INTRODUCTION
The need for animal protein in Indonesia has increased every year. One of the affordable sources of animal protein for all levels of society is poultry. The area with the largest broiler in Indonesia is West Java, namely in the districts of Bogor and Sukabumi (Hardiati et al., 2021; Livestock and Animal Health Statistics, 2020). Broilers have the advantage of having a high level of productivity, but the disadvantage is that they have a low level of disease immunity. Chickens’ immunity can be boosted in a variety of methods, including the administration of vitamins, immunizations, and antibiotics (Alagawan et al., 2020; Dibner and Richards, 2005). Antibiotics such as β-lactams, enramycin, bacitracin, oxytetracycline (OT), streptomycin, and chlortetracycline were often used as growth promoters before being banned by the Indonesian government (Department of Nutrition Science and Feed Technology, 2018).
The systematic and excessive use of antibiotics in the poultry farming sector, without regard for the guidelines for usage, has a negative impact. The growth of antibiotic resistance is one of the effects. Antibiotic use generates stress, which can trigger resistance in DNA, resulting in bacterial cell mutations and genetic alterations (Huddleston, 2014). According to the World Health Organization (WHO, 2014), microorganisms resistant to antibiotics cause infection in animals, resulting in higher rates of morbidity and mortality. Both harmful bacteria and normal flora can develop antibiotic resistance. Klebsiella pneumoniae is one of the bacteria that is at risk of antibiotic resistance.
Bacterium K. pneumoniae is Gram-negative and is found in the skin, mouth, and intestines as part of the normal flora. Inhalation transmission of K. pneumoniae can result in pneumonia, bacteremia, and nosocomial infections. Chicken meat is a potential reservoir for the transmission of antibiotic-resistant K. pneumoniae from animals to humans (Davis et al., 2015). The CTXM gene is produced by K. pneumoniae, an extended-spectrum β-lactamase (ESBL) generating bacteria. This gene can be passed down directly and indirectly (Mahanti et al., 2018). Resistant Gram-negative and Gram-positive bacteria will be difficult to treat, if not impossible to treat, with antimicrobials (Akova, 2016).
According to some reports, K. pneumoniae has developed drug resistance. Antibiotic resistance was found in K. pneumoniae isolated from chicken meat in Surabaya, Indonesia, with 61.54% resistance to tetracycline (TET), 15.38% resistance to gentamicin (CN), 7.69% resistance to cefoxitin, 38.46% resistance to sulfametizole, 53.85% resistance to nalidixic acid (NA), and 15.38% resistance to chloramphenicol (Yulistiani et al., 2017). In a 2005 survey, at a hospital in Surabaya, Indonesia, broad-spectrum β-lactamases were detected in 20% of Escherichia coli and 28% of K. pneumoniae (Lestari et al., 2008). Antibiotic resistance and the discovery of antibiotic resistance coding genes in K. pneumoniae bacteria from hens in Indonesia are currently understudied. As a result, antibiotic resistance and antibiotic resistance coding genes in K. pneumoniae bacteria isolated from broilers in Bogor should be researched.
MATERIALS AND METHODS
Procedures
Isolation and sample identification
In this study, 200 cloaca swab samples from broiler farms in Bogor and Sukabumi were cultured on MacConkey agar (MCA) (Oxoid) in a selective medium for K. pneumoniae bacteria. For 18–24 hours, the culture was incubated at 37°C. Gram staining was used to identify Klebsiella colonies. The Indole, Methyl Red, Voges Proskauer and Citrate (IMViC) test, methyl red/Voges–Proskauer test (Oxoid), Simmons citrate test (Oxoid), triple sugar iron agar, urease test, motility test, and carbohydrate test in the form of fermentation of glucose, sucrose, lactose, maltose, and Mannitol were used to test positive isolates.
Confirming K. pneumoniae sample using polymerase chain reaction (PCR)
Identification of K. pneumoniae bacteria was carried out by detecting the RNA polymerase β subunit (rpoB) gene with an amplicon length of 1,090 bp. Klebsiella bacteria extraction carried out using the PrestoTM Mini gDNA Bacteria Kit protocol (Geneaid). The primers used in this study were forward 5’-AAC CAG TTC CGC GTT GGC CTG G-3 ‘and reverse 5’-CCT GAA CAA CAC GCT CGG A-3’ (Alves et al., 2006). The reagent used for amplification was MyTaqTM HS Red Mix (Bioline) with a total PCR of 50 µl. The amplification condition began with predenaturation at 94°C for 2 minutes; then 30 cycles with denaturation at a temperature of 94°C for 30 seconds; annealing at 54°C for 1 minute; extension at 72°C for 4 minutes; and final extension at 72°Cand final extension at 72 n condition al ard 5’-AAC CAG TTCK. pneumoniae ATCC 700603.
Antibiotics susceptibility test
Bacterial isolates positive for the rpoB gene were then tested for antibiotic resistance. The antibiotic resistance test followed the Kirby–Bauer disk diffusion method using Mueller–Hinton agar based on the guidelines (CLSI, 2018). The antibiotics used were TET 30 µg, OT 30 µg, erythromycin (E) 15 µg, ciprofloxacin (CIP) 5 µg, enrofloxacin (EN) 5 µg, NA 30 µg, CN 10 µg, chloramphenicol (C) 30 µg, and ampicillin (AMP) 10 µg (Table 1). The selection of antibiotics was based on the results of a questionnaire, namely the top antibiotics most often used in chicken farms in West Java (Nilasari et al., 2018).
Molecular detection of antibiotic resistance gene by PCR amplification
Antibiotic resistance coding genes were detected using primary target genes gyrA (quinolone), tetA (TET), blaTEM (β-lactam), and ermB (macrolide). MyTaqTM HS Red Mix (Bioline) was used in the application of this study. The total volume of the PCR was 50 µl containing forward primers (2 µl), reversed primers (2 µl), DNA templates (3 µl), MyTaqTM Red Mix (25 µl); then H2O was added so that the total volume would reach 50 µl. The following primers were used in the study (Table 2).
 | Table 1. Constraint zone diameter standard (CLSI, 2018 ). [Click here to view] |
 | Table 2. Primer lists of antibiotic resistance coding genes. [Click here to view] |
The amplification process began with predenaturation at 95°C for 3 minutes, then 95°C for 30 seconds, annealing at 53°C–62°C (Table 2), extension at 72°C for 1 minute, and final extension at 72°C for 5-minute amplification cycles of 30 times. The amplified samples were then visualized by electrophoresis on 1.0% agarose gel in a 1× Tris acetate-ethylenediaminetetraacetic acid buffer.
RESULTS
Isolation and identification of samples
Klebsiella pneumoniae colonies in MCA media were pink, round, convex, and mucoid (Fig. 1). Klebsiella pneumoniae bacteria have a rod-shaped, pink Gram stain, which is typical of Gram-negative bacteria (Fig. 2). The findings of the sample identification biochemical testing revealed 45 positive isolates (22.5%) as Klebsiella colonies (Table 3).
Confirmation of K. pneumoniae isolates using PCR
A total of 45 isolates were molecularly examined, with 40 of them having the rpoB gene (Table 3). This gene plays an essential role in transcribing DNA into RNA (Mosaei and Harbottle, 2019). The isolates showed a band with a length of 1,090 bp, according to the results of molecular identification (Fig. 3).
Sensitivity test
The sensitivity test was carried out by calculating the diameter of the antibiotic inhibition zone formed on the Mueller–Hinton agar. The area appears clear because it is not overgrown by bacteria (Fig. 4). Klebsiella pneumoniae bacterial resistance to antibiotics is extremely high in 40 samples. Among other antibiotics, erythromycin resistance is the highest (100%), followed by OT (97.5%), AMP (97.5%), and TETs (95%) (Table 4).
Detection of antibiotic resistance coding genes
Resistance genes were discovered in K. pneumoniae isolates with phenotypic intermediates and resistance (40 samples) utilizing the genes tetA (TETs and OTs), ermB (erythromycin), gyrA (CIP, NA, and EN), and blaTEM (AMP). Figures 5 and 6 show the electrophoresis of PCR amplification products using a UV transilluminator. The gyrA and blaTEM genes were found in all 40 samples analyzed (Table 5).
 | Figure 1. Klebsiella colonies on MCA medium. [Click here to view] |
 | Figure 2. Gram stain of Klebsiella bacteria. [Click here to view] |
 | Table 3. Results of isolation and identification K. pneumoniae. [Click here to view] |
 | Figure 3. Amplification of the rpoB gene (1,090 bp) in K. pneumoniae isolates isolated from broilers in West Java. M: marker, ATCC: K. pneumoniae positive control, K1–K12: K. pneumoniae positive isolates. [Click here to view] |
 | Figure 4. Measurement of the inhibition zone diameter on the diffusion disk. The black arrow indicates the diameter of the inhibition zone that is formed. [Click here to view] |
DISCUSSION
The bacteria K. pneumoniae were isolated from the cloaca of West Javan broilers, in accordance with the findings of this research. In East Java, Indonesia, 7.8% of cloacal swab samples from broiler chickens were positive for Klebsiella bacteria (Hayati et al., 2019), while in Nigeria 60% of poultry cloacal swab samples were positive for Klebsiella bacteria (Chika et al., 2017).
According to the findings, K. pneumoniae was found in 40 out of the 45 samples. Because the isolates were not K. pneumoniae species, but other Klebsiella genus species, such as K. oxytoca or K. variicola, five negative samples were suspected. The difference in positive biochemical and molecular test results is thought to be due to the fact that biochemical testing did not employ all criteria; hence, some isolates were positive in biochemical tests but not in K. pneumoniae species tests.
The presence of K. pneumoniae in chicken cloacal swabs suggests that this bacterium can be found in the digestive tract of chickens, as well as in the feces and environment of chicken farms. Klebsiella pneumoniae was also found in poultry drinking water and the feces of veterinarians and farm employees (Hamza et al., 2016). Klebsiella can also be found in livestock waste, hospital waste, hospital worker gloves and clothing, and contaminated river water (Muraleedharana et al., 2019; Rock et al., 2014; Runcharoen et al., 2017). As a result, more research with a different sample source from this study is required. Although Klebsiella bacteria are still considered low pathogenic microorganisms, they can induce primary infections in poultry that are immunosuppressed. Klebsiella in chicken can contaminate the carcass, and if not properly treated, it can cause human infection. Because the bacteria are resistant to antibiotics, this bacterial infection becomes increasingly difficult to cure (Kowalczyk et al., 2017).
Resistance to erythromycin, AMP, OT, TET, NA, EN, CIP, and CN was found in more than 50% of the K. pneumoniae strains studied (Table 4). Klebsiella spp. isolated from broilers in Shandong province, China, was resistant to AMP 98.9%, CIP 80.0%, TET 78.9%, and chloramphenicol 92.2%, according to a similar study (Wu et al., 2016).
Antibiotic resistance can develop for a variety of reasons, including the length of antibiotic use (Moini et al., 2015). Antibiotic misuse has resulted in a global antibiotic resistance epidemic (Bartlett et al., 2013). The overuse of antibiotics in chickens is linked to the high rate of antibiotic resistance, increasing the danger of normal gut flora becoming resistant to antibiotics (Chishimba et al., 2016). Antibiotics like those employed in this study are often utilized in farms in West Java. As a result, K. pneumoniae has a high level of resistance and is resistant to multiple antibiotic classes.
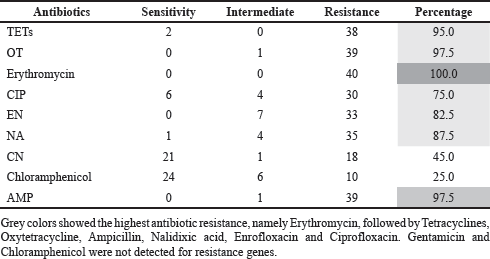 | Table 4. Percentage of antibiotic resistance in K. pneumoniae bacteria (n = 40). [Click here to view] |
 | Figure 5. Amplification of the gyrA (626 bp), blaTEM (516 bp), and tetA (965 bp) genes. M: marker, K13–K24: K. pneumoniae isolates. [Click here to view] |
 | Figure 6. Amplification of gene ermB (636 bp). M: marker, K13–K24: K. pneumoniae isolate. [Click here to view] |
Klebsiella pneumoniae isolate was resistant to drugs from three different classes (multidrug-resistant). Bacteria are not sensitive to the majority of TET, AMP, and erythromycin classes. Klebsiella was found on a turkey farm in Oklahoma, and it possesses multidrug resistance to medicines such as AMP, TET, streptomycin, CN, and kanamycin (Kim et al., 2005). This has a devastating effect on the health of both animals and humans. WHO (2014) has identified bacteria that are the main focus of antimicrobial resistance research. Klebsiella pneumoniae is one of the bacteria that is the main focus in the international community. These bacteria are the source of the spread of resistance to antibiotics is shown in Figure 7 (Venezia et al., 2017).
Bacterial resistance to chloramphenicol antibiotics was only 25% in this study (Table 4), which could be because chloramphenicol in chicken has been banned in Indonesia since 1994. Chloramphenicol is included in the list of harsh drugs that are not authorized to be used for animals, according to Ditjen PP KemenKumHAM (1994) Decree of the Minister of Agriculture Number: 806/KFpts/TN.260/12/94 concerning veterinary pharmaceuticals classification regulation. Furthermore, the government notes that chloramphenicol is one of Indonesia’s nine illegal food additives via Permenkes Number: 1168/Menkes/PER/X/1999 about food additives (Ditjenpkh, 2018). Despite the fact that chloramphenicol was outlawed 27 years ago, this study indicates that bacterial isolates resistant to it still exist. Until now, it was thought that the gene coding for antibiotic resistance of the chloramphenicol type was still present in K. pneumoniae.
 | Table 5. The result of amplification of resistance coding genes in K. pneumoniae isolates (n = 40). [Click here to view] |
 | Figure 7. The mechanism of emergence and spread of K. pneumoniae is MDR and extensively drug-resistant (XDR) and the accumulation of Antibiotic resistance gene (ARG) (Venezia et al., 2017 ). [Click here to view] |
Quinolone phenotypic resistance is extremely common. Bacterial resistance to quinolones is induced by mutations in the genes gyrase and topoisomerase IV, which impair the interaction between quinolones and enzymes, causing quinolones to become toxic and destroy bacterial chromosomes (Aldred et al., 2014). A gene mutation in the quinolone resistance determining regions—gyrA and parC, gyrA or parC only, or both genes—is responsible for the high level of resistance to the quinolone group and its derivatives (Hooper and Jacoby, 2015). In the molecular test, all of the isolates tested positive for the gyrA gene. In Enterobacteriaceae, the gyrA gene has a significant impact on the high level of resistance. Resistance is caused by mutations in DNA gyrA at amino acid positions 83 (serine tyrosine/leucine/isoleucine/treonin) or 87 (aspartate asparagine) (Nawaz et al., 2012). According to another study, the gyrA mutation causes alterations at positions 83 (serine), 84 (alanine), and 87 (aspartate) in the protein (Piekarska et al., 2015).
In this investigation, the TETs’ sensitivity was quite poor. Efflux pumps, ribosomal protection, degradation, and rRNA mutation are the four basic methods by which bacteria become resistant to TETs. The efflux pump is the most common source of TET resistance; so far, 28 genes have been identified as the efflux pump’s cause. And tetA is one of them (Nguyen et al., 2014). In this study, the tetA gene was found in 83.33% of the participants. Previous research has found that 100% of clinical K. pneumoniae isolates carry the tetA gene (Bokaeian et al., 2014).
Because it may be spread via plasmids, the tetA gene is frequently found in bacteria. The tetA coding gene in Klebsiella bacteria can be used to transfer TET resistance to other bacteria such as E. coli via plasmid mediation (Wang et al., 2014). The tetA gene in bacteria obtained from layer chickens can be transmitted to other bacteria, and the tetA gene can spread faster in the environment than the tetB gene (Balasubramaniam et al., 2014). The efflux pump is encoded by the tetA gene (Akiyama et al., 2013). TET efflux pumps are regulated by the TET repressor “tetR,” which tightly controls TET mRNA expression (Møller et al., 2016). TET-resistant Klebsiella pneumoniae isolates from Kenya had 18% of the tetA gene; tetD, tetB, tetG, and maybe more coding genes made up the rest. So it is possible that the isolates that were not detected by the tetA gene have alternative resistance coding genes or resistance mechanisms in addition to efflux pumps in this investigation (Taitt et al., 2017).
Erythromycin resistance was phenotypically present in all isolates. However, not all isolates possessed the ermB gene following molecular testing. Plasmids are used to carry the ermB gene (Dong et al., 2018). By altering 23S rRNA from bacterial ribosomes and establishing cross-resistance to macrolides, the erm gene acts as a methyltransferase (Dzyubakand Yap, 2016). Like cross-resistance, cross-sensitivity to antibiotics is another major problem where antibiotics of same class or antibiotics having similar structure become reactive/allergic (Chaudhary et al., 2021). The erm gene has around 30 variants, with ermA, ermB, ermC, and ermF being the 4 primary classes found in pathogenic microbes. In Staphylococcus, the ermA and erm genes are commonly detected. The ermB class genes are mostly found in Streptococcus and Enterococcus bacteria (Hoek et al., 2011). The ermB gene responds to erythromycin resistance in Enterococcus. Isolates that are negative for the ermB gene may carry additional genes, according to research findings (Iweriebor et al., 2016). Furthermore, because the permeability of the outer membrane of Enterobacteriaceae bacteria is poor, it is innately resistant to the macrolide group (Gomes et al., 2016).
This study also detected the ESBL coding gene. Detection of the ESBL coding gene needs to be carried out because Klebsiella is a bacterium that produces ESBL (Muraleedharana et al., 2019). In India, 60.3% of K. pneumoniae isolates produced ESBL (Kaur et al., 2013). ESBL is an enzyme produced by Gram-negative bacteria which can cause β-lactam antibiotics. Since 2018, the bla gene has been identified in the description, and 223 TEM types, 193 SHV types, and 172 CTX-M types have been listed in the database (https://www.lahey.org/Studies) (Ramadan et al., 2018).
The ESBL coding gene was also discovered in this investigation. Because Klebsiella is a bacterium that produces ESBL, detection of the ESBL coding gene is required (Muraleedharana et al., 2019). 60.3% of K. pneumoniae isolates in India developed ESBL (Kaur et al., 2013). Gram-negative bacteria develop an enzyme called ESBL, which can make β-lactamase antibiotics ineffective. The bla gene has been found in 223 TEM types, 193 SHV types, and 172 CTX-M types since 2018 (Ramadan et al., 2018). The temoniera (TEM) gene was identified as an ESBL gene in this investigation. When compared to other forms of ESBL, the presence of the blaTEM gene was the highest (Pishtiwan and Khadija, 2019). The blaTEM gene is a β-lactamase (bla) gene that was found in Klebsiella isolates for the first time. This gene is a forerunner in the evolution of the bla gene, paving the way for the appearance of other ESBL coding genes, which later underwent changes. The bla gene can help antibiotics spread horizontally between strains via plasmids and transposons (Barguigua et al., 2011). The blaTEM gene is an antibiotic resistance gene found in plasmids that is most commonly found in Gram-negative bacteria in clinical settings (Wilopo et al., 2015).
Clinical disease is more usually linked to research on the bacterium K. pneumoniae. Environmental isolates are still rarely explored. Both phenotypically and genotypically, these isolates are remarkably similar to clinical isolates (Runcharoen et al., 2017). However, the two groups differed in terms of virulence characteristics (Davis et al., 2015). Only cloacal swab samples were used in this investigation, and only a few genes coding for antibiotic resistance were discovered. As a result, more research into K. pneumoniae samples from the environment and other chicken organs is required, as well as the utilization of more resistance coding genes for identification.
CONCLUSION
Klebsiella pneumoniae can be isolated and identified from broilers in West Java, Indonesia, according to this study. Resistance to more than three antibiotic classes indicates that the isolates are multidrug-resistant. In West Java, resistance coding genes such gyrA, tetA, ermB, and blaTEM were found in K. pneumoniae from broiler cloaca swabs.
ACKNOWLEDGMENTS
The authors appreciate the financial support from the Ministry of Research, Technology, and Higher Education, as well as the assistance provided by the Division of Medical Microbiology Research Laboratory workers during the research.
CONFLICT OF INTEREST
The authors declare no conflict of interest for this manuscript.
FUNDING
This research was fund by PENELITIAN DASAR UNGGULAN PERGURUAN TINGGI (PDUPT) TAHUN ANGGARAN 2021, The Ministry of Education, Culture, Research, and Technology of the Republic of Indonesia. Contract number: 12/INS-1/PPK/ES/2018.
AUTHORS’ CONTRIBUTIONS
All the authors have equally contributed to study design, collection, analysis, and interpretation of the data, writing, and manuscript drafting. All the authors have read and approve the final version.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included within this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Akiyama T, Presedo J, Khan AA. The tetA gene decreases tigecycline sensitivity of Salmonella enterica isolates. Int J Antimicrob Agents, 2013; 42(2):133–40. CrossRef
Akova M. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence, 2016; 7(3):252–66. CrossRef
Alagawany M, Elnesr SS, Farag MR, Tiwari R, Yatoo MI, Karthik K, Michalak I, Dhama K. Nutritional significance of amino acids, vitamins and minerals as nutraceuticals in poultry production and health—a comprehensive review. Vet Q, 2020; 41(1):1–29. CrossRef
Aldred KJ, Kerns RJ, Osheroff N. Mechanism of quinolone action and resistance. Biochemistry, 2014; 53(10):1565–74. CrossRef
Alves MS, Dias RCDS, Castro ACDD, Riley LW, Moreira BM. Identification of clinical isolates of indole-positive and indole negative Klebsiella spp. J Clin Microbiol, 2006; 44(10):3640–6. CrossRef
Balasubramaniam A, Eswaran MA, Suresh P, Sukumar K. Detection of tetracycline resistance determinant tetA gene and antimicrobial resistance pattern in Escherichia coli isolates recovered from healthy layer chickens. Vet World, 2014; 7(9):635–8. CrossRef
Barguigua A, Otmani EI, Talmi M, Bourjilat F, Haouzane F, Timinouni M, Zerouali K. Characterization of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates from the community in Morocco. J Med Microbiol, 2011; 60(Pt 9):1344–52. CrossRef
Bartlett JG, GilbertDN, SpellbergB. Seven ways to preserve the miracle of antibiotics. Clin. Infect. Dis, 2013; 56(10):1445-1450. CrossRef
Bokaeian M, Saedi S, Shahi Z, Kadaei. tetA and tetB genes in Klebsiella pneumoniae isolated from clinical samples. Gene Cell Tissue, 2014; 1(2):e18152. CrossRef
Chaudhary RK, Metgudmath AR, Bhandari R, Karoli SS, Uday Kumar R. Fixed drug eruptions secondary to fixed drug combination (ofloxacin/ornidazole): a cross sensitivity case report. Curr Drug Ther, 2021; 16(5):448–53. CrossRef
Chika E, Ifeanyichukwu I, Benigna O, Loveday OO, Stanley E, Collins O, Kenneth O, Chika E. Emerging multi-drug resistant metallo-β-lactamases (MBLs) positive Klebsiella species from cloacal swabs of poultry birds. J Bacteriol Parasitol, 2017; 8(1):305. CrossRef
Chishimba K, Hang’ombe BM, Muzandu K, Mshana SE, Matee MI, Nakajima C, Suzuk Y. Detection of extended-spectrum beta-lactamase-producing Escherichia coli in market-ready chickens in Zambia. Int J Microbiol, 2016; 2016:5275724. CrossRef
Chuah LO, Syuhada AS, Suhaimi IM, Hanim TF, Rusula. Genetic relatedness antimicrobial resistance and biofilm formation of Salmonella isolated from naturally contaminated poultry and their processing environment in northern Malaysia. Food res Int, 2018; 105:743–51. CrossRef
CLSI. Performance standards for antimicrobial susceptibility testing. 26th edition, Wayne, NJ, pp 30–7, 2018.
Colom K, Perez J, Alonso R, Aranguiz AF, Larino E, Cisterna R. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol Latt, 2003; 223(2):147–51. CrossRef
Davis GS, Horwinski J, Waits K, Nordstrom L, Porter S, Weaver B, Stegger M, Aziz M, Johnson JR, Gauld L, Liu CM, Grande H, Bigler R, Price LB. Intermingled Klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clin Infect Dis, 2015; 61(6):892–9. CrossRef
Department of Nutrition Science and Feed Technology. Antibiotic growth promoter/AGP—D-INTP. 2018. Available via http://intp.fapet.ipb.ac.id (Accessed 18 July 2018).
Dibner JJ, Richards JD. Antibiotic growth promoters in agriculture: history and mode of action. Poult Sci, 2005; 84(4):634–43. CrossRef
Ditjenpkh. Permentan-Klasifikasi-Obat-Hewan. 2018. Available at Permentan-14-2017-Klasifikasi-Obat-Hewan-1.pdf (pertanian.go.id)[Accessed 08 April 2020].
Dong N, Yang X, Zhang R, Chan EWCC, Chen S. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg Microbes Infect, 2018; 7(1):146–53. CrossRef
Dzyubak E, Yap MNF. The expression of antibiotic resistance methyl transferase correlates with mRNA stability independently of ribosome stalling. Antimicrob Agents Chemother, 2016; 60(12):7178–88. CrossRef
Gomes C, Martínez-Puchol S, Palma N, Horna G, Ruiz-Roldán L, Pons MJ, Ruiz J. Macrolide resistance mechanisms in Enterobacteriaceae: focus on azithromycin. Crit Rev Microbiol, 2016; 43(1):1–30. CrossRef
Hamza E, Dorgham SM, Hamza DA. Carbapenemase-producing Klebsiella pneumoniae in broiler poultry farming in Egypt. J Glob Antimicrob Resist, 2016; 7:8–10. CrossRef
Hardiati A, Safika S, Wibawan IWT, Indrawati A, Pasaribu FH. Isolation and detection of antibiotics resistance genes of Escherichia coli from broiler farms in Sukabumi, Indonesia. J Adv Vet Anim Res, 2021; 8(1):84–90. CrossRef
Hayati M, Indrawati A, Mayasari NLPI, Istiyaningsih I, Atikah N. Molecular detection of extended-spectrum ß-lactamase-producing Klebsiella pneumoniae isolates of chicken origin from East Java, Indonesia. Vet World, 2019; 12(4):578–83. CrossRef
Hoek AHAMV, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJM. Acquired antibiotic resistance genes: an overview. Front Microbiol, 2011; 2:1–27. CrossRef
Hooper DC, Jacoby GA. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci, 2015; 1354(1):12–31. CrossRef
Huddleston JR. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist, 2014; 7:167–76. CrossRef
Iweriebor BC, Obi LC, Okoh AI. Macrolide, glycopeptide resistance and virulence genes in Enterococcus species isolates from dairy cattle. J Med Microbiol, 2016; 65(7):641–8. CrossRef
Kaur J, Chopra S, Sheevani, Mahajan G. Modified double-disc synergy test to detect ESBL production in urinary isolates of Escherichia coli and Klebsiella pneumoniae. J Clin Diagn Res, 2013; 7(2):229–33. CrossRef
Kim SH, Wei CI, Tzou YM, An H. Multidrug-resistant Klebsiella pneumoniae isolated from farm environments and retail products in Oklahoma. J Food Prot, 2005; 68(10):2022–9. CrossRef
Kowalczyk J, ?mia?ek M, Tyka?owski B, Koncicki A. Klebsiella spp. in the pathology of poultry and their role in epidemiology of human foodborne diseases. Med Water, 2017; 73(9):528–31. CrossRef
Lestari ES, Severin JA, Filius PM, Kuntaman K, Duerink DO, Hadi U, Wahjono H, Verbrugh HA, Antimicrobial Resistance in Indonesia: Prevalence and Prevention (AMRIN). Antimicrobial resistance among commensal isolates of Escherichia coli and Staphylococcus aureus in the Indonesian population inside and outside hospitals. Eur J Clin Microbiol Infect Dis, 2008; 27(1):45–51. CrossRef
Livestock and Animal Health Statistics. Direktorat Jenderal Peternakan dan Kesehatan Hewan Kementerian Pertanian RI. 2020. Available via http://ditjenpkh.pertanian.go.id/userfiles/file/Buku_Statistik_2021.pdf?time=1633686831406 (Accessed 8 July 2021).
Mahanti A, Ghosh P, Bandyopadhyay S, Batabyal S, Samanta I, Bhattacharyya D, Sar TK, Joardar SN, Banerjee J, Dutta TK. Prevalence of CTX-M-producing Klebsiella spp. in broiler, kuroiler, and indigenous poultry in West Bengal State, India. Microb Drug Resist, 2018; 24(3):299–306. CrossRef
Moini AS, Soltani B, Ardakani AT, Moravveji A, Erami A, Rezae MH, Namazi M. Multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolated from patients in Kashan, Iran. Jundishapur J Microbiol, 2015; 8(10):e27517. CrossRef
Mosaei H, Harbottle J. Mechanisms of antibiotics inhibiting bacterial RNA polymerase. Biochem Soc Trans, 2019; 47(1):339–50. CrossRef
Møller TSB, Overgaard M, Nielsen SS, Bortolaia V, Sommer MOA, Guardabassi L, Olsen JE. Relation between tetR and tetA expression in tetracycline resistant Escherichia coli. BMC Microbiol, 2016; 16:39–47. CrossRef
Muraleedharana C, Talrejaab D, Kanwar M, Kumar A, Walia SK. Occurrence of extended-spectrum β-lactamase-producing bacteria in urban Clinton River habitat. J Glob Antimicrob Resist, 2019; 16:225–35. CrossRef
Nawaz M, Khan SA, Tran Q, Sung K, Khan AA, Adamu I, Steele RS. Isolation and characterization of multidrug-resistant Klebsiella spp. isolated from shrimp imported from Thailand. Int J Food Microbiol, 2012; 155(3):179–84. CrossRef
Nguyen F, Starosta AL, Arenz S, Sohmen D, Dönhöfer A, Wilson DN. Tetracycline antibiotics and resistance mechanisms. Biol Chem, 2014; 395(5):559–75. CrossRef
Nilasari Z, Safika S, Pasaribu FH. Antibiotic resistance of Klebsiella species isolated from broiler chickens in Sukabumi and Bogor areas. Proceedings of the 20th FAVA CONGRESS & the 15th KIVNAS PDHI, Bali, Indonesia, pp 91–3, 2018.
Piekarska K, Wo?kowicz T, Zacharczuk K, Rzeczkowska M, Chróst A, Bareja A, Olak M, Gierczy?ski R.Co-existence of plasmid-mediated quinolone resistance determinants and mutations in gyrA and parC among fluoroquinolone-resistant clinical Enterobacteriaceae isolated in a tertiary hospital in Warsaw, Poland. Int J Antimicrob Agents, 2015; 45(3):238–43. CrossRef
Pishtiwan AH, Khadija KM. Prevalence of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing Klebsiella pneumoniae and Escherichia coli isolated from thalassemia patients in Erbil, Iraq. Mediterr J Hematol, 2019; 11(1):e2019041. CrossRef
Ramadan AA, Abdelaziz NA, Amin MA, Aziz RK. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase producing Escherichia coli. Sci Rep, 2018; 9(1):4224. CrossRef
Rock C, Thom KA, Masnick M, Johnson JK, Harris AD, Morgan DJ. Frequency of Klebsiella pneumonia carbapenemase (KPC) and non-KPC-producing Klebsiella contamination of healthcare workers and the environment. Infect Control Hosp Epidemiol, 2014; 35(4):426–9. CrossRef
Runcharoen C, Moradigaravand D, Blane B, Paksanont S, Thammachote J, Anun S, Parkhill J, Chantratita N, Peacock SJ. Whole-genome sequencing reveals high-resolution epidemiological links between clinical and environmental Klebsiella pneumoniae. Genome Med, 2017; 9(1):2–10. CrossRef
Song JH, Chang HH, Suh JY, Ko KS, Jung Si, Oh WS, Pack KR, Lee NY, Yang Y, Chongthaleong A. Macrolide resistance and genotypic characterization of Streptococcus pneumonia in Asian countries: a study of the asian network for surveillance of resistant pathogens (ANSORP). J Antimocrob Chemotherap, 2004; 53:457–63. CrossRef
Taitt CR, Leski TA, Erwin DP, Odundo EA, Kipkemoi NC, Ndonye JN, Kirera RK, Ombogo AN, Walson JL, Pavlinac PB, Hulseberg C, Vora GJ. Antimicrobial resistance of Klebsiella pneumoniae stool isolates circulating in Kenya. PLoS One, 2017; 12(6):e0178880. CrossRef
Venezia SN, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev, 2017; 41(3):252–75. CrossRef
Wang W, Ye X, Guo Q, Wang M, Xu X, Sheng Z. High-level tetracycline resistance mediated by efflux pumps Tet(A) and Tet(A)-1 with two start codons. J Med Microbiol, 2014; 63(Pt 11):1454–9. CrossRef
Wilopo BAP, Sudigdoadi S, Sahiratmadja E, Dewi IMW. Loop-mediated isothermal Amplification untuk Mendeteksi Gen blaTEM sebagai penyandi extended-spectrum beta-lactamase pada isolat Enterobacteriaceae. MKB, 2015; 47(4):242–9. CrossRef
WHO. WHO antimicrobial resistance: global report on surveillance 2014. p 257, 2014. Available via https://www.who.int/antimicrobial-resistance/publications/surveillancereport/en/ (Accessed 18 August 2021).
Wu H, Wang M, Liu Y, Wang X, Wang Y, Lu J, Xu H. Characterization of antimicrobial resistance in Klebsiella species isolated from chicken broilers. Int. J Food Microbiol, 2016; 232:95–102. CrossRef
Yulistiani R, Praseptiangga D, Supyani, Sudibya D, Raharjo, Shirakawa T. Prevalence of antibiotic-resistance Enterobacteriaceae strains isolated from chicken meat at traditional markets in Surabaya, Indonesia. ICFSE, 2017; 193:012007. CrossRef