INTRODUCTION
Tth DNA polymerase is a thermostable DNA polymerase enzyme derived from the thermophilic bacterium Thermus thermophilus, which exhibits dual activity as a DNA polymerase and reverse transcriptase so that it can be used in Reverse transcription-polymerase chain reaction (RT-PCR). It catalyzes the polymerization of DNA in the presence of Mg2+ ions, thereby aiding the formation of cDNA from RNA (Auer and Landre, 1995; Moreno et al., 2005; Myers and Gelfand, 1991). Tth DNA polymerase has 5?-3? exonuclease activity but does not have 3?-5? exonuclease activity. Tth DNA polymerase has also been reported to have RNAse-H-like activity that plays a role in the degradation of DNA-RNA duplexes (Auer and Landre, 1995).
Recombinant protein technology is typically used for large-scale enzyme production (Tripathi, 2016). Among the many expression systems producing recombinant protein, Escherichia coli is commonly used due to its high density on an inexpensive substrate and relatively inexpensive cultivation costs. Moreover, its genetics has been well characterized, and there is a wide availability of mutant host strains (Sriwidodo et al., 2019). Common protocols for protein production in E. coli cells include cloning of target genes in expression plasmids, transformation in the host strain, and then induction of target protein synthesis (Chae et al., 2017). Protein production in E. coli can use two approaches, extracellular and intracellular (Maksum et al., 2017; Silaban et al., 2019; Sriwidodo et al., 2017). Intracellular protein expression occurs in the cytoplasm of E. coli, while extracellular expression will direct protein expression to the culture medium by adding signal peptides. The level of protein expression intracellularly has a higher rate than extracellularly (Choi and Lee, 2004; Su et al., 2006).
Various types of enzymes were successfully produced in the host E. coli, including the recombinant Tth DNA polymerase enzyme. The recombinant Tth DNA polymerase enzyme was successfully expressed in the host E. coli (Moreno et al., 2005). There have been previous studies on the recombinant DNA polymerase Tth enzyme successfully expressed in both T. thermophilus and E. coli hosts (Dabrowski and Kur, 1998; Moreno et al., 2005). In this study, protease-deficient strains, such as BL21, were used. BL21 is an E. coli strain lacking in ompT and lon, two proteases that can interfere with the isolation of intact recombinant proteins as an approach to increase target protein expression (Maksum et al., 2019). In addition, E. coli BL21 is also able to live on media with limited nutrition. Therefore, with the use of the E. coli BL21 strain, protein recombinants can be expressed with low protease levels (Jeong et al., 2009).
Although E. coli expression systems are cost-effective and suitable for large-scale recombinant protein production, the lack of molecular chaperons and the reducing environment of the E. coli cytoplasm can lead to the formation of insoluble aggregates. Their formation is often undesirable because they generally lack biological activity (Gomes et al., 2016; Singh et al., 2015). The formation of inclusion bodies was observed in the expression of the recombinant Tth DNA polymerase enzyme in E. coli, so efforts are needed to increase the acquisition of the active enzyme in the cytoplasm (Moreno et al., 2005). To address this issue, a solubility enhancer is required to improve the yield of soluble recombinant proteins, such as maltose-binding protein (MBP). Previously, it was shown that MBP-tagged fusion protein is more abundantly expressed in the soluble fraction than His-tagged and GST-tagged fusion protein, indicating that MBP is a very efficient solubility enhancer for the soluble expression of recombinant protein (Raran-Kurussi and Waugh, 2012). For these reasons, the E. coli expression system and MBP were chosen for this work to express recombinant Tth DNA polymerase in E. coli BL21(DE3).
In this report, we evaluated the expression of recombinant Tth DNA polymerase in E. coli BL21(DE3). The Tth DNA polymerase gene was optimized using the preference codon of E. coli, then constructed on the expression plasmid. Escherichia coli BL21(DE3) was transformed with pD861-His-Tth DNA polymerase and pD861-MBP-Tth DNA polymerase plasmids. His-Tth DNA polymerase and MBP-Tth DNA polymerase were expressed as soluble forms in E. coli BL21(DE3). Therefore, it is highly expected that the production of the recombinant Tth DNA polymerase enzyme in the host E. coli BL21(DE3) is expected to be developed as a component of the RT-PCR kit for the detection of SARS-CoV-2.
MATERIALS AND METHODS
Bacterial strain, plasmids, and materials
Escherichia coli BL21(DE3) was obtained from our laboratory stock. Thermus thermophilus (Tth) DNA polymerase I gene (GeneBank accession No. AP008226.1) was inserted into the pD861-His and pD861-MBP plasmids. Plasmids pD861-His-Tth DNA Pol and pD861-MBP-Tth DNA Pol were synthesized and purchased from ATUM (Newark, CA). A bacterial protein extraction kit was purchased from Thermo-Fisher Scientific (USA). Bacto agar and tryptone were from 1’st Base (Singapore). Bovine serum albumin, kanamycin sulfate, and L-rhamnose were purchased from Sigma-Aldrich (Singapore). Bradford reagent was purchased from HiMedia (India). Calcium chloride, hydrochloric acid, tris-base, urea, and β-mercaptoethanol were from Merck (Kenilworth, NJ), and yeast extract was purchased from Oxoid (UK).
Transformation of E. coli BL21(DE3)
Escherichia coli BL21(DE3) was grown overnight in Luria Betani (LB) media at 37°C before 1% of the culture was inoculated into 50 ml LB media containing 75 μg/ml kanamycin. Cells were grown until the OD600 reached 0.4–0.5 and then placed in an ice bath for 30 minutes before centrifugation at 4,000 g for 10 minutes at 4°C. The cell pellet was resuspended in 25 ml of 0.1 M cold CaCl2 and incubated for 60 minutes before centrifugation at 4,000 g for 10 minutes at 4°C. The cell pellet was resuspended in 1 ml of 0.1 M cold CaCl2. Escherichia coli BL21(DE3) was transformed with pD861-His-Tth DNA polymerase and pD861-MBP-Tth DNA polymerase by the heat shock method. The plasmid (1 μl) was added into a microtube containing 100 μl competent cells and incubated in an ice bath for 30 seconds and then placed in a water bath at 42°C for 45 seconds before cooling in an ice bath for 2 minutes. Subsequently, 900 μl of LB media was added to the microtube and incubated at 37°C for 1 hour. A 100 μl aliquot of transformed cells was spread on LB agar supplemented with 75 μg/ml kanamycin.
Optimization of induction time
Escherichia coli BL21(DE3) cells harboring pD861-His-Tth DNA Pol and pD861-MBP-Tth DNA Pol plasmids were grown overnight in LB media at 37°C. The starter culture was then transferred into three flasks containing 20 ml LB medium supplemented with 75 μg/ml kanamycin and incubated at 37°C with shaking at 200 rpm. Cells were grown until the OD600 reached 0.4 (early log phase), 0.6 (mid-log phase), and 1.0 (late log phase). Protein expression was induced by the addition of 4 mM L-rhamnose and incubation at 37°C. Cells were harvested after 4 hours of incubation by centrifugation at 6,000 g for 20 minutes at 4°C. For each 1 g of pellet, a 4 ml bacterial protein extraction buffer containing 2 μl DNase and 2 μl lysozyme was added and the suspension was incubated for 25 minutes at room temperature before centrifugation at 15,000 g for 5 minutes at 4°C. The supernatant was collected as a soluble fraction, and 8 ml of solubilization buffer (8 M urea, 20 mM Tris-HCl, and 5 mM β-mercaptoethanol, pH 8.5) was added for each 1 g of pellet. The suspension was incubated at 100 rpm for 1 hour at room temperature and centrifuged at 12,000 g for 20 minutes at 4°C. The supernatant was collected as solubilized inclusion bodies and stored at −20°C for further analysis.
Optimization of postinduction incubation time
Escherichia coli BL21(DE3) cells harboring pD861-His-Tth DNA Pol and pD861-MBP-Tth DNA Pol plasmids were grown overnight in LB media at 37°C. The starter culture was inoculated into 50 ml LB media containing 75 μg/ml kanamycin. For His-Tth DNA polymerase, cells were grown until the OD600 reached 0.4 and 1.0 for MBP-Tth DNA polymerase, and then 4 mM L-rhamnose was added to induce protein expression at 37°C. After induction, cells were harvested at different times (4, 5, 6, 7, and 8 hours of incubation) by centrifugation at 6,000 g for 20 minutes at 4°C. Soluble protein and inclusion bodies were isolated following the previously described methods.
Overexpression of recombinant His-Tth DNA polymerase and MBP-Tth DNA polymerase under optimized conditions
Escherichia coli BL21(DE3) cells harboring pD861-His-Tth DNA Pol and pD861-MBP-Tth DNA Pol plasmids were grown overnight in LB media at 37°C. The starter culture was inoculated into a flask containing 50 ml LB media. For His-Tth DNA polymerase, cells were grown until the OD600 reached 0.4 and then induced with 4 mM L-rhamnose at 37°C. Cells were harvested after 4 hours of incubation. For MBP-Tth DNA polymerase, cells were grown until OD600 reached 1.0 and induced with 4 mM L-rhamnose at 37°C. Cells were harvested after 8 hours of incubation by centrifugation for 20 minutes at 6,000 g at 4°C. Protein extraction and inclusion bodies solubilization procedures were repeated to collect the soluble fraction and inclusion bodies.
Direct western blotting
Whole-cell extracts were fractionated by SDS-PAGE and transferred to a nitrocellulose membrane using a transfer apparatus according to the manufacturer’s protocol (Bio-Rad). The membrane was immersed in 25 ml of blocking solution [5% nonfat dry milk in Tris Buffered Saline with Tween (TBST) (50 mM Tris-HCl pH 7.5, 150 mM NaCl, and 0.1% Tween 20)] and incubated for 15 minutes at room temperature. After incubation, the membrane was washed once with 10 ml of primary antibodies (penta-His antibody QIAGEN) for 10–20 minutes. The membrane was rinsed in 50 ml of wash buffer for 3 minutes before the addition of 10 ml of the substrate and incubated for 10–30 minutes.
Total protein quantification
The total protein yield was measured by the Bradford assay. A dilution series of standards were prepared using the Bovine Serum Albumin (BSA) solution (0.2–2 mg/ml). The Bradford reagent (1 ml) was added to 100 μl of the standards and samples, and the absorbance was measured at 595 nm after 10 minutes.
RESULTS AND DISCUSSION
Construction of the recombinant plasmid
The Tth DNA polymerase synthetic gene was obtained from GeneBank (Accession No. AP008226.1), consisting of 2,502 nucleotides. Tth DNA polymerase gene shares approximately 50% similarity to the codon preference of E. coli. A high percentage of rare codons on the gene of interest will decrease recombinant protein expression (Gomes et al., 2016); therefore, the gene should be optimized to achieve a high codon adaptive index (CAI) and 40%–70% GC content (Parret et al., 2016; Silaban et al., 2019; Sriwidodo et al., 2017). The Tth DNA polymerase gene was optimized using the OPTIMIZER software and the E. coli B preference codon (Puigbò et al., 2007) to achieve a CAI of 1.000% and 60.64% GC content. The optimized Tth DNA polymerase gene was inserted into the expression plasmids pD861-His and pD861-MBP, then synthesized by ATUM. pD861-His-Tth DNA polymerase contains the N-terminal His-tag, while pD861-MBP-Tth DNA polymerase contains the N-terminal MBP tag. The His-tag was used as an affinity tag for purification purposes, and the MBP tag was used as a solubility enhancer. The plasmid map of pD861-His-Tth DNA polymerase and pD861-MBP-Tth DNA polymerase is shown in Figure 1.
 | Figure 1. Detailed map of (a) pD861-His-Tth DNA polymerase and (b) pD861-MBP-Tth DNA polymerase. Both plasmids contain the rhaBAD promoter, kanamycin selection marker, ORI pUC, and strong RBS. [Click here to view] |
 | Figure 2. Escherichia coli BL21(DE3) transformed cells harboring the (a) pD861-His-Tth DNA polymerase and (b) pD861-MBP-Tth DNA polymerase plasmids. [Click here to view] |
 | Figure 3. SDS-PAGE analysis of recombinant Tth DNA polymerase expression at different induction times: (a) His-Tth DNA polymerase and (b) MBP-Tth DNA polymerase. Lane M: protein marker; Lanes 1, 3, and 5: soluble fraction of induced cells at different induction times (0.4, 0.6, and 1.0, respectively); Lanes 2, 4, and 6: inclusion bodies of induced cells at different induction times (0.4, 0.6, and 1.0, respectively). The red and black arrows correspond to His-Tth DNA polymerase and MBP-Tth DNA polymerase, respectively. [Click here to view] |
Transformation of E. coli BL21(DE3)
The competent E. coli BL21(DE3) cells were chemically transformed using the heat shock method as it is a relatively inexpensive, simple method that does not require special equipment (Liu et al., 2014). Escherichia coli BL21(DE3) transformed cells were observed on LB agar containing kanamycin as a selection marker (Fig. 2). The plasmid contains gene encoding neomycin phosphotransferase II which is responsible for kanamycin resistance; therefore, the transformant cell harboring pD861-His-Tth DNA polymerase and pD861-MBP-Tth DNA polymerase will survive in the presence of kanamycin (Jang and Magnuson, 2013; Wegerer et al., 2008).
Optimization of the induction time
Induction time in different growth phases of E. coli plays a significant role in the production of soluble recombinant protein (Ahmad et al., 2018; Kaur et al., 2018). The results showed that His-Tth DNA polymerase and MBP-Tth DNA polymerase were expressed in soluble form when the induction was performed between the early exponential to late exponential growth phase (Fig. 3). To determine the optimal expression of the soluble fraction, semiquantitative densitometry analysis was carried out using ImageJ (Yosua et al., 2021), indicating that the highest yield of His-Tth DNA polymerase was achieved when the induction was carried out in the mid-log phase (Fig. 4a), whereas induction in the mid-log phase provided a high yield due to the rapid growth of E. coli leading to a higher rate of protein expression (Yosua et al., 2021). Meanwhile, the optimal induction of MBP-Tth DNA polymerase was in the late log phase (Fig. 4b), in line with a previous study that reported that the level of recombinant protein increased up to the late log phase (Noi and Chung, 2017).
Optimization of postinduction incubation time
To investigate the effect of postincubation time on Tth DNA polymerase expression, five different incubation times (4, 5, 6, 7, and 8 hours) were evaluated, as a previous study suggested that postinduction incubation time can affect the recombinant protein yield (Fazaeli et al., 2019). The duration of postinduction incubation time is affected by several factors such as the strength of the promoter, concentration of inducer, solubility of recombinant protein, and intrinsic properties of the recombinant protein (Noi and Chung, 2017). The different incubation times after induction resulted in different yields of soluble protein (Fig. 5), with the highest expression of the His-Tth DNA polymerase reached within 4 hours of incubation, whereas MBP-Tth DNA polymerase reached maximal expression within 8 hours of incubation (Fig. 6).
 | Figure 4. Densitometry analysis of the protein bands at different induction times (0.4, 0.6, and 1.0). [Click here to view] |
 | Figure 5. SDS-PAGE analysis of recombinant Tth DNA polymerase expression at different incubation times: (a) His-Tth DNA polymerase and (b) MBP-Tth DNA polymerase. Lane M: protein marker; Lanes 1, 2, 4, 6, and 8: soluble fraction of induced cells at different incubation times (4, 5, 6, 7, and 8 hours, respectively); Lanes 3, 5, 7, and 9: inclusion bodies of induced cells at different incubation times (5, 6, 7, and 8 hours, respectively). The red and black arrows correspond to His-Tth DNA polymerase and MBP-Tth DNA polymerase, respectively. [Click here to view] |
Expression of recombinant His-Tth DNA polymerase and MBP-Tth DNA polymerase under optimized conditions
Recombinant His-Tth DNA polymerase and MBP-Tth DNA polymerase were successfully expressed as 95 and 135 kDa protein in E. coli, respectively. The expression of recombinant His-Tth DNA polymerase and MBP-Tth DNA polymerase was conducted under optimized induction and postinduction incubation time with the soluble fractions obtained before optimization used for comparison. As shown in Figure 7, His-Tth DNA polymerase was mainly expressed in the insoluble form when MBP-Tth DNA polymerase was expressed in soluble form, with the amount of both proteins increasing after optimization in Figure 8. Also, the expression of MBP-Tth DNA polymerase produced more soluble protein than His-Tth DNA polymerase. This is in line with a previous study that reported that all MBP-tagged protein fusions yielded more soluble protein than His-tagged protein fusions (Raran-Kurussi and Waugh, 2012), as MBP serves as a “holdase” to maintain the passenger protein in a soluble form until it reaches its native conformation either by spontaneous folding or with the assistance of endogenous chaperones (Raran-Kurussi and Waugh, 2012). This mechanism may involve molecular interaction between MBP and hydrophobic amino acid residues existing on the incompletely folded passenger protein, thereby preventing aggregation (Guo et al., 2018; Raran-Kurussi and Waugh, 2012).
 | Figure 6. Densitometry analysis of protein bands at different incubation times (4, 5, 6, 7, and 8 hours). [Click here to view] |
 | Figure 7. SDS-PAGE analysis of recombinant Tth DNA polymerase expression in optimized conditions: (a) His-Tth DNA polymerase and (b) MBP-Tth DNA polymerase. Lane M: protein marker; Lane 1: soluble fraction of induced cells without optimization; Lane 2: soluble fraction of induced cells with optimization. The red and black arrows correspond to His-Tth DNA polymerase and MBP-Tth DNA polymerase, respectively. [Click here to view] |
Western blotting
The expressions of pD861-His-Tth DNA polymerase and pD861-HMBP-Tth DNA polymerase were confirmed by Western blotting using Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the internal loading control (Fig. 9).
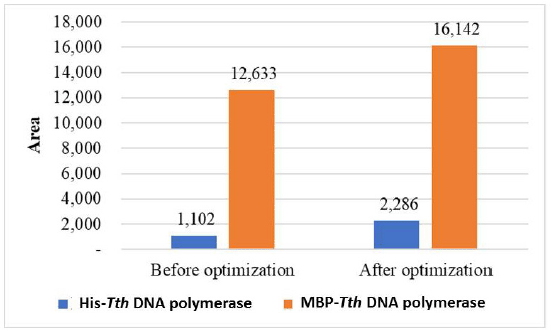 | Figure 8. Densitometry analysis of protein bands under optimized and nonoptimized conditions. [Click here to view] |
 | Figure 9. Characterization of pD861-His-Tth DNA polymerase and pD861-HMBP-Tth DNA polymerase by Western blotting: (a) the primary penta-His monoclonal antibody (band at 15 kDa) and (b) GAPDH as the internal loading control (band 38 kDa). [Click here to view] |
Total protein quantification
The total protein concentration of the His-Tth DNA polymerase soluble fraction before and after optimization was 3.7002 and 3.9095 mg/ml, respectively. Meanwhile, the total protein concentration of the MBP-Tth DNA polymerase soluble fraction before and after optimization was 30.097 and 33.541 mg/ml, respectively. These results indicate that the optimization process increased the total protein concentration. The concentration of total protein in the soluble fraction of MBP was eight times higher than the concentration of total protein in the soluble fraction of His.
CONCLUSION
In summary, soluble Tth DNA polymerase was expressed in E. coli BL21(DE3), with MBP-Tth DNA polymerase producing more soluble protein than His-Tth DNA polymerase due to its ability as a solubility enhancer.
ACKNOWLEDGMENTS
This work was supported by Grant No. 74/FI/P-KCOVID-19.2B3/IX/2020 from the National Innovation Research Agency, Ministry of Technology Research and Higher Education.
CONFLICTS OF INTEREST
The authors have nothing to disclose.
AUTHORS’ CONTRIBUTIONS
All authors made significant contributions to the work reported, in the conception and design, acquisition and interpretation of data, statistical analysis, drafting, revising, or critically reviewing the article and gave final approval of the version to be published.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Ahmad I, Nawaz N, Darwesh NM, Rahman SU, Mustafa MZ, Khan SB, Patching SG. Overcoming challenges for amplified expression of recombinant proteins using Escherichia coli. Protein Expr Purif, 2018; 144:12–8. CrossRef
Auer T, Landre PA. Properties of the 5′→3′ exonuclease/ribonuclease H activity of Thermus thermophilus DNA polymerase. Biochemistry, 1995; 34(15):4994–5002. CrossRef
Chae YK, Kim SH, Markley JL. Relationship between recombinant protein expression and host metabolome as determined by two-dimensional NMR spectroscopy. PLoS One, 2017; 12(5):1–12. CrossRef
Choi JH, Lee SY. Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl Microbiol Biotechnol, 2004; 64(5):625–35. CrossRef
Dabrowski S, Kur J. Recombinant His-tagged DNA polymerase. I. Cloning, purification and partial characterization of Thermus thermophilus recombinant DNA polymerase. Acta biochimica Polonica. 1998; 45:653–660. CrossRef
Guo Y, Yu M, Jing N, Zhang S. Production of soluble bioactive mouse leukemia inhibitory factor from Escherichia coli using MBP tag. Protein Expression and Purification. 2018; 150: 86-91. CrossRef
Fazaeli A, Golestani A, Lakzaei M, Varaei SSR, Aminian M. Expression optimization, purification, and functional characterization of cholesterol oxidase from Chromobacterium sp. DS1. PLoS One, 2019; 14(2):1–15. CrossRef
Gomes A, Byregowda S, Veeregowda B, Vinayagamurthy BA. An overview of heterologous expression host systems for the production of recombinant proteins. Adv Anim Vet Sci, 2016; 4(4):346–56. CrossRef
Jang CW, Magnuson T. A novel selection marker for efficient DNA cloning and recombineering in E. coli. PLoS One, 2013; 8(2):1–7. CrossRef
Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi SH, Couloux A, Lee SW, Yoon SH, Cattolico L, Hur CG, Park HS, Ségurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol, 2009; 394(4):644–52. CrossRef
Kaur J, Kumar A, Kaur J. Strategies for optimization of heterologous protein expression in E. coli: roadblocks and reinforcements. Int J Biol Macromol, 2018; 106:803–22. CrossRef
Liu X, Liu L, Wang Y, Wang X, Ma Y, Li Y. The study on the factors affecting transformation efficiency of E. coli competent cells. Pak J Pharm Sci, 2014; 27(3):679–84.
Maksum IP, Lestari A, Fauzia RP, Rachman DS, Soedjanaatmadja UMS. Escherichia coli BL21(DE3) expression system using TorA signal peptide for recombinant human albumin (rHA) secretion. Int J Res Pharm Sci, 2019; 10(4):3319–24. CrossRef
Maksum IP, Utama E, Sriwidodo, Subroto T. Extracellular secretion of recombinant human epidermal growth factor by using trimethylamine N-Oxide reductase a (TORA) signal peptide in escherichia coli BL21 (DE3). Journal of Pharmaceutical Sciences and Research. 2017; 9(6):1007–1016.
Moreno R, Haro A, Castellanos A, Berenguer J. High-level overproduction of his-tagged Tth DNA polymerase in Thermus thermophilus. Appl Environ Microbiol, 2005; 71(1):591–3. CrossRef
Myers TW, Gelfand DH. Reverse transcription and DNA amplification by a Thermus thermophilus DNA polymerase. Biochemistry, 1991; 30(31):7661–6. CrossRef
Noi NV, Chung YC. Optimization of expression and purification of recombinant S1 domain of the porcine epidemic diarrhea virus spike (PEDV- S1) protein in Escherichia coli. Biotechnol Biotechnol Equip, 2017; 31(3):619–29. CrossRef
Parret AH, Besir H, Meijers R. Critical reflections on synthetic gene design for recombinant protein expression. Curr Opin Struct Biol, 2016; 38:155–62. CrossRef
Puigbò P, Guzmán E, Romeu A, Garcia-Vallvé S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res, 2007; 35:126–31. CrossRef
Raran-Kurussi S, Waugh DS. The ability to enhance the solubility of its fusion partners is an intrinsic property of maltose-binding protein but their folding is either spontaneous or chaperone-mediated. PLoS One, 2012; 7(11):1–10. CrossRef
Silaban S, Gaffar S, Simorangkir M, Maksum IP, Subroto T. Construction and optimization of prethrombin-2 human genes in E. coli for the production of active thrombin. J Phys Conf Ser, 2019; 1374:012047. CrossRef
Singh A, Upadhyay V, Upadhyay AK, Singh SM, Panda AK. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb Cell Factories, 2015; 14(1):1–10. CrossRef
Sriwidodo S, Maksum IP, Riswanto N, Rostinawati T, Subroto T. Extracellular secretion recombinant of human epidermal growth factor (hEGF) using pectate lyase B (PelB) signal peptide in Escherichia coli BL21 (DE3). Int J Res Pharm Sci, 2017; 8:33–40.
Sriwidodo S, Subroto T, Maksum IP, Wathoni N, Rostinawati T, Ulya H. Optimization of secreted recombinant human epidermal growth factor production using pectate lyase B from Escherichia coli BL21(DE3) by central composite design and its production in high cell density culture. J Pharm Bioallied Sci, 2019; 11(Suppl 4):S562–6. CrossRef
Su Z, Huang Y, Zhou Q, Wu Z, Wu X, Zheng Q, Ding C, Li X. High-level expression and purification of human epidermal growth factor with SUMO fusion in Escherichia coli. Protein Peptide Lett, 2006; 13(8):785–92. CrossRef
Tripathi NK. Production and purification of recombinant proteins from Escherichia coli. ChemBioEng Rev, 2016; 3(3):116–33. CrossRef
Wegerer A, Sun T, Altenbuchner J. Optimization of an E. coli L-rhamnose-inducible expression vector: test of various genetic module combinations. BMC Biotechnol, 2008; 8:1–12. CrossRef
Yosua Y, Melati R, Sriwidodo S, Maksum IP. Expression of human epidermal growth factor using Ssp DnaB mini-intein as fusion partner in Escherichia coli BL21(DE3). J Appl Pharm Sci, 2021; 11(04):40–5. CrossRef