INTRODUCTION
Resistance to antimicrobial agents among numerous microbes (pathogens) has increased at an alarming rate around the world, posing a serious threat to human health. Because of the emergence of new resistant mechanisms and a decline in the efficacy of infectious disease therapy, microbial responses to routine treatment fail have resulted in longer sickness, higher healthcare costs, and a significant risk of mortality (Arsene et al., 2022; Rihana et al., 2021). In 2011, the WHO declared “combat drug resistance: no action today, no cure tomorrow.” Recently, certain strains of multidrug-resistant microorganisms have quadrupled worldwide (Cohen et al., 2020; Sharma et al., 2011). Antimicrobial resistance (AMR) is currently posing a greater threat to humans by increasing hospital stays, morbidity, mortality, and severe economic loss to the patient and the nation (Morales et al., 2012; Rihana et al., 2021; Rosenberger et al., 2011). Over the most recent 20 years, there was exceptional expansion in microbial diseases that the norm of overall public health in many regions of the world is currently like those of the pre-antibiotic period (Magiorakos et al., 2012). According to the standardized global definitions created by the Centre for Disease Control and Prevention (CDC) and European Centre for Disease Control, the pandrug-resistant (PDR), extensively drug-resistant (XDR), and multidrug-resistant (MDR) microbes have been fairly described (Basak et al., 2016). MDR microbes are strains which acquire resistance to a minimum of one drug in several kinds of antibiotic groups (Divya and Vijey, 2017). XDR microbes are strains which acquire resistance to a minimum of one drug in all but two or fewer antibiotic groups (i.e., these strains remain vulnerable to only one or two antibiotic groups). PDR microbes are strains resistant to all antimicrobial drugs in all antibiotic groups (Pelluri et al., 2022).
It is well implicit that microbes are one of the smartest organisms which can not only stimulate recent techniques incessantly to circumvent the immunological system and antimicrobial agents, but also become accustomed to numerous circumstances to safeguard its existence and growth (Tacconelli et al., 2018). By prospecting this, it is essential to understand their mechanism and to come up with a clear knowledge of how they do this and through which kind of function. Nevertheless, it would seem sensible to address all these attempts to certain alarming microorganisms which are resistant by ordering them according to specific conditions. To accomplish this, the WHO issued a list of drug-resistant microorganisms, which has worldwide main concern to enable a way that will guide investigators around the globe and where the necessity of acquiring innovative therapies is essential. Along with the support of expert belief and established statistics, the WHO’s microbial list of international importance established a multi-criteria decision analysis procedure for focusing on the research and development of innovative and successful antibiotic therapies. A list of the 12 most dangerous and resistant bacteria families was given with the help of researchers all around the globe based on wherever necessary new therapies are required. The three foremost bacteria were considered a critical priority, and they are Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii. The later six bacteria were considered high-ranking priority, and those are Enterococcus faecium, Staphylococcus aureus, Helicobacter pylori, Salmonella spp., Campylobacter, and Neisseria gonorrhoeae (Luepke et al., 2016). The final three bacteria were considered moderate priority, and they include Haemophilus influenzae, Staphylococcus pneumoniae, and Shigella spp (Toit et al., 2014). These microorganisms were selected based on 10 criteria, which included “mortality, healthcare and community burden, a prevalence of resistance, a 10-year trend of resistance, transmissibility, preventability in the hospital and community settings, treatability and current pipeline” (Luepke et al., 2016). The goal of this review is to highlight the current knowledge of the resistance mechanisms acquired in the Streptococcus species. This will emphasize mainly on the multidrug-resistant Streptococcus species and discuss alternatives or advancements to current antibiotic treatments for the Streptococcus species.
ABOUT STREPTOCOCCUS SPECIES
The strains of Streptococcus are microorganisms belonging to the Firmicutes phylum below the order of Lactobacillales and belonging to the Streptococcaceae family (Anon et al., 2018). Triad genera existing in the Streptococcaceae family include Streptococcus, Lactovum, and Lactococcus, and among these, Streptococcus species is extremely distinctive, comprising around 79 species (Anon et al., 2018). Few of the species of Streptococcus are infective to animals and humans, with S. pyrogens and S. pneumoniae being the extremely crucial pathogens. The strains of Streptococcus are Gram-positive microbes that mostly become visible as chains or pairs and ovoid to spherical in appearance, with a nutritionally demanding fermentative metabolism, and several of these varieties form capsules (Hayes et al., 2001).
The Streptococcus strains are usually found in the nasopharynx and oral cavity and make up a crucial portion of the normal microbiota of individuals and animals (Davis, 1996; Hayes et al., 2001). In healthy individuals, the microbiota is harmless; however, they can cause infections in specific instances, for example, immunocompromised stage (Hayes et al., 2001; Mitchell, 2003). The strains of Streptococcus species (e.g., S. pneumoniae, Streptococcus pyogenes, and S. agalactiae) can be categorized serologically by considering carbohydrates and glycoproteins present in the cell wall into various groups, like Group A to Group V (Hayes et al., 2001; Nobbs et al., 2009; Patterson, 1996). Streptococci are categorize based on structural variations, pili-associated protein with cell wall, biochemical reactions, type of hemolysis using blood agar media, and the presence of a capsule made of polysaccharide (particular for group B Streptococci) (Facklam et al., 2002). Figure 1 shows the Lancefield classification of Streptococcus species. Till now, there are more than 85 capsule antigenic types of S. pneumoniae, around 124 serotypes strains of S. pyogenes, and 9 CPS of S. agalactiae have been recommended (Boyer, 2016; Facklam et al., 2002; Zapun et al., 2008). The cell wall of Streptococci is among the highly researched bacterial cell walls (Cole et al., 2008; Facklam et al., 2002).
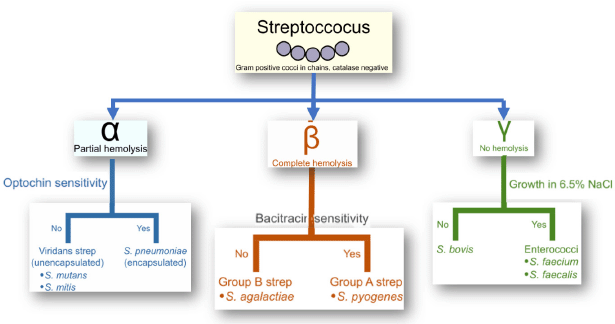 | Figure 1. Lancefield classification of the Streptococcus species. [Click here to view] |
Streptococcal genus—pathogenicity
Different species of the Streptococcus genus consist of a substantial number (more than a hundred) of bacteria residing in mucous membranes of human beings and animals. These species appear as biological flora in the buccal cavity and bowels. In supplement to this, these organisms frequently populate the upper respiratory tract, skin, and throat. However, various strains of Streptococci occur as unscrupulous pathogens, triggering infectious deceases in the host which have weak immunological reaction. Pathogenic Streptococci can be divided into three categories, which are those frequently affecting humans, commensal, and epizootic species that trigger symptoms of infection under circumstances (WHO, 2013).
Corresponding to the opinions of the WHO, nearly 1.2 million newborns die every year because of pneumonia, and the major cause of infection is S. pneumoniae, which represents 18% of infant deaths (Sharma et al., 2012). The successive major frequent cause of contaminations and global mortality are intrusive group A streptococcal (GAS) diseases. However, S. pyogenes is accountable for about 700 million infections every year, leading to the mortality of about 0.5 million people (Cunningham, 2020). Group B streptococcal (GBS) infections, S. agalactiae, develop as an equivalently critical microorganism that is accountable for miscarriages, and might as well represent a possibility of early births and neonatal infections of pneumonia type, meningitis, or sepsis (WHO, 2013). Group B Streptococcus infections are identified in about 5,000 neonates every year, with an estimated mortality of 5%. Table 1 presents the various pathogenic species of Streptococcus and their clinical manifestations of human infections.
RESISTANCE MECHANISM IN STREPTOCOCCUS SPECIES
AMR pathways are complex and several mechanisms may exist in the same strain, producing a multidrug-resistant phenotype. The following are some of the basic biochemical processes of AMR: (i) enzymatic inactivation of antibiotics, such as lactamases; (ii) changes of the antibacterial target, such as genome and RNA alterations, which hinder efficient antibiotic binding (e.g., rRNA mutations associated with resistance to several antibiotics); and (iii) limiting drug access to targets, for example, by reducing cell absorption via a decrease in outer membrane permeability in Gram-negative bacteria and/or increasing clearance from within the cell via active efflux pumps.
Various plasmids found in Streptococcus are linked to the transmission of antibiotic resistance and pathogenicity. A large number of transposons, including Tn3-family transposons, composite, and conjugative transposons, have been found in streptococci in addition to plasmids. Tn916, which encodes tetM, a ribosomal protection protein, has been linked to the independent resistance transfer between a variety of strains via plasmids, including Enterococcus faecalis, S. aureus, S. pneumoniae, S. agalactiae, and Streptococcus dysgalactiae subspecies dysgalactiae isolates, which act as reservoirs of functional antibiotic resistance genes.
Various mechanisms of AMR have already been reported in pyogenic streptococci, the major mechanisms of action and associated resistances of which will be briefly reviewed (Alves et al., 2020). The primary mechanisms of resistance in Streptococcus species are shown in Figure 2.
RESISTANCE PATTERNS IN STREPTOCOCCUS SPECIES
In this section, the most relevant Streptococcus species responsible for the resistance to selected antibiotics, like macrolides, streptogramins B tetracyclines, lincosamides, β-lactams, fluoroquinolone, and integrative and conjugative elements, are discussed.
GROUP A: In this group, the strains of Streptococcus have distinct environmental sources, and some of these species are differently tailored to a distinctive host as characterized by β-hemolytic S. pyogenes, which is believed as the utmost infective kind of Streptococcus species. Together with S. pneumoniae, it is accountable for infections like erysipelas, pharyngitis, and other invasive infections, like soft rheumatic fever, tissue infection, streptococcal toxic shock syndrome (STSS), and glomerulonephritis (Carapetis et al., 2005; Gillespie, 1998).
Resistance patterns of S. pyogenes
Despite the fact S. pyogenes has remained commonly vulnerable to β-lactams ever since the 1940s, a considerable number of ineffective treatments have been recorded worldwide (Markowitz et al., 1993). A meta-analysis on microbiological rates of ineffective treatment in S. pharyngotonsillitis was found to be around 12% during 1953–1993 (Brook et al., 2013). During the last two decades, the percentage of penicillin resistance has significantly boosted to about 40% in some regions of the world (Kebede et al., 2021). The major justifications for treatment failures with penicillin include: (i) perseverance in intracellular region of S. pyogenes because of poor diffusion of this drug into tonsillar tissues spaces, with tonsillar epithelial cells; (ii) coaggregation between S. pyogenes and M. catarrhalis, which may develop colonization of S. pyogenes through with the help of its adherence epithelial cells in human beings; (iii) protection of S. pyogenes by β-lactamase-producing microorganisms (specifically Haemophilus spp, Moraxella catarrhalis, Staphylococcus aureus, and some anaerobes) that are generally a part of the human microbiota; and (iv) changes of the commensal microbiota, which can strive for nutrients. S. pyogenes resistance to macrolides is primarily caused by Erm(B) or Mef(A). Erm(B) is the most important sign of high-level macrolide resistance, whereas Erm(A) solely reflects limited macrolide resistance. S. pyogenes resistance to macrolides varies from 2% to 19%, depending on the location. Although other investigations from 2002 to 2012 found no GAS resistance to ceftriaxone, two isolates (5.3%) of GAS in some studies had a higher MIC to this antibiotic, which could be attributable to ceftriaxone overuse (Berwal et al., 2019). The AMR patterns of S. pyogenes for various antibiotics are shown graphically in Figure 3, among which S. pyogenes attained more resistance to Clindamycin (around 50%) and less resistance to Chloramphenicol (about 7.1%) (Ishida et al., 2008).
Resistance patterns of S. pneumoniae
Among all Streptococcus species, S. pneumoniae is the utmost frequent basis for community-acquired respiratory tract infections, namely sinusitis, pneumonias, and otitis media (Mulholland, 1999). Worldwide, pneumococcal infections account for 1–2 million casualties yearly in both extremes of time (Collignon et al., 1996). Once it was believed to be a most susceptible bacteria to standard antibiotics, especially to penicillin. Nevertheless, with the identification of the initial clinically important penicillin-resistant pneumococcus (PRP) appearing in 1967, various investigations from various regions of the globe have registered an elevated occurrence of PRP infections (Lalitha et al., 2002). Presently, along with the resistant strains of S. pneumoniae to the β-lactam group of antibiotics, there is a remarkable emergence of multidrug-resistant strains (Thomas et al., 1999).
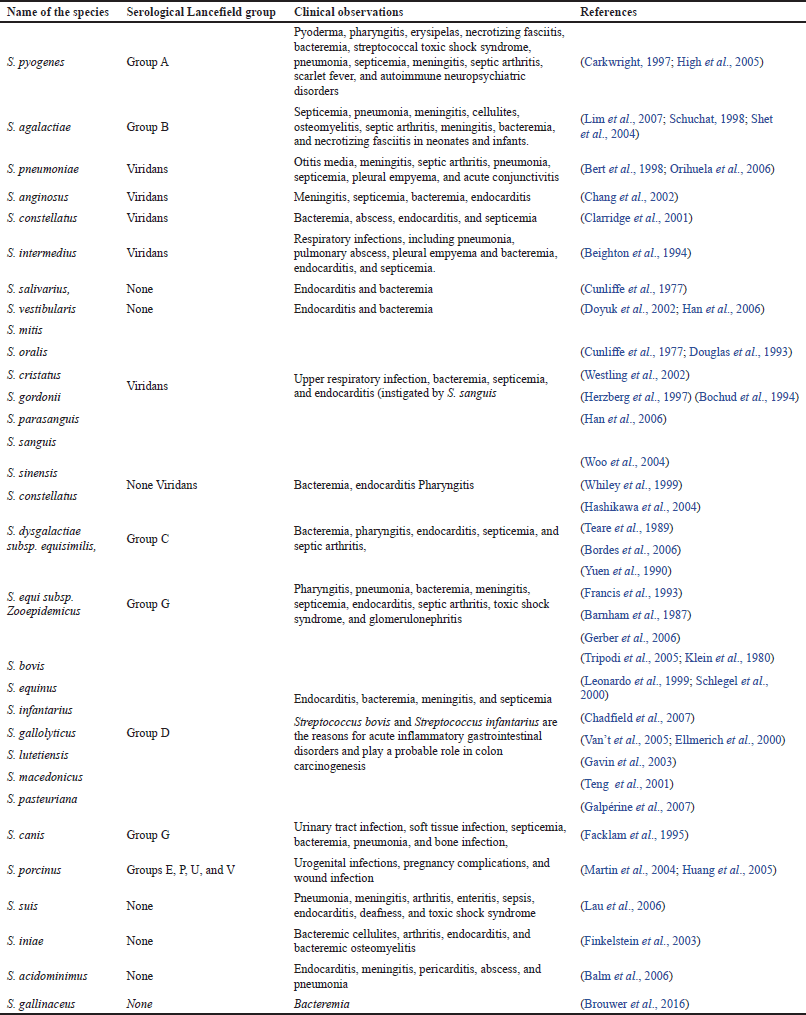 | Table 1. Pathogenic Streptococcus species and its clinical manifestations. [Click here to view] |
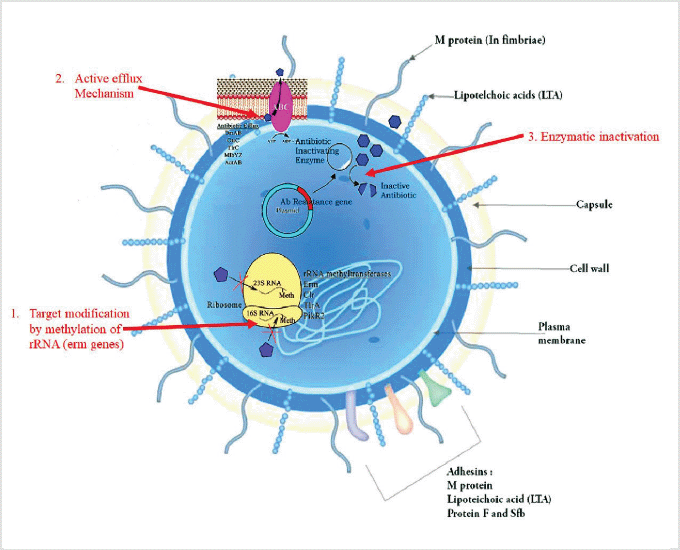 | Figure 2. Possible mechanisms of resistance in Streptococcus species. [Click here to view] |
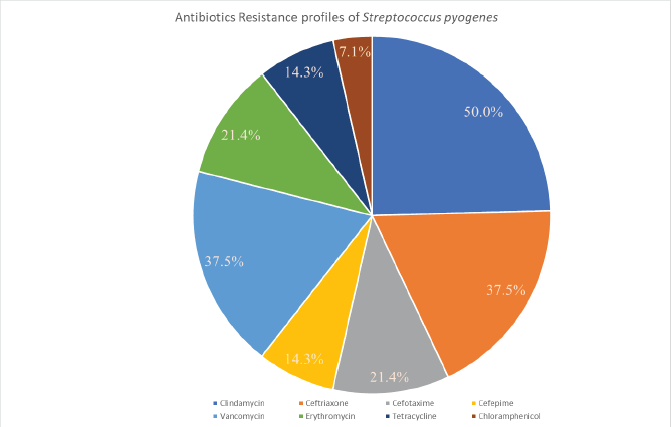 | Figure 3. Graphical representation of antibiotics resistance profiles of Streptococcus pyogenes. [Click here to view] |
Streptococcus pneumoniae can produce invasive disorders like meningitis and bacteremia in addition to respiratory infections. The prevalence of invasive pneumococcal disease (IPD) varies greatly by nation; the CDC stated in 2016 that 90% of IPD in the United States occurred in adults, but the WHO estimates that 75% of IPD occurs in children below the age of 2. Pneumococcal bacteremia is a common consequence of pneumococcal pneumonia, occurring in 25%–30% of adult cases and 12–16% of IPD in children. Since the widespread adoption of the H. influenzae type vaccination reduced Hib invasive illness, pneumococcal meningitis has become the most frequent kind of meningitis in children (Golden et al., 2019).
There are now 97 different capsular forms of S. pneumoniae having a capsule is required for S. pneumoniae to survive in the bloodstream and is linked to its potential to produce invasive illness. The presence of the capsule lowers bacterial entrapment in mucus, allowing for easier invasion and inhibiting complement activity and phagocytosis. Streptococcus pneumoniae is a highly adaptable pathogen that is prone to genetic recombination as a result of frequent environmental stressors like antibiotic usage. IPD episodes are frequently transitory, with fast and direct therapy; as a result, the organism has little opportunity to adjust to antimicrobial pressure. Pneumococcal carriage, on the other hand, is typically long-term. Carriage serotypes are exposed to extended antimicrobial pressure as a result of their long residence of the nasopharynx, which can lead to the selection of antimicrobial-resistant strains. The frequency of serotype isolation from the nasopharynx, the duration of carriage, and the possibility of antibiotic resistance have all been linked in studies (Straume et al., 2015).
Considering India, there are just a small number of findings which demonstrates the resistance pattern in S. pneumoniae. Kanungo et al.’s (2001) investigations to recognize the resistance patterns of S. pneumoniae strains have observed the upsurgence of intermediary sensitivity from Christian Medical College, Vellore, South India. However, research conducted by Goyal et al. (2007) in North India indicated 2.3% resistance. A similar investigation from South India has described low-altitude resistance, even though they did not find any strain exhibiting absolute resistance (Farley et al., 2001). However, one more cooperative research from 8 Asian nations together with India has shown 35.1% of the total resistance in strains of S. pneumoniae (Chawla et al., 2010). Table 2 presents the antibiotic resistance patterns of S. pneumoniae from various studies conducted at different geographical location over the time.
GROUP B: Group B Streptococcus, also known as β-hemolytic S. agalactiae, is a pathogenic bacterium that causes infections such as pneumonia, meningitis, and sepsis in pregnant women and newborns (Johri et al., 2006); more recently, the infective status of these strains in immunocompromised and elderly patients has been re-evaluated (Le Doare et al., 2017).
Resistance patterns of S. agalactiae
Two sets of antimicrobial drugs, aminopenicillins and penicillin, are suggested as first choice of treatment against Group B Streptococcus infections; whereas Lincosamide (Clindamycin) and macrolides (Erythromycin) correspond to the second choice of antibiotics which are commonly recommended for the patients who are allergic to β-lactams antibiotics. Penicillin G (PEN) is the medicine of first option and is widely utilized in the dealing and stoppage of Group B Streptococcus infections, like intrapartum antimicrobial prophylaxis in pregnant women to avoid early inception of Group B Streptococcus infections (Orand, 2012). β-lactams and penicillin G are also the most used drug of choice in households, agricultural animals, and aquaculture for preventive or infection management purposes (Simoni et al., 2018). Accordingly, penicillin nonsusceptibility (PEN-NS) is a significant burden and could involve alternatives in medication guidelines. Group B Streptococcus-associated multidrug resistance, including fluoroquinolone resistance, was identified owing to the outflow mutations or mechanisms in the quinolone resistance determining genes. DNA gyrase (gyrA/gyrB) and topoisomerase IV (parC/parE) are two types of II topoisomerase enzymes (Wessman, 1986).
GROUP D: The strains of Streptococcus in Group D were categorized in two varying types in the early 1980s: i.e., Streptococcus faecium and Streptococcus faecalis. Later, they were retitled as E. faecium and Enterococcus faecalis. Ever after that, various new strains have been listed in the Enterococcus genus (Facklam et al., 1995).
GROUP E: According to Lancefield’s classification, the strains of Group E Streptococcus, namely S. porcinus, is normally associated with pneumonia, sepsis, lymphadenitis, and endocarditis in swine. Around 87 types of infections caused by S. porcinus have also reported in human beings also (Le Bouguenec et al., 1990).
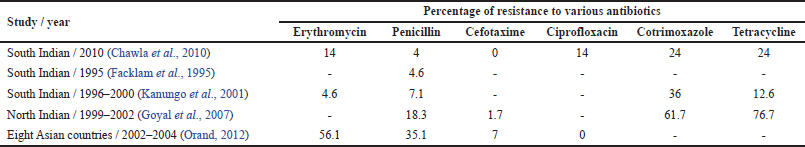 | Table 2. Resistance patterns of S. pneumoniae. [Click here to view] |
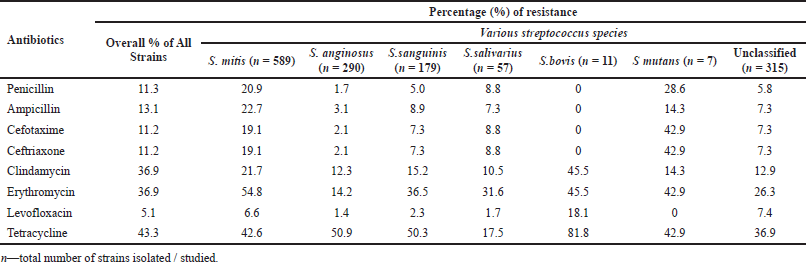 | Table 3. Resistance patterns shown by the viridans group Streptococci (Angeletti et al., 2015; Chun et al., 2016; Woo et al., 2004). [Click here to view] |
Resistance patterns in S. porcinus:
An indication which strengthens the role of S. porcinus as a recently evolving pathogenic microbe is correlated with the reality that this pathogen has only been inaccessible from mankind in the last two decades. The elevated incidence of Tetracycline resistance among S. porcinus bacteria may be associated with the existence of conventional genetic factors in chromosomal DNA of these bacteria, for example, transposons Tn3701 and Tn916, which code for resistance to Tetracycline are extensively spread among other members of the Streptococcus genus (Jonsson et al., 1991).
GROUPS C AND G: As per Lancefield’s classification, Streptococcus dysgalactiae subsp. dysgalactiae goes to Group C and G Streptococcus and shows a key position in mastitis (Rojo et al., 2021). Based on unusual instances, this strain was reported in humans and in fish necrotic tissues, as being accountable for several infections. The strain S. dysgalactiae subsp. Equisimilis from the same group is a β-hemolytic microbe responsible for STSS and sepsis infections in humans.
Resistance patterns S. dysgalactiae
Streptococcus dysgalactiae caused diseases in older people who had malignancy or diabetes. The percentage of bacteremia and the number of deaths were slightly higher. Various M-protein genes in the emm or emm-ST sequences were associated with either bacteremia or both Lincosamide and macrolide resistance. Resistance levels to the groups of this group and tetracycline were analogous, but the dissemination of antibiotic resistance strains differed, representing distinct resistance mechanisms. Consequently, there were variations in the epidemiological results, clinical data, and antibiotic resistance genotypes between different regions (Goyette et al., 2014).
GROUP R: The Streptococcus suis belonging to Group R is accountable for STSS and meningitis. It is a perfectly zoonotic strain, and in fact, human beings nearby swine and/or consuming swine derived food are a cause for Streptococcus suis infections (Segura et al., 2016). The antimicrobial resistant patterns of various species of viridans groups of Streptococci group are presented in Table 3, in which the first column lists the various antibiotics, whereas the second column shows the overall resistance percentage of all the strains together. This table clearly indicates that these viridans groups got high resistance to antimicrobials like tetracycline, clindamycin, and erythromycin in comparison with other antibiotics (Chun et al., 2015).
CONCLUSION
Undiscriminating usage of antimicrobial substances at an unsuitable dosage might be the probable cause of resistance. Consequently, there should be restrictions for the indiscrete use of antimicrobial agents to reduce the development of resistant microorganisms. The development of drug-resistant species and the MDR strains of Streptococcus species require uninterrupted national and international monitoring of susceptibility, to develop the best line of treatment. Antimicrobial drug resistance among Streptococcus species arising from earlier sensitive inhabitants resulted in parallel gene transfer or point mutations in chromosomes due to the unnecessary use of antibiotics. The strains of Streptococcus were recognized as producers of biofilm. The intensified resistance to antibiotics by biofilms among Streptococcus species promotes frequent infections, which comprise roughly 80% of microbial diseases in people. The antibiotic resistance in Streptococcus species has become the major problem of worry that is categorized and prioritized by the WHO.
These statistics indicate the significance of clinical studies in various geographical regions before recommending certain antimicrobial drugs to various infections to minimize the resistant strains among Streptococcus species. An entirely distinct problem is how to slow down the resistance of antimicrobial drugs by these Streptococcus species among adolescent children and older persons in long-term care facilities. Answers may include decreasing antibiotic utilization, which is the most important driver of recently gained resistance. Constant observation to measure streptococcal resistance is additionally required to identify the occurrence of new strains showing high-level penicillin resistance and more drug resistance. Furthermore, everyone should clearly recognize the scientific importance and influence of antimicrobial drug resistance on streptococcal infections as there is no constantly clear relationship between resistance and medication failure.
Attempts to diminish antibiotic utilization should be promoted by knowledge-sharing programs and healthcare guidelines for professionals. The most excellent approach to prevent streptococcal infection is by the application of conjugate vaccinations. Also, it is essential to examine the development of streptococcal infections and resistance, concentrating on serotype replacement. Research aiming at the improvement of new vaccine designs must be adopted to avoid emerging resistant strains.
ACKNOWLEDGMENTS
The authors thank JSS College of Pharmacy, Ooty, and JSS Academy of Higher Education & Research, Mysuru, for providing all the necessary facilities and support to write this article. They also thank the physicians/consultants of the Govt. Medical College & Hospital, Ooty, for providing the necessary input and corrections for this manuscript.
AUTHORS’ CONTRIBUTIONS
All authors contributed significantly to the conception and design, data acquisition, and data analysis and interpretation; participated in the drafting of the article or critically revised it for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. According to the requirements/guidelines of the International Committee of Medical Journal Editors, all authors are entitled to be authors.
FUNDING
They are thankful to the Department of Science and Technology (New Delhi) for providing financial support (DST/ WOS-B/HN-5/2021).
CONFLICTS OF INTEREST
There are no financial or other conflicts of interest reported by the authors in this review work.
ETHICAL APPROVAL
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Alves-Barroco C, Rivas-García L, Fernandes AR, Baptista PV. Tackling multidrug resistance in Streptococci–From novel biotherapeutic strategies to nanomedicines. Front Microbiol, 2020; 11:579916. CrossRef
Angeletti S, Dicuonzo G, Avola A, Crea F, Dedej E, Vailati F, Farina C, De Florio L. Viridans Group Streptococci clinical isolates: MALDI-TOF mass spectrometry versus gene sequence-based identification. PLoS One, 2015; 10(3):e0120502. CrossRef
Anon, Flesh-eating bacteria thrive on pain. Nature, 2018; 557(7705):283. CrossRef
Arsene MMJ, Jorelle ABJ, Sarra S, Viktorovna PI, Davares AKL, Ingrid NKC, Steve AAF, Andreevna SL, Yashina NV, Carime BZ. Short review on the potential alternatives to antibiotics in the era of antibiotic resistance. J Appl Pharm Sci, 2022; 12(01):029–40.
Balm MND, Truong HT, Choudhary AS, Robinson GM, Blackmore TK. Streptococcus gallinaceus bacteraemia in an abattoir worker presenting with a febrile illness. J Medi Microbiol, 2006; 55(7), 957–9. CrossRef
Barnham M, Cole G, Efstratiou A, Tagg JR, Skjold SA. Characterization of Streptococcus zooepidemicus (Lancefield group C) from human and selected animal infections. Epidemiol Infect, 1987; 98(2):171–82. CrossRef
Basak S, Singh P, Rajurkar M. Multidrug resistant and extensively drug resistant bacteria: a study. J Pathog, 2016; 2016:4065603. CrossRef
Beighton D, Carr AD, Oppenheim BA. Identification of viridans streptococci associated with bacteraemia in neutropenic cancer patients. J Medi Microbiol, 1994; 40(3):202–4. CrossRef
Bert F, Lancelin MB, Zechovsky NL. Clinical significance of bacteremia involving the “Streptococcus milleri” group: 51 cases and review. Clin Infec Dis, 1998; 27(2):385–7. CrossRef
Berwal A, Chawla K, Shetty S, Gupta A. Trend of antibiotic susceptibility of Streptococcus pyogenes isolated from respiratory tract infections in tertiary care hospital in south Karnataka. Iran J Microbiol. 2019; 11(1):13–8. CrossRef
Bochud PY, Calandra T, Francioli P. Bacteremia due to viridans streptococci in neutropenic patients: a review. Am J Med, 1994; 97(3):256–64. CrossRef
Bordes-Benitez A, Sanchez-Onoro M, Suárez-Bordón P, García-Rojas AJ, Saéz-Nieto JA, González-García A, Bolaños-Rivero M. Outbreak of Streptococcus equi subsp. zooepidemicus infections on the island of Gran Canaria associated with the consumption of inadequately pasteurized cheese. Eur J Clin Microbiol Infect Dis, 2006; 25(4):242–6. CrossRef
Boyer K. National Institute of Environmental Health Sciences (NIEHS). Encyclopedia of Global Health [Internet]. SAGE Publications, Inc., 2008. doi:10.4135/9781412963855.n833
Brook I. Penicillin failure in the treatment of Streptococcal pharyngo-tonsillitis. Curr Infect Dis Rep, 2013; 15(3):232–5. CrossRef
Brouwer S, Barnett TC, Rivera-Hernandez T, Rohde M, Walker MJ. Streptococcus pyogenes adhesion and colonization. FEBS Lett, 2016; 590(21):3739–57. CrossRef
Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis, 2005; 5(11):685–94. CrossRef
Chadfield MS, Christensen JP, Decostere A, Christensen H, Bisgaard M. Geno-and phenotypic diversity of avian isolates of Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis) and associated diagnostic problems. J Clin Microbiol, 2007; 45(3):822–7. CrossRef
Chang W, Wu J, Huang C, Tsai Y, Chien C, Lu C. Identification of viridans streptococcal species causing bacterial meningitis in adults in Taiwan. Eur J Clin Microbiol Infect Dis, 2002; 21(5):393–6. CrossRef
Chawla K, Gurung B, Mukhopadhyay C, Bairy I. Reporting emerging resistance of Streptococcus pneumoniae from India. J Glob Infect Dis, 2010; 2(1):10. CrossRef
Chun S, Huh HJ, Lee NY. Species-specific difference in antimicrobial susceptibility among viridans group streptococci. Ann Lab Med, 2015; 35(2):205–11. CrossRef
Clarridge JE, Attorri S, Musher DM, Hebert J, Dunbar S. Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus (“Streptococcus milleri Group”) are of different clinical importance and are not equally associated with abscess. Clin Infect Dis, 2001; 32(10):1511–5. CrossRef
Cohen ML. Changing patterns of infectious disease. Nature, 2000; 406(6797):762–7. CrossRef
Cole JN, Henningham A, Gillen CM, Ramachandran V, Walker MJ. Human pathogenic streptococcal proteomics and vaccine development. Proteomics—Clin App, 2008; 2(3):387–410. CrossRef
Collignon PJ, Bell JM. Australian Group on Antimicrobial Resistance (AGAR). Drug-resistant Streptococcus pneumoniae: the beginning of the end for many antibiotics? Med J Aust, 1996; 164(2), 64-67. CrossRef
Cunliffe N. Jacob A. Bacteraemia. J Infect, 1997; 34(1):85.
Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev, 2000; 13(3):470–511. CrossRef
Davis CP. Normal flora. Medical Microbiology, 4th edition, University of Texas Medical Branch, Galveston, TX, 1996.
Divya MJ, Vijey AM. An overview on antibiotic use and resistance. Res J Pharm Tech, 2017; 10(8):2793–6. CrossRef
Douglas CWI, Heath J, Hampton KK, Preston FE. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol, 1993; 39(3):179–82. CrossRef
Doyuk E, Ormerod OJ, Bowler I. Native valve endocarditis due to Streptococcus vestibularis and Streptococcus oralis. J Infect, 2002; 45(1):39–41. CrossRef
Ellmerich S, Scholler M, Duranton B, Gosse F, Galluser M, Klein JP, Raul F. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis, 2000; 21(4):753–6. CrossRef
Facklam R, Elliott J, Pigott N, Franklin AR. Identification of Streptococcus porcinus from human sources. J Clin Microbiol, 1995; 33(2):385–8. CrossRef
Facklam RF, Martin DR, Marguerite L, Dwight RJ, Efstratiou A, Thompson T, Gowan S, Kriz P, Tyrrell GJ, Kaplan E, Beall B. Extension of the lancefield classification for group A Streptococci by addition of 22 new m protein gene sequence types from clinical isolates: emm103 to emm124. Clin Infect Dis, 2002; 34(1);28–38. CrossRef
Farley MM, Strasbaugh LJ. Group B streptococcal disease in nonpregnant adults. Clin Infect Dis, 2001; 33(4):556–61. CrossRef
Finkelstein Y, Marcus N, Mosseri R, Bar-Sever Z, Garty BZ. Streptococcus acidominimus infection in a child causing Gradenigo syndrome. Int J Pedi Otorhinolaryngol, 2003; 67(7):815–7. CrossRef
Francis AJ, Nimmo GR, Efstratiou A, Galanis V, Nuttall N. Investigation of milk-borne Streptococcus zooepidemicus infection associated with glomerulonephritis in Australia. J Infect, 1993; 27(3):317–23. CrossRef
Galpérine T, Cazorla C, Blanchard E, Boineau F, Ragnaud JM, Neau D. Streptococcus canis infections in humans: retrospective study of 54 patients. J Infect, 2007; 55(1):23–6. CrossRef
Gavin PJ, Thomson RB, Horng SJ, Yogev R. Neonatal sepsis caused by Streptococcus bovis variant (Biotype II/2): report of a case and review. J Clin Microbiol, 2003; 41(7):3433–5. CrossRef
Gerber JS, Glas M, Frank G, Shah SS. Streptococcus bovis Infection in Young Infants. Ped Infect Dis J, 2006; 25(11):1069–73. CrossRef
Gillespie S. Failure of penicillin in Streptococcus pyogenes pharyngeal infection. Lancet, 1998; 352(9145):1954–6. CrossRef
Golden AR, Baxter MR, Davidson RJ, Martin I, Demczuk W, Mulvey MR, Karlowsky JA, Hoban DJ, Zhanel GG, Adam HJ. Comparison of antimicrobial resistance patterns in Streptococcus pneumoniae from respiratory and blood cultures in Canadian hospitals from 2007–16. J Antimicro Chemo, 2019; 1;74(Supplement_4):iv39–47. CrossRef
Goyal R, Singh NP, Kaur M, Talwar V. Antimicrobial resistance in invasive and colonising Streptococcus pneumoniae in North India. Ind J Med Microbiol, 2007; 25(3):256–9. CrossRef
Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect, 2014; 3(1):1–20. CrossRef
Han XY, Kamana M, Rolston KVI, Viridans Streptococci isolated by culture from blood of cancer patients: clinical and microbiologic analysis of 50 cases. J Clin Microbiol, 2006; 44(1):160–5. CrossRef
Hashikawa S, Iinuma Y, Furushita M, Ohkura T, Nada T, Torii K, Hasegawa T, Ohta M. Characterization of group C and G streptococcal strains that cause streptococcal toxic shock syndrome. J Clin Microbiol, 2004; 42(1):186–92. CrossRef
Hayes CS, Williamson Jr HA. Management of group A beta-hemolytic streptococcal pharyngitis. Am Fam Phys, 2001; 63(8):1557.
Herzberg MC, Meyer MW, Kiliç A, Tao L. Host-pathogen interactions in bacterial endocarditis: streptococcal virulence in the host. Adv Dent Rese, 1997; 11(1):69–74. CrossRef
High KP, Edwards MS, Baker CJ. Group B streptococcal infections in elderly adults. Clin Infect Dis, 2005; 41(6):839–47. CrossRef
Huang YT. Teng LJ. Ho SW. Hsueh PR. Streptococcus suis infection. J Microbiol Imm Infect. 2005; 38(5):306–13.
Ishida T, Maniwa K, Kagioka H, Hirabayashi M, Tomioka H, Hayashi M, Tomii K, Gohma I, Ito Y, Hirai T, Ito I, Mishima M. Antimicrobial susceptibilities of Streptococcus pneumoniae isolated from adult patients with community-acquired pneumonia in Japan. Respirology, 2008; 13(2):240–6. CrossRef
Johri AK, Paoletti LC, Glaser P, Dua M, Sharma PK, Grandi G, Rappuoli R. Group B Streptococcus: global incidence and vaccine development. Nat Rev Microbiol, 2006; 4(12):932–42. CrossRef
Jonsson P, Olsson SO, Olofson AS, Fälth C, Holmberg O, Funke H. Bacteriological investigations of clinical mastitis in heifers in Sweden. J Dairy Res, 1991; 58(2):179–85. CrossRef
Kanungo R, Rajalakshmi B. Serotype distribution & antimicrobial resistance in Streptococcus pneumoniae causing invasive & other infections in south India. Ind J Med Res, 2001; 114:127.
Kebede D, Admas A, Mekonnen D. Prevalence and antibiotics susceptibility profiles of Streptococcus pyogenes among pediatric patients with acute pharyngitis at Felege Hiwot Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Microbiol, 2021; 21(1):1–10. CrossRef
Klein RS, Catalano MT, Edberg SC, Casey JI. Streptococcus equinus septicemia: report of two cases and review of the literature. Am J Med Sci, 1980; 279(2):99–103. CrossRef
Lalitha M, Pai R, Manoharan A. Multidrug-resistant Streptococcus pneumoniae from India. Lancet, 2002; 359(9304):445. CrossRef
Lau SKP, Woo PCY, Luk W, Fung AMY, Hui WT, Fong AH, Chow CW, Wong SS, Yuen KY. Clinical isolates of Streptococcus iniae from Asia are more mucoid and β-hemolytic than those from North America. Diag Microbiol Infect Dis, 2006; 54(3):177–81. CrossRef
Le Bouguenec C, De Cespedes G, Horaud T. Presence of chromosomal elements resembling the composite structure Tn3701 in streptococci. J Bacteriol, 1990; 172(2):727–34. CrossRef
Le Doare K, O’Driscoll M, Turner K, Seedat F, Russell NJ, Seale AC, Heath PT, Lawn JE, Baker CJ, Bartlett L, Cutland C, Gravett MG, Ip M, Madhi SA, Rubens CE, Saha SK, Schrag S, Sobanjo-Ter Meulen A, Vekemans J, Kampmann B; GBS Intrapartum Antibiotic Investigator Group. Intrapartum antibiotic chemoprophylaxis policies for the prevention of group B streptococcal disease worldwide: systematic review. Clin Infect Dis, 2017; 65(suppl_2):S143–51. CrossRef
Leonardo A, Sechi RC. Streptococcus equinus endocarditis in a patient with pulmonary histiocytosis X. Scand J Infect Dis, 1999; 31(6), 598–600. CrossRef
Lim LH, Lee WS, Parasakthi N. Childhood invasive pneumococcal disease: A hospital-based study from Malaysia. J Pead Child Health, 2007; 43(5):366–9. CrossRef
Luepke KH, Suda KJ, Boucher H, Russo RL, Bonney MW, Hunt TD, Mohr JF. Past, present, and future of antibacterial economics: increasing bacterial resistance, limited antibiotic pipeline, and societal implications. Pharmacotherapy: J Hum Pharmacal Drug Ther, 2016; 37(1), 71–84. CrossRef
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect, 2012; 18(3):268–81. CrossRef
Markowitz M, Gerber MA, Kaplan EL. Treatment of streptococcal pharyngotonsillitis: reports of penicillin’s demise are premature. J Pediatr, 1993; 123(5):679–85. CrossRef
Martin C, Fermeau V, Eyraud JL, Aubard Y. Streptococcus porcinus as a Cause of spontaneous preterm human stillbirth. J Clin Microbiol, 2004; 42(9):4396–8.
Mitchell TJ. The pathogenesis of streptococcal infections: from Tooth decay to meningitis. Nature Rev Microbiol, 2003; 1(3):219–30. CrossRef
Morales E, Cots F, Sala M, Comas M, Belvis F, Riu M, Castells X. Hospital costs of nosocomial multi-drug resistant Pseudomonas aeruginosa acquisition. BMC Health Ser Res, 2012; 12(1):122. CrossRef
Mulholland K. Strategies for the control of pneumococcal diseases. Vaccine, 1999; 17:S79–84. CrossRef
Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev, 2009; 73(3):407–50. CrossRef
Orand JP. Antimicrobial resistance and the standards of the World Organisation for Animal Health. Rev Scient Tech (Int Off Epizootics), 2012; 31(1):335–42. CrossRef
Orihuela CJ, Tuomanen EI. Models of pneumococcal disease. Drug Disc Today: Dis Models, 2006; 3(1):69–75. CrossRef
Patterson In: Baron S (ed.). Medical microbiology. 4th edition, University of Texas Medical Branch at Galveston, Galveston, TX, Chapter 13, 1996. CrossRef
Pelluri R, Monika P, Paritala H, Annapareddy CR, Kotha B, Meenavilli S, Angadi SR, Rayapati G, Puttagunta S. Antibiotics susceptibility pattern and prevalence of isolated uropathogens in inpatient and out patients with lower urinary tract infections. J Appl Pharm Sci, 2022; 12(01):159–64.
Rihana BP, Wadhwani A, Balasubramaniam V, Ponnusankar S, Need for the implementation of antibiotic policy in India: An Overview. Int J Cur Res Rev, 2021; 13(05):168–78. CrossRef
Rojo-Bezares B, Toca L, Azcona-Gutiérrez JM, Ortega-Unanue N, Toledano P, Sáenz Y. Streptococcus dysgalactiae subsp. equisimilis from invasive and non-invasive infections in Spain: combining epidemiology, molecular characterization, and genetic diversity. Eur J Clin Microbiol Infect Dis, 2021; 40(5):1013–21. CrossRef
Rosenberger LH, Hranjec T, Politano AD, Swenson BR, Metzger R, Bonatti H, Sawyer RG. Effective Cohorting and “Superisolation” in a single intensive care unit in response to an outbreak of diverse multi-drug-resistant organisms. Surg Infect, 2011; 12(5):345–50. CrossRef
Schlegel L, Grimont F, Collins MD, Regnault B, Grimont PA, Bouvet A. Streptococcus infantarius sp. nov., Streptococcus infantarius subsp. infantarius subsp. nov. and Streptococcus infantarius subsp. coli subsp. nov., isolated from humans and food. Int J Syst Evol Microbiol, 2000; 50(4):1425–34. CrossRef
Schuchat A. Epidemiology of Group B Streptococcal Disease in the United States: Shifting Paradigms. Clin Microbiol Rev, 1998; 11(3):497–513. CrossRef
Segura M, Calzas C, Grenier D, Gottschalk M. Initial steps of the pathogenesis of the infection caused by Streptococcus suis: fighting against nonspecific defenses. FEBS Lett, 2016; 590(21):3772–99. CrossRef
Sharma A. Antimicrobial resistance: no action today, no cure tomorrow. Ind J Med Microbiol, 2011; 29(2):91–2. CrossRef
Sharma A, Arya DK, Sagar V, Bergmann R, Chhatwal GS, Johri AK. Identification of potential universal vaccine candidates against group A Streptococcus by using high throughput in silico and proteomics approach. J Proteomic Res, 2012; 12(1):336–46. CrossRef
Shet A, Ferrieri P, Neonatal & maternal group B streptococcal infections: a comprehensive review. Ind J Med Res, 2004; 120:141–50.
Simoni S, Vincenzi C, Brenciani A, Morroni G, Bagnarelli P, Giovanetti E, Varaldo PE, Mingoia M. Molecular characterization of Italian isolates of fluoroquinolone-resistant Streptococcus agalactiae and relationships with chloramphenicol resistance. Micro Drug Resist, 2018; 24(3):225–31. CrossRef
Straume D, Stamsås GA, Håvarstein LS. Natural transformation and genome evolution in Streptococcus pneumoniae. Infect Genet Evol 2015; 33: 371–80. CrossRef
Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N; WHO Pathogens Priority List Working Group. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis, 2018; 18(3):318–27. CrossRef
Teare EL, Smithson RD, Efstratiou A, Devenish WR, Noah ND. An outbreak of puerperal fever caused by group C streptococci. J Hosp Infect, 1989; 13(4):337–47. CrossRef
Teng LJ, Hsueh PR, Ho SW, Luh KT. High Prevalence of Inducible Erythromycin Resistance among Streptococcus bovis Isolates in Taiwan. Antimicrob Agents Chemother, 2001; 45(12):3362–5. CrossRef
Thomas K, Group IBISI, Network ICE. Prospective multicentre hospital surveillance of Streptococcus pneumoniae disease in India. Lancet, 1999; 353(9160):1216–21. CrossRef
Toit M du, Huch M, Cho GS, Franz CMAP. The family Streptococcaceae. Lactic Acid Bact, 2014; 445–6. CrossRef
Tripodi MF, Fortunato R, Utili R, Triassi M, Zarrilli R. Molecular epidemiology of Streptococcus bovis causing endocarditis and bacteraemia in Italian patients. Clin Microbiol Infect, 2005; 11(10):814–9. CrossRef
Van’t Wout JW, Bijlmer HA. Bacteremia Due to Streptococcus gallolyticus, or the Perils of Revised Nomenclature in Bacteriology. Clin Infect Dis, 2005; 40(7):1070–1. CrossRef
Wessman GE. Biology of the group E streptococci: a review. Vet Microbiol, 1986; 12(4):297–328. CrossRef
Westling K. Ljungman P. Thalme A. Julander I. Streptococcus viridans Septicaemia: a comparison study in patients admitted to the Departments of Infectious Diseases and Haematology in a University Hospital. Scandinavian J Infect Dis, 2002; 34(4):316–9. CrossRef
Whiley RA, Hall LMC, Hardie JM, Beighton D. A study of small-colony, β-haemolytic, Lancefield group C streptococci within the anginosus group: description of Streptococcus constellatus subsp. pharyngis subsp. nov., associated with the human throat and pharyngitis. Int J Sys Evol Microbiol, 1999; 49(4):1443–9. CrossRef
Woo PC, Teng JL, Leung K, Lau SK, Tse H, Wong BH, Yuen K. Streptococcus sinensis may react with Lancefield group F antiserum. J Med Microbiol, 2004; 53(11):1083–8. CrossRef
World Health Organization. Weekly Epidemiological Record. Relive épidémiologique hebdomadaire, 2013; 88(31):321–36.
Yuen KY, Seto WH, Choi CH, Ng W, Ho SW, Chau PY. Streptococcus zooepidemicus (Lancefield group C) septicaemia in Hong Kong. J Infect, 1990; 21(3):241–50. CrossRef
Zapun A. Vernet T. Pinho MG. The different shapes of cocci. FEMS Microbiol Rev, 2008; 32(2):345–60. CrossRef