AND NONCODING RNA
Classical conditioning showed an impact on the noncoding RNA’s expression in Drosophila (Maniatis, 2015). There are few other reports that showed the same principle of the involvement of noncoding RNA in higher cognitive processes and classical conditioning in other animal models (Salta and De Strooper, 2012; Schmidt et al., 2015). One report showed clear evidence of the impact of the classical conditioning on the noncoding RNA’s expression in Lymnaea, where antiNOS-2 RNA is axonally trafficked and regulated by classical conditioning (Korneev et al., 2013) and critically a single conditioning trial changes the amount of antiNOS-2 RNA transported along the axon. The previous study concluded that gene regulation networks get altered by conditioning; a specific behavior therefore comes out from the organism. Thus, such data might summarize the fact that the specific behavior is regulated while the noncoding RNA is involved as a part of the regulatory system in the specific organism.
The core difference between neural and nonneural organisms in acquiring a new behavior is briefed in Figure 2. As in animal models, the new behavior is acquired by associative learning via creating or modifying a neural network (Veit et al., 2015). This is different in plants since a recent report showed that they could learn also by association, as the plant behavior is acquired via regulating the metabolic pathways, which are subsequently regulated by gene regulation networks (Gagliano et al., 2016). However, evidence for learning in nonneural organisms is scant, and only a few unequivocal reports of learning have been described in single-cell organisms (Andersson, 2016; Ginsburg and Jablonka, 2009). Several studies showed that the nonneural organism Physarum polycephalum could be classically conditioned (Boisseau et al., 2016; Nakagaki et al., 2000, 2004;Saigusa et al., 2008), although P. polycephalum is considered a multicellular organism.
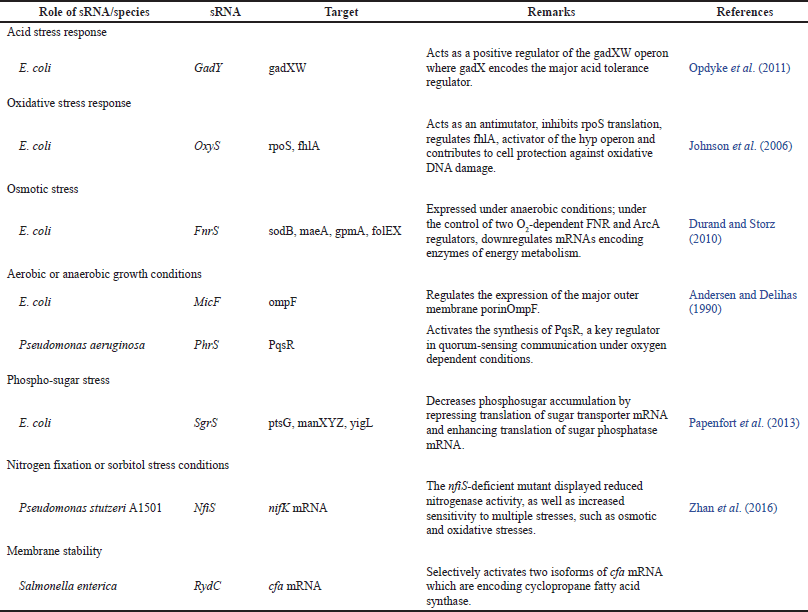 | Table 2. Examples of bacterial noncoding RNA involved in stress response regulation. [Click here to view] |
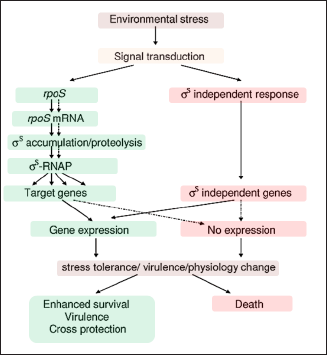 | Figure 1. A diagram of the model for general stress response in E. coli. Solid line: positive control, dotted line: negative control (Chung et al., 2006). [Click here to view] |
FUTURE RESEARCH AND APPLICATIONS
Ability to predict
Serial studies since 2009 have been showing the ability of the bacterial cells to memorize and plan for the future events by developing a biochemical-based timer (Brunke and Hube, 2014; Mandli and Modak, 2014; Mitchell et al., 2009; Mitchell and Pilpel, 2011). The predictive behavior of the bacterial cells could be considered as associative learning (Fernando et al., 2009; Tagkopoulos et al., 2008). According to an original study (Mitchell et al., 2009), they found that one type of sugar, “lactose,” is followed by a second sugar, “maltose,” in the human intestine. The team checked E. coli’s genetic response to lactose and found that the gene network for utilizing maltose was partially activated alongside the genes that enable the same bacterial cells to digest lactose. When they reversed the order of the sugars, maltose at first, they found no corresponding activation of lactose genes, ensuring that bacteria have naturally adapted (or learned) to prepare for utilizing maltose after lactose. Generally speaking, bacterial evolution for adaptive purposes could be considered also as long-term learning by association. Several bacterial evolution experiments make it evident that bacteria are capable of hanging and choosing between several carbon sources (Feldgarden et al., 2003; Görke and Stülke, 2008; Hua et al., 2007; Mazurie et al., 2010; Trevors, 1997).
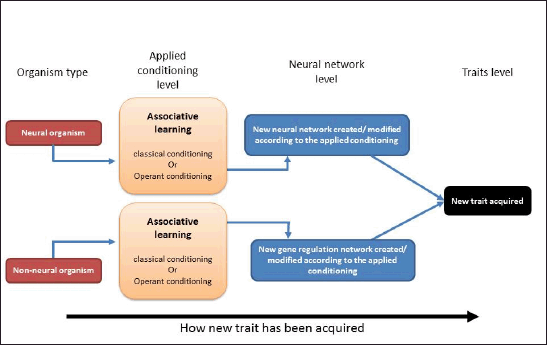 | Figure 2. A diagram briefing the difference between neural and nonneural organisms while learning a new behavior. [Click here to view] |
Object sensing
In a series of tests, James Shapiro reported that Myxococcus xanthus worms were guided toward three-dimensional objects. Worms were directed toward either biologically noticed (a clump of prey cells) or chemically inert (sterilized glass beads) objects. The mechanisms should be studied through replicated experiments (Dworkin, 1983).
Bistability
Bistability to cognition in higher organisms, as well as its involvement in perception, memory, and learning is important.
Endogenous activity
The discovery of endogenous, oscillatory activity in the brain that is independent of external stimuli is one of the classic views of cognition. This oscillatory activity in prokaryotic regulatory circuits can serve as a guide to the processes related to endogenous brain activity (Bechtel, 2012).
Nanobrain
The cluster of chemoreceptors that mainly drives motility in E. coli may provide a tractable model for explaining the integration of signals from numerous sources into a behavioral response, which still represents a challenge.
Valence
The mechanisms of integration of external stimuli combinations of varying valence with interoceptive cues to form a coherent behavioral response in either bacteria or humans are still unknown. Except for circadian (24-hour) frequency, little information is available on the power spectrum (i.e., intensity and frequency) of signals to which each microorganism in any natural habitat is exposed. Stress responses in prokaryotes have been well investigated, especially those with diverse behavioral alternatives that represent potentially good research platforms (Lyon, 2015).
Memory and learning
Although the mechanisms by which the bacterial memory occurs have been characterized in many species, they need to be further investigated. In prokaryotes, the possibility of associative learning is likewise unknown. Given the large number of genes involved in generating serine/threonine protein kinases (STPKs) in this diversified predator, M. xanthus could be a viable candidate for such research. CaMKII is an autophosphorylating STPK that is engaged in numerous signaling pathways in animal homeostasis and is also hypothesized to play a role in human memory and learning in the reisany sequence homology between CaMKII and any of the myxobacterial STPKs (Goldman et al., 2006).
Communication
The ability of quorum sensing molecules to influence the behavior of dispersed cells, particularly in response to stress, suggests that they may be precursors of hormone messengers, and certain homologies, such as between Acyl-homomserine lactones (AHLs) and ghrelin, have been discovered (Lyon, 2015).
CONCLUSION AND FUTURE ASPECTS
There would be a relation between the degree of development of an organism and the repetition of the stimulus to achieve the desired conditioning. It may be suggested that the higher organisms could get the stimulus from the first few repetitions, but the nonneural organisms sometimes need years of conditioning to achieve the specific behavior. However, a condensed number of events on a regular basis with a focus on the bacterial generations could lead to the solution of how to teach bacteria a new behavior. Psychologically informed studies are urgently needed to facilitate change and evaluate the effectiveness of theory-based interventions targeting reducing antibiotics, including the promotion of biological alternatives.
Since the concept of microbial learning is, scientifically, acceptable as shown previously, then the future of training bacteria is limitless. Numerous possible future applications could be invented in regard to that aspect of bacterial learning, which could represent a breakthrough in future cellular communication. A review in 2004 titled “Teaching bacteria a new language” (Gerchman and Weiss, 2004) derived the inspiration of many synthetic biology scientists as they were wondering about the new ways of bacterial communications other than the known chemical or physical signaling and recently electrical signaling (Bassler, 2002; Farhadi, 2014; Humphries et al., 2017; Marx, 2014; Von Bodman et al., 2008). The first thought that came to our mind was electromagnetic (EM) signaling. Few resources have discussed cellular communication based on EM waves, especially radiofrequency (RF). In general, the studies on EM and its impact on bacterial cells are very limited (Farhadi, 2014; Ku?era and Cifra, 2013; Montagnier et al., 2009; Soghomonyan et al., 2016; Tessaro et al., 2015; Trushin, 2003). Besides limited resources, no biological receptor has been discovered yet to receive only a specific range of RF. However, the magnetic locomotor mechanism present in magnetotactic bacteria would imply the presence of EM cellular signaling in bacterial cells (Chen et al., 2017). Thus, here comes the importance of microbial psychology and classical conditioning; it could be possible to train the bacterial cell to evolve by developing a reception system for RF using stress as a stimulus. However, there are many unanswered questions and unexplained avenues of research to be explored.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FUNDING
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Andersen J, Delihas N. micF RNA binds to the 5′ end of ompF mRNA and to a protein from Escherichia coli. Biochemistry, 1990; 29:9249–56; doi:10.1021/bi00491a020 CrossRef
Andersson SGE. Stress management strategies in single bacterial cells. Proc Natl Acad Sci U S A, 2016; 113:3921–3; doi:10.1073/ pnas.1603151113 CrossRef
Azhikina TL, Ignatov DV, Salina EG, Fursov MV, Kaprelyants AS. Role of small noncoding RNAs in bacterial metabolism. Biochemistry (Mosc), 2015; 80:1633–46; doi:10.1134/S0006297915130015 CrossRef
Baker DB. The Oxford handbook of the history of psychology global perspectives. Oxford University Press, New York, NY, 2012; doi:10.1093/oxfordhb/9780195366556.001.0001 CrossRef
Bassler BL. Small talk: cell-to-cell communication in bacteria. Cell, 2002; 109:421–4; doi:10.1016/S0092-8674(02)00749-3 CrossRef
Bechtel W. Understanding endogenously active mechanisms: a scientific and philosophical challenge. Eur J Philos Sci, 2012; 2:233–48; doi:10.1007/s13194-012-0046-x CrossRef
Ben Jacob E, Becker I, Shapira Y, Levine H. Bacterial linguistic communication and social intelligence. Trends Microbiol, 2004; 12:366– 72; doi:10.1016/j.tim.2004.06.006 CrossRef
Ben-Jacob E. Learning from bacteria about natural information processing. Ann N Y Acad Sci, 2009; 1178:78–90; doi:10.1111/j.17496632.2009.05022.x CrossRef
Ben-Jacob E, Coffey DS, Levine H. Bacterial survival strategies suggest rethinking cancer cooperativity. Trends Microbiol, 2012; 20:403– 10; doi:10.1016/j.tim.2012.06.001 CrossRef
Ben-jacob E, Shapira Y. Meaning-Based Natural Intelligence Vs. Information-Based Artificial Intelligence. Cradlse Creat Shaarey Mishpat, Jerusalem 2005:1–67.
Bitterman ME. Classical conditioning since Pavlov. Rev Gen Psychol, 2006; 10:365–76; doi:10.1037/1089-2680.10.4.365 CrossRef
Boisseau RP, Vogel D, Dussutour A. Habituation in non-neural organisms: evidence from slime moulds. Proc R Soc B Biol Sci, 2016; 283:20160446; doi:10.1098/rspb.2016.0446 CrossRef
Brennan FX. Genetic differences in leverpress escape/avoidance conditioning in seven mouse strains. Genes Brain Behav, 2004; 3:110–4; doi:10.1111/j.1601-183X.2003.0057.x CrossRef
Brooks AN, Turkarslan S, Beer KD, Yin Lo F, Baliga NS. Adaptation of cells to new environments. Wiley Interdiscip Rev Syst Biol Med, 2011; 3:544–61; doi:10.1002/wsbm.136 CrossRef
Brunke S, Hube B. Adaptive prediction as a strategy in microbial infections. PLoS Pathog, 2014; 10:e1004356; doi:10.1371/journal.ppat.1004356 CrossRef
Canli T. The emergence of genomic psychology. Insights from genomic analyses might allow psychologists to understand, predict and modify human behaviour. EMBO Rep, 2007; 8:S30–4; doi:10.1038/sj.embor.7400938 CrossRef
Carew TJ, Sahley CL. Invertebrate learning and memory: from behavior to molecules. Annu Rev Neurosci, 1986; 9:435–87; doi:10.1146/ annurev.ne.09.030186.002251 CrossRef
Chambers JA, Crumlish M, Comerford DA, O’carroll RE. Antimicrobial resistance in humans and animals: rapid review of psychological and behavioral determinants. Antibiotics, 2020; 9:1–20; doi:10.3390/antibiotics9060285 CrossRef
Chen L, Chen C, Wang P, Song T. Mechanisms of Cellular Effects Directly Induced by Magnetic Nanoparticles under Magnetic Fields. J Nanomater 2017;2017:1–22. https://doi.org/10.1155/2017/1564634. CrossRef
Chen Y, Li Z, Chen X, Zhang S. Long non-coding RNAs : from disease code to drug role. Acta Pharm Sin B, 2021; 11:340–54; doi:10.1016/j.apsb.2020.10.001 CrossRef
Chung HJ, Bang W, Drake MA. Stress response of Escherichia coli. Compr Rev Food Sci Food Saf, 2006; 5:52–64. CrossRef
Ciabuschi F, Baraldi E, Lindahl O, Callegari S. Supporting innovation against the threat of antibiotic resistance: exploring the impact of public incentives on firm performance and entrepreneurial orientation. J Bus Res, 2020; 112:271–80; doi:10.1016/j.jbusres.2019.12.021 CrossRef
Clark RE, Manns JR, Squire LR. Classical conditioning, awareness, and brain systems. Trends Cogn Sci, 2002; 6:524–31; doi:10.1016/S1364-6613(02)02041-7 CrossRef
Craddock N, Owen MJ. Modern molecular genetic approaches to psychiatric disease. Br Med Bull, 1996; 52:434–52. CrossRef
Crespi BJ. The evolution of social behavior in microorganisms. Trends Ecol Evol, 2001; 16:178–83; doi:10.1016/ S0169-5347(01)02115-2 CrossRef
Croucher NJ, Thomson NR. Studying bacterial transcriptomes using RNA-seq. Curr Opin Microbiol, 2010; 13:619–24; doi:10.1016/j. mib.2010.09.009 CrossRef
Davidson EH, Peter IS. Gene regulatory networks. Academic Press, Cambridge, MA, 2015; doi:10.1016/b978-0-12-404729-7.00002-2 CrossRef
Dayan P, Kakade S, Read Montague P. Learning and selective attention. Nat Neurosci, 2000; 3:1218–23; doi:10.1038/81504 CrossRef
De Bruijn FJ. Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. vol. 1. Hoboken, New Jersey, Canada: Wiley Blackwell; 2016. https://doi.org/10.1002/9781119004813 CrossRef
De La Fuente M, Martínez-Guitarte JL. Thermal stress noncoding RNAs in prokaryotes and eukaryotes: a comparative approach. In: De Bruijn FJ (ed.). Stress and environmental regulation of gene expression and adaptation in bacteria, John Wiley & Sons, Inc., Hoboken, NJ, vol. 1, pp 412–21, 2016; doi:10.1002/9781119004813.ch37 CrossRef
Demkow U, Wola?czyk T. Genetic tests in major psychiatric disorders—integrating molecular medicine with clinical psychiatry—why is it so difficult? Transl Psychiatry, 2017; 7:e1151; doi:10.1038/tp.2017.106 CrossRef
Durand S, Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol, 2010; 75:1215–31; doi:10.1111/ j.1365-2958.2010.07044.x CrossRef
Dutta T, Srivastava S. Small RNA-mediated regulation in bacteria: a growing palette of diverse mechanisms. Gene, 2018; 656:60–72; doi:10.1016/j.gene.2018.02.068 CrossRef
Dworkin M. Tactic behavior of Myxococcus xanthus. J Bacteriol, 1983; 154:452–9; doi:10.1128/jb.154.1.452-459.1983 CrossRef
Farhadi A. Non-chemical distant cellular interactions as a potential confounder of cell biology experiments. Front Physiol, 2014; 5:405; doi:10.3389/fphys.2014.00405 CrossRef
Feldgarden M, Byrd N, Cohan FM. Gradual evolution in bacteria: evidence from Bacillus systematics. Microbiology, 2003; 149:3565–73; doi:10.1099/mic.0.26457-0 CrossRef
Fernando CT, Liekens AML, Bingle LEH, Beck C, Lenser T, Stekel DJ, Rowe JE. Molecular circuits for associative learning in single-celled organisms. J R Soc Interface, 2009; 6:463–9; doi:10.1098/ rsif.2008.0344 CrossRef
Flint J. The genetic basis of cognition. Brain, 1999; 122:2015– 31; doi:10.1093/brain/122.11.2015 CrossRef
Follette WC, Dalto G. Classical conditioning methods in psychotherapy. Int Encycl Soc Behav Sci, 2015:764–70; doi:10.1016/B9780-08-097086-8.21052-0 CrossRef
Ford BJ. On intelligence in cells: the case for whole cell biology. Interdiscip Sci Rev, 2009; 34:350–65; doi:10.1179/03080180 9X12529269201282 CrossRef
Ford BJ. Revealing the ingenuity of the living cell. Biologist, 2006; 53:221–4.
Fröhlich F, Christiano R, Walther TC. Native SILAC: metabolic labeling of proteins in prototroph microorganisms based on lysine synthesis regulation. Mol Cell Proteomics, 2013; 12:1995–2005; doi:10.1074/mcp. M112.025742 CrossRef
Gagliano M, Vyazovskiy VV, Borbély AA, Grimonprez M, Depczynski M. Learning by association in plants. Sci Rep, 2016; 6:38427; doi:10.1038/srep38427 CrossRef
Gandhi N, Ashkenasy G, Tannenbaum E. Associative learning in biochemical networks. J Theor Biol, 2007; 249:58–66; doi:10.1016/j. jtbi.2007.07.004 CrossRef
Gao Q, Tan GY, Xia X, Zhang L. Learn from microbial intelligence for avermectins overproduction. Curr Opin Biotechnol, 2017; 48:251–7; doi:10.1016/j.copbio.2017.08.016 CrossRef
Gerchman Y, Weiss R. Teaching bacteria a new language. Proc Natl Acad Sci U S A, 2004; 101:2221–2; doi:10.1073/ pnas.0400473101 CrossRef
Gershman SJ, Balbi PEM, Gallistel CR, Gunawardena J. Reconsidering the evidence for learning in single cells. Elife, 2021; 10:1– 15; doi:10.7554/eLife.61907 CrossRef
Ginsburg S, Jablonka E. Epigenetic learning in non-neural organisms. J Biosci, 2009; 34:633–46; doi:10.1007/s12038-009-0081-8 CrossRef
Giurfa M, Sandoz JC. Invertebrate learning and memory: fifty years of olfactory conditioning of the proboscis extension response in honeybees. Learn Mem, 2012; 19:54–66; doi:10.1101/ lm.024711.111 CrossRef
Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen J, Ronning CM, Barbazuk WB, Blanchard M, Field C, Halling C, Hinkle G, Iartchuk O, Kim HS, Mackenzie C, Madupu R, Miller N, Shvartsbeyn A, Sullivan SA, Vaudin M, Wiegand R, Kaplan HB. Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci U S A, 2006; 103:15200–5; doi:10.1073/ pnas.0607335103 CrossRef
Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol, 2008; 6:613–24; doi:10.1038/nrmicro1932 CrossRef
Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol, 2006; 71:1–11; doi:10.1101/ sqb.2006.71.016 CrossRef
Guentzel MN. Escherichia, Klebsiella, Enterobacter, Serratia, Citrobacter, and Proteus. University of Texas Medical Branch, Galveston, TX, 1996; doi:NBK8035 [bookaccession].
Hellingwerf KJ. Bacterial observations: a rudimentary form of intelligence? Trends Microbiol, 2005; 13:152–8; doi:10.1016/j. tim.2005.02.001 CrossRef
Hesslow G, Yeo C. A Neuroscientist’s guide to classical conditioning. Springer New York, New York, NY, 2002; doi:10.1007/9781-4419-8558-3
Hettema JM, Annas P, Neale MC, Kendler KS, Fredrikson M. A twin study of the genetics of fear conditioning. Arch Gen Psychiatry, 2003; 60:702–8; doi:10.1001/archpsyc.60.7.702 CrossRef
Hua Q, Joyce AR, Palsson B, Fong SS. Metabolic characterization of Escherichia coli strains adapted to growth on lactate. Appl Environ Microbiol, 2007; 73:4639–47; doi:10.1128/AEM.00527-07 CrossRef
Huang S, Kauffman SA. Complex gene regulatory networks— from structure to biological observables: cell fate determination. In: Meyers R (ed.). Computational complexity, Springer, New York, NY, pp 527–60, 2013; doi:10.1007/978-1-4614-1800-9_35 CrossRef
Humphries J, Xiong L, Liu J, Prindle A, Yuan F, Arjes HA, Tsimring L, Süel GM. Species-independent attraction to biofilms through electrical signaling. Cell, 2017; 168:200–9.e12; doi:10.1016/j. cell.2016.12.014 CrossRef
Iliadi KG. The genetic basis of emotional behavior: has the time come for a Drosophila model? J Neurogenet, 2009; 23:136–46; doi:10.1080/01677060802471650 CrossRef
Ismail S, Essawi M. Genetic polymorphism studies in humans. Middle East J Med Genet, 2012; 1:57–63; doi:10.1097/01. mxe.0000415225.85003.47 CrossRef
Jennings HS. Behavior of the lower organisms. Columbia University Press, The Macmillan Company, Agents, New York, NY, 1906. CrossRef
Johnson JR, Clabots C, Rosen H. Effect of inactivation of the global oxidative stress regulator oxyR on the colonization ability of Escherichia coli O1:K1:H7 in a mouse model of ascending urinary tract infection. Infect Immun, 2006; 74:461–8; doi:10.1128/IAI.74.1.461-468.2006 CrossRef
Karakas D, Ozpolat B. The role of LncRNAs in translation. Noncoding RNA, 2021; 7:16; doi:10.3390/ncrna7010016 CrossRef
Korneev SA, Kemenes I, Bettini NL, Kemenes G, Staras K, Benjamin PR, O’Shea M. Axonal trafficking of an antisense RNA transcribed from a pseudogene is regulated by classical conditioning. Sci Rep, 2013; 3:1027; doi:10.1038/srep01027 CrossRef
Ku?era O, Cifra M. Cell-to-cell signaling through light: just a ghost of chance? Cell Commun Signal, 2013; 11:87; doi:10.1186/1478811X-11-87
Li X. Using the conditioned fear stress (CFS) animal model to understand the neurobiological mechanisms and pharmacological treatment of anxiety. Shanghai Arch Psychiatry, 2012; 24:241–9; doi:10.3969/j. issn.1002-0829.2012.05.001 CrossRef
Lorenzetti FD, Baxter DA, Byrne JH. Classical conditioning analog enhanced acetylcholine responses but reduced excitability of an identified neuron. J Neurosci, 2011; 31:14789–93; doi:10.1523/ JNEUROSCI.1256-11.2011 CrossRef
Lozada-Chávez I, Janga SC, Collado-Vides J. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res, 2006; 34:3434–45; doi:10.1093/nar/gkl423 CrossRef
Lyon P. The cognitive cell: bacterial behavior reconsidered. Front Microbiol, 2015; 6:1–18; doi:10.3389/fmicb.2015.00264
Malik BR, Hodge JJL. Drosophila adult olfactory shock learning. J Vis Exp. 2014;90:e50107. doi:10.3791/50107 CrossRef
Mandli AR, Modak JM. Cybernetic modeling of adaptive prediction of environmental changes by microorganisms. Math Biosci, 2014; 248:40–5; doi:10.1016/j.mbs.2013.11.005
Maniatis SD. Classical conditioning alters short noncoding RNA expression in Drosophila. Harvard University, Cambridge, MA, 2015. CrossRef
Marx V. Cell communication: stop the microbial chatter. Nature, 2014; 511:493–7; doi:10.1038/511493a CrossRef
Mazurie A, Bonchev D, Schwikowski B, Buck GA. Evolution of metabolic network organization. BMC Syst Biol, 2010; 4:59; doi:10.1186/1752-0509-4-59 CrossRef
McAdams HH, Srinivasan B, Arkin AP. The evolution of genetic regulatory systems in bacteria. Nat Rev Genet, 2004; 5:169–78; doi:10.1038/nrg1292 CrossRef
Menzel R, Benjamin PR. Beyond the cellular alphabet of learning and memory in invertebrates. Handb Behav Neurosci, 2013; 22:3– 5; doi:10.1016/B978-0-12-415823-8.00001-0 CrossRef
Michaux C, Verneuil N, Hartke A, Giard JC. Physiological roles of small RNA molecules. Microbiology (Reading), 2014; 160:1007–19; doi:10.1099/mic.0.076208-0 CrossRef
Miller SC, Porcella SF, Raffel SJ, Schwan TG, Barboura AG. Large linear plasmids of Borrelia species that cause relapsing fever. J Bacteriol, 2013; 195:3629–39; doi:10.1128/JB.00347-13 CrossRef
Mitchell A, Pilpel Y. A mathematical model for adaptive prediction of environmental changes by microorganisms. Proc Natl Acad Sci U S A, 2011; 108:7271–6; doi:10.1073/pnas.1019754108 CrossRef
Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. Adaptive prediction of environmental changes by microorganisms. Nature, 2009; 460:220–4; doi:10.1038/nature08112 CrossRef
Montagnier L, Aïssa J, Ferris S, Montagnier JL, Lavalléee C. Electromagnetic signals are produced by aqueous nanostructures derived from bacterial DNA sequences. Interdiscip Sci Comput Life Sci, 2009; 1:81–90; doi:10.1007/s12539-009-0036-7 CrossRef
Moreira PSA, Volpato GL. Conditioning of stress in Nile tilapia. J Fish Biol, 2004; 64:961–9; doi:10.1111/j.1095-8649.2004.00362.x CrossRef
Nakagaki T, Kobayashi R, Nishiura Y, Ueda T. Obtaining multiple separate food sources: behavioural intelligence in the Physarum plasmodium. Proc R Soc B Biol Sci, 2004; 271:2305–10; doi:10.1098/ rspb.2004.2856 CrossRef
Nakagaki T, Yamada H, Tóth Á. Maze-solving by an amoeboid organism. Nature, 2000; 407:470; doi:10.1038/35035159 CrossRef
Niu B, Wang H. Bacterial colony optimization. Discret Dyn Nat Soc, 2012; 2012:1–28; doi:10.1155/2012/698057 CrossRef
Opdyke JA, Fozo EM, Hemm MR, Storz G. RNase III participates in gadY-dependent cleavage of the gadX-gadW mRNA. J Mol Biol, 2011; 406:29–43; doi:10.1016/j.jmb.2010.12.009 CrossRef
Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell, 2013; 153:426–37; doi:10.1016/j. cell.2013.03.003 CrossRef
Pinto D, Mascher T. (Actino) Bacterial “intelligence”: using comparative genomics to unravel the information processing capacities of microbes. Curr Genet, 2016; 62:487–98; doi:10.1007/s00294-0160569-3 CrossRef
Plomin R. Molecular genetics and psychology. Curr Dir Psychol Sci, 1995; 4:114–7; doi:10.1111/1467-8721.ep10772416 CrossRef
Plomin R, Spinath FM. Intelligence: genetics, genes, and genomics. J Pers Soc Psychol, 2004; 86:112–29; doi:10.1037/00223514.86.1.112 CrossRef
Richardson K. Heritability lost; intelligence found. EMBO Rep, 2012; 13:591–5; doi: 10.1038/embor.2012.83 CrossRef
Saigusa T, Tero A, Nakagaki T, Kuramoto Y. Amoebae anticipate periodic events. Phys Rev Lett, 2008; 100:018101; doi:10.1103/ PhysRevLett.100.018101 CrossRef
Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol, 2012; 11:189–200; doi:10.1016/S1474-4422(11)70286-1 CrossRef
Schmidt U, Keck ME, Buell DR. MiRNAs and other non-coding RNAs in posttraumatic stress disorder: a systematic review of clinical and animal studies. J Psychiatr Res, 2015; 65:1–8; doi:10.1016/j. jpsychires.2015.03.014 CrossRef
Schreurs BG, Burhans LB. Eyeblink classical conditioning and post-traumatic stress disorder—a model systems approach. Front Psychiatry, 2015; 6:50; doi:10.3389/fpsyt.2015.00050 CrossRef
Shors T, Weiss C, Thompson R. Stress-induced facilitation of classical conditioning. Science, 1992; 257:537–9; doi:10.1126/science.1636089 CrossRef
Soghomonyan D, Trchounian K, Trchounian A. Millimeter waves or extremely high frequency electromagnetic fields in the environment: what are their effects on bacteria? Appl Microbiol Biotechnol, 2016; 100:4761–71; doi:10.1007/s00253-016-7538-0 CrossRef
Somogyi R, Sniegoski CA. Modeling the complexity of genetic networks: understanding multigenic and pleiotropic regulation. Complexity, 1996; 1:45–63; doi:10.1002/cplx.6130010612 CrossRef
Song Z, Lin J, Li Z, Huang C. Non-coding RNA research the nuclear functions of long noncoding RNAs come into focus. Noncoding RNA Res, 2021; 6:70–9; doi:10.1016/j.ncrna.2021.03.002 CrossRef
Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, Williams P, Hüttenhofer A, Haas D, Bläsi U. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol, 2011; 80:868–85; doi:10.1111/j.1365-2958.2011.07620.x CrossRef
Tagkopoulos I, Liu YC, Tavazoie S. Predictive behavior within microbial genetic networks. Science, 2008; 320:1313–7; doi:10.1126/ science.1154456 CrossRef
Tarnita CE. The ecology and evolution of social behavior in microbes. J Exp Biol, 2017; 220:18–24; doi:10.1242/jeb.145631 CrossRef
Tessaro LWE, Murugan NJ, Persinger MA. Bacterial growth rates are influenced by cellular characteristics of individual species when immersed in electromagnetic fields. Microbiol Res, 2015; 172:26–33; doi:10.1016/j.micres.2014.12.008 CrossRef
The Desk Encyclopedia of Microbiology. Choice Rev Online, 2004; 42:42-1294-42–1294; doi:10.5860/choice.42-1294 CrossRef
Trevors JT. Bacterial evolution and metabolism. Antonie van Leeuwenhoek, 1997; 71:257–63; doi:10.1023/A:1000175217677
Trushin M V. The possible role of electromagnetic fields in bacterial communication. J Microbiol Immunol Infect, 2003; 36:153–60. CrossRef
Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A, 1985; 157:263–77; doi:10.1007/BF01350033Veit L, Pidpruzhnykova G, Nieder A. Associative learning rapidly establishes neuronal representations of upcoming behavioral choices in crows. Proc Natl Acad Sci U S A, 2015; 112:15208–13; doi:10.1073/pnas.1509760112 CrossRef
Von Bodman SB, Willey JM, Diggle SP. Cell-cell communication in bacteria: united we stand. J Bacteriol, 2008; 190:4377–91; doi:10.1128/ JB.00486-08 CrossRef
Wagner EGH, Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv Genet, 2015; 90:133– 208; doi:10.1016/bs.adgen.2015.05.001 CrossRef
Walhout AJM. Gene-centered regulatory network mapping. Methods Cell Biol, 2011; 106:271–88; doi:10.1016/B978-0-12-5441728.00010-4 CrossRef
Waters LS, Storz G. Regulatory RNAs in bacteria. Cell, 2009; 136:615–28; doi:10.1016/j.cell.2009.01.043 CrossRef
West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. The social lives of microbes. Annu Rev Ecol Evol Syst, 2007; 38:53–77; doi:10.1146/annurev.ecolsys.38.091206.095740 CrossRef
Wood GE, Shors TJ. Stress facilitates classical conditioning in males, but impairs classical conditioning in females through activational effects of ovarian hormones. Proc Natl Acad Sci U S A, 1998; 95:4066–71; doi:10.1073/pnas.95.7.4066 CrossRef
Zhan Y, Yan Y, Deng Z, Chen M, Lu W, Lu C, Shang L, Yang Z, Zhang W, Wang W, Li Y, Ke Q, Lu J, Xu Y, Zhang L, Xie Z, Cheng Q, Elmerich C, Lin M. The novel regulatory ncRNA, NfiS, optimizes nitrogen fixation via base pairing with the nitrogenase gene nifK mRNA in Pseudomonas stutzeri A1501. Proc Natl Acad Sci, 2016; 113:E4348–56; doi:10.1073/pnas.1604514113 CrossRef