INTRODUCTION
For hospitalized patients with comorbidities and mortality risk factors, nosocomial infection was considered the leading reason for mortality and morbidity, and it was linked to increased healthcare and financial expenses (Hirsch and Tam, 2010).
Pseudomonas aeruginosa is an opportunistic Gram-negative organism accountable for 51,000 healthcare-associated infections and leads to serious and potentially fatal nosocomial infections, such as urinary tract infections, respiratory tract infections, and bacteremia, in immunocompromised patients (Ruppé et al., 2015).
The innate and acquired machineries of antibiotics resistance were leading risk factors for spreading this organism in healthcare places (Landman et al., 2002; Lister et al., 2009).
Pseudomonas aeruginosa has developed resistance to various antibiotic families, including carbapenems, which are considered trustworthy and efficient antibiotics that are only used as a last resort for the treatment of serious diseases till now (Sekiguchi et al., 2005).
Metallo-ß-lactamase (MBL) assembly is the leading source of extraordinary imipenem resistance in between P. aeruginosa isolates (Franco et al., 2010). This study aimed to isolate and identify P. aeruginosa from Al-Azhar University Hospital, identify the measurement of antibacterial activity of different groups of antibiotics on P. aeruginosa, determine the incidence of multi-drug resistant (MDR) P. aeruginosa in infected patients, and detect MBLs genes among MDR P. aeruginosa strains using polymerase chain reaction (PCR).
MATERIALS AND METHODS
Isolation and identification P. aeruginosa
This study included 74 P. aeruginosa isolates recovered from 412 clinical isolates taken from patients urine admitted to wards/intensive care units (ICUs) and who acquired nosocomial infections after excluding signs or symptoms of infection at the time of admission. This cross-sectional observational study was carried out from November 2015 to April 2017 in Al-Azhar University Hospital and Molecular Biology Research Unit, Assiut University. All recovered isolates were identified using conventional protocols and validated by growing them on Cetrimide agar and Pseudomonas agar and examining the morphology of their colonies (Govan, 2007).
General information was collected, including age, sex, managing unit, and reason for ICU admission, risk factors as diabetes mellitus, previous operations, catheterizations, total parenteral nutrition and antibiotics used during the hospital stay, and drugs used for prolonged periods.
Antimicrobial susceptibility of P. aeruginosa
Three to five visually identical pure colonies were suspended with 3–4 ml of sterile saline. The turbidity was adjusted to a 0.5 McFarland turbidity standard (approximately 108 colony forming unit/ml). A sterile cotton swab was immersed into the bacterial solution, spinning it firmly and evenly distributing bacteria over the whole agar plate. A disk dispenser was used to insert antibiotic disks into the agar plates, and the plates were incubated at 37°C for 18 hours. The zone of inhibition diameters was recorded, and the results were interpreted using Clinical and Laboratory Standards Institute guidelines (Wayne, 2011). For each isolate, a resistance score (R score) was calculated based on the number of antibiotics that were resistant (a score of 0.5 considered intermediately resistant while a score of 1 was resistant).
Exploring biofilm formation using microtiter plate assay technique
Microtiter plate assay was accomplished to evaluate biofilm formation for MDR P. aeruginosa urine isolates, according to Stewart et al. (2004). Briefly, P. aeruginosa was cultured overnight in trypticase soy broth (TSB), and then 200 µl of 1% diluted bacteria was applied to 96-well flat-bottom polystyrene microtiter plates. The inoculated plate was incubated at 30°C for 22–24 hours and phosphate-buffered saline (PBS, pH 7.2) was used in three successive washing steps to allow the non- or loosely attached bacteria to be discarded.
A 100 µl of a 0.1% crystal violet was applied to wells of the microtiter plate, and the plate was then incubated at room temperature for 30 minutes. After several washing steps, 200 µl of 95% ethanol was used to solubilize the crystal violet staining of the biofilm, and 125 µl of the extracted color was transferred to a new microtiter plate. The absorbance was measured at 450 nm using a plate reader. Measurements were carried out in triplicate. According to the criteria of Stepanovic et al. (2007), the nonbiofilm producer is indicated when optical density (OD) ≤ ODc, weak biofilm producer is defined as OD > ODc and ≤ 2× ODc, moderate biofilm producer is defined as OD > 2× ODc and ≤ 4× ODc, and strong biofilm producer is indicated when OD > 4× ODc (Stepanovic et al., 2007).
Detection of blaIMP-1 and blaVIM-1 genes by PCR
The bacterial colonies were subcultured into TSB for 24 hours at 37°C. To extract the genomic DNA, the cells grown in suspension were collected by centrifugation for 5 minutes at 8,000 rpm in a 1.5 ml Eppendorf tube. Cell pellets were resuspended in 200 µl PBS, and an equal volume of QIAGEN protease was added. A 200 µl buffer AL was added, followed by vortexing for 15 seconds. After incubating the samples at 56°C for 10 minutes, 200 µl of 96% ethanol was added and mixed by vortexing for 15 seconds. The mixture was centrifuged at 8,000 rpm for 1 minute using the QIAamp Mini spin column. The washing protocol was followed according to the manufacturer’s instructions before the final elution step and the collection of the extracted DNA.
PCR amplification was accomplished using specified primers (Table 1), using the thermocycler (Personal Thermocycler Model, Biometra, Germany). An amplification program for the detection of blaIMP-1 and blaVIM-1 genes was conducted according to Rodrigues et al. (2011) and Shibata et al. (2003).
Statistical analysis
Various data visualization, graphing, statistical calculations, data entry, and data analysis systems and software were used as Statistical Package for Social Science (Inc., Chicago, version 16). Statistical significance was recognized when p < 0.05. Using R (https://www.r-project.org) and the ggplot2 package, data visualization was carrieed out. The unweighted pair group technique with the arithmetic mean (UPGMA) clustering (http://genomes.urv.cat/UPGMA) algorithm was employed for constructing a distance matrix and a tree, which was produced with the free software application FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
RESULTS
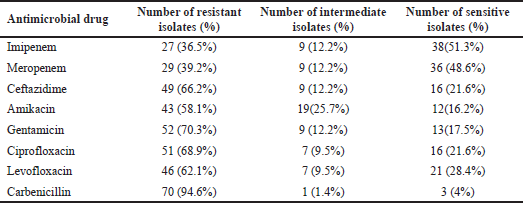
Pseudomonas aeruginosa isolates were found in 18% (74/412) of clinical isolates. Of them, the resistance rate to imipenem, carbenicillin, gentamicin, ciprofloxacin, ceftazidime, levofloxacin, and amikacin was 36.5%, 94.6%, 70.3%, 68.9%, 66.2%, 62.2%, and 58.1%, respectively (Supplementary Table S1). A heatmap was conducted to compare the antibiotic resistance among 74 isolates of P. aeruginosa, depicting the relative resistance of each isolate (Fig. 1A and B). The clustering dendrogram was created using the UPGMA (Fig. 2) to show relations between different isolates phenotypes. However, the tree has the benefit of allowing one to see the more obscure phenotypes, like that of isolate P59 (sensitive to four antibiotics).
 | Table 1. Nucleotide sequences of primers used for the detection of IMP and VIM genes. [Click here to view] |
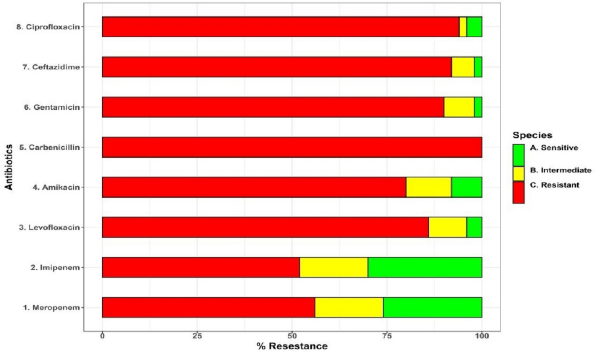 | Figure 1. (A) A stacked bar plot that depicts the 74 P. aeruginosa isolates’ distribution of resistance to eight antibiotics (Y-axis). (B) A heatmap that represents resistance phenotypes of 74 P. aeruginosa isolates (red = resistant, yellow = intermediate, and green = sensitive). The X-axis describes isolates, whereas the Y-axis shows antibiotics. The horizontal and vertical trees reflect hierarchical clustering, which is used to order isolates and antibiotics, respectively. [Click here to view] |
Out of the 74 P. aeruginosa isolates, 51 (69%) strains were MDR (resistance to at least three antipseudomonal antibiotic groups including β-lactams, aminoglycosides, and fluoroquinolones), and 23 (32%) strains were non-MDR strains. Isolates were classified into four classes according to the OD of the bacterial biofilms (ODc was calculated as 0.064): nonbiofilm forming (if OD ≤ 0.064), weak biofilm forming (if OD > 0.064, but ≤ 0.128), moderate biofilm forming (if OD > 0.128, but ≤ 0.256), and strong biofilm forming (if OD > 0.256).
A 44/51 (86.27%) of MDR strains were biofilm producers; 33.33% of them were strong biofilm producers, 29.41% of them were moderate, and 23.53% of strains were weak biofilm producers.
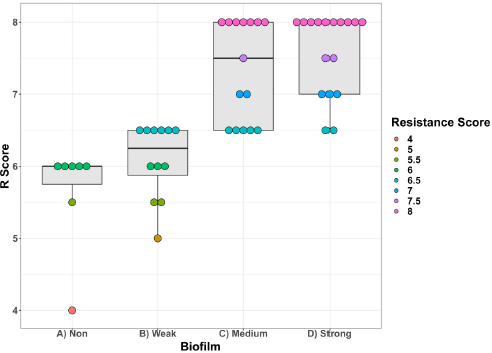
There was a relationship between biofilm formation and R score, with significant biofilm formation associated with isolates with high R scores contrasted to poor biofilm formation associated with isolates with low R scores, as seen in (Fig. 3).
PCR showed that eight strains of imipenem-resistant P. aeruginosa contained blaVIM, while blaIMP gene was not detected, as shown in (Fig. 4).
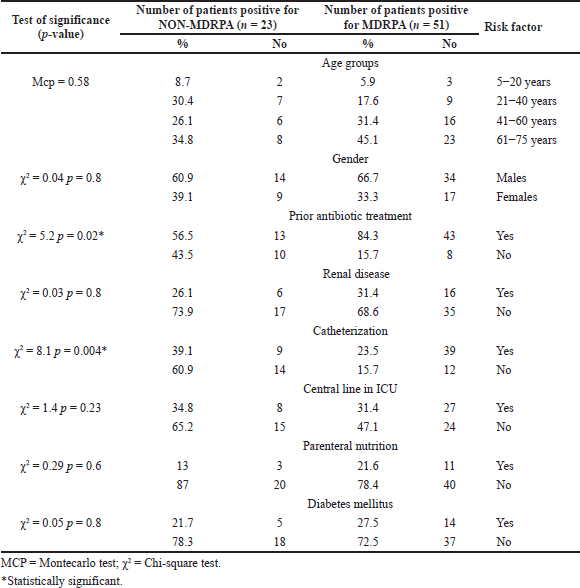
Patients positive for MDR P. aeruginosa (n = 51) were compared with patients positive for non-MDR P. aeruginosa (n = 23) to determine factors associated with the acquisition of multidrug-resistant phenotype. Several variables were studied, including age, gender, prior antibiotic treatment, catheterization, central line in ICU, parenteral nutrition, renal diseases, and diabetes mellitus. p-value was calculated for each variable, and significance was determined when the p-value was lower than 0.05.
Table 2 illustrates the risk factor for the nosocomial multi-drug resistant P. aeruginosa (MDRPA) and non-MDRPA infection. Possession of nosocomial MDRPA infections was significantly associated with past antibiotic therapy or catheterization (p < 0.05). The other risk factors, including age, sex, mechanical ventilation, central line in ICU, parenteral nutrition, and diabetes mellitus, were not significant with MDRPA infection.
DISCUSSION
During the study period, nosocomial P. aeruginosa accounted for 18% of all isolated uropathogens. This finding agreed to some extent with that reported by Gad et al. (2007), who identified P. aeruginosa isolates in 18.2% of the clinical samples, and Wassef et al. (2015), who identified P. aeruginosa isolates in 20.7% of different clinical specimens. Furthermore, it was higher than that obtained in Iran, where the rate of incidence of P. aeruginosa in UTI was 13.2% (Aminizadeh and Kashi, 2011). However, these results were less than those found by Mansour et al. (2013), who reported that the frequency of isolation of P. aeruginosa from clinical specimens in Egypt and Saudi Arabia was 32.8% and 30.0%, respectively.
In the current study, urinary isolated P. aeruginosa had a resistance rate of 94.59% to carbenicillin and 66.2% to ceftazidime. The same was reported by Kotsakis et al. (2010), who revealed that 65.4% of P. aeruginosa were resistant to ceftazidime. Our result was lower than that of Arora et al. (2011), who reported that 73.2% of P. aeruginosa were ceftazidime-resistant. At the same time, Manjunath et al. (2011) reported higher rates (69.3%) of carbenicillin.
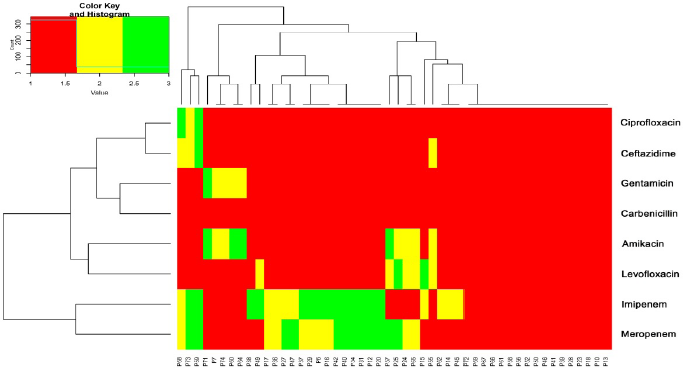 | Figure 2. Circular tree depicting the relationships between 74 P. aeruginosa isolates based on antibiotic susceptibility phenotypes. [Click here to view] |
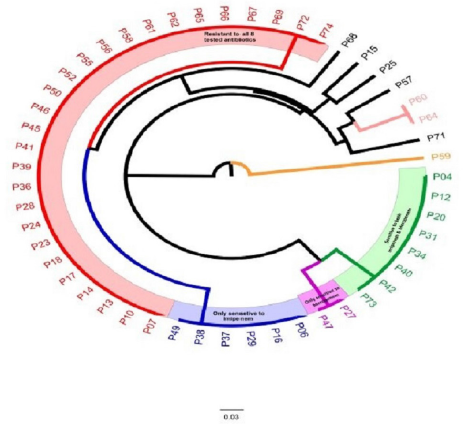 | Figure 3. Boxplots depicting the R scores of P. aeruginosa isolates with relationship to their biofilm formation. [Click here to view] |
In our study, the resistance rate of P. aeruginosa to imipenem was 36.5% and to meropenem was 39.2%. Our results were in agreement with those reported by Mahmoud et al. (2013), who showed that 33.3% of P. aeruginosa strains were resistant to imipenem. Our results were higher than those described by Fazlul et al. (2011), who showed that imipenem and meropenem were found to be more effective agents, where the resistance of the P. aeruginosa isolates toward these antibacterial agents was 18.5% and 20.4%, respectively. Carbapenem resistance rates can vary according to local antibiotic regulations, strain origin, and geographic location.
 | Figure 4. Gel electrophoresis of blaVIM-1 gene PCR product. Lane M is the 1 kb ladder; lanes 1, 3,–8 show blaVIM-1-specific band size product. [Click here to view] |
 | Table 2. Risk factors for nosocomial MDRPA infection in the 51 case. [Click here to view] |
The misuse of antimicrobial agents can explain this rate of carbapenem resistance in the last decade, especially in developing countries which allow the bacteria to modify the resistant mechanisms (Picao et al., 2009). Acinetobacter and Pseudomonas bacteria had the highest resistance levels to imipenem in Egypt (37.03%) (Ashour and El-Sharif, 2009). The emergence of imipenem-resistant strains of P. aeruginosa in the Middle East is concerning, particularly in Saudi Arabia, where the resistance rate of P. aeruginosa to imipenem has risen to 38.57% (Al-Agamy et al., 2011).
The antibiotic resistance pattern in P. aeruginosa isolates to aminoglycoside was relatively high (58.1% to amikacin and 70.3% gentamicin). These results are close to those obtained by Mohanasoundaram (2011). Arora et al. (2011) found that 79% and 41.5% of P. aeruginosa isolates were gentamicin- and amikacin-resistant, respectively.
Fluoroquinolones are most commonly used in urinary tract infections. The resistance to ciprofloxacin and levofloxacin was 68.9% and 62.2%, respectively. These rates were lower than those reported in India, where the rate of ciprofloxacin resistance was 85% (Manjunath et al., 2011).
In our study, the prevalence of MDR strains among P. aeruginosa urine isolates was 51 (68.92%). Our findings are consistent with previous research, which found that the percentage of MDR in P. aeruginosa had grown from 64% in 2008 to 71% in 2011 (Mohanasoundaram, 2011). Our findings were greater than those of Fazlul et al. (2011), who reported that the prevalence of MDR in P. aeruginosa isolated from catheterized patients’ urine was 40%. However, the result was 91.6% lower than that reported by Paranjothi and Dheepa (2010). This correlates with the fact that a growing number of places of the world are witnessing a rise in the MDR of P. aeruginosa, which raises a critical treatment challenge.
In our study, biofilm production of nosocomial MDR P. aeruginosa strains was estimated by 44/51 (86.27%) of MDR strains.
Our results are comparable to those of Jain and Agarwal (2009) and with a study undertaken by Bendouah et al. (2006), who reported that biofilm production was noted in 6/10 P. aeruginosa isolates. Yang et al. (2005) also supported our results, who declared that biofilm formation is an important phenotype associated with chronic P. aeruginosa infections. On the other hand, our results disagree with Delissalde and Amabile-Cuevas (2004), who found that only 14% of their P. aeruginosa isolates produced biofilm after 8 hours and 8% after 24 hours incubation.
The appropriate treatment of infected patients and the management of nosocomial spread of resistance are both dependent on reliable detection of the MBL-producing strains (Walsh et al., 2005). The prevalence of MBL-producing strains has increased significantly in Latin America (Sader et al., 2005). In Brazil, the prevalence of MBL-producing P. aeruginosa is ranging from 7.5% to 44% in different hospitals (Mendes et al., 2006).
In the present study, 8 out of 27 imipenem-resistant P. aeruginosa isolates were found to harbor blaVIM, while blaIMP was not discovered in any of them. This finding was in harmony with the results of Walsh et al. (2005), demonstrating that blaVIM was the most dominant MBL that mediated the imipenem-resistant properties of P. aeruginosa and constituted as the greatest clinical threat (Walsh et al., 2005). In 2012, 100 P. aeruginosa isolates from Shahid Motahari Hospital in Teheran were checked to detect blaIMP and blaVIM and forty-48 out of 83 (57.9%) imipenem-resistant P. aeruginosa showed MBL activity while 12% of them had only blaVIM gene (Fallah et al., 2013).
On the other hand, our results disagree with a study from Iran, which reported that 8 (9.75%) and 10 (12.19%) of MBL-producing P. aeruginosa isolates were positive for blaIMP-1 and blaVIM-1 genes, respectively (Sarhangi et al., 2013).
The presence of MBL genes was the widely accepted explanation for carbapenem resistance by hydrolyzing most beta-lactams such as imipenem and meropenem (Lagatolla et al., 2004). So, the detection of the MBL-producing strains was necessary for the proper treatment of infected patients and to decrease and prevent the nosocomial spread of resistance strains (Walsh et al., 2005).
In the present study, the age of patients with nosocomial MDRPsA infection ranged from 5 years to 75 years with male predominance (66.7%). This means a high prevalence of nosocomial MDRPA infection in old age. These results follow those obtained by Mahmoud et al. (2013) and Bashir et al. (2011), who reported that 54.8% of MDR P. aeruginosa strains were isolated from male patients.
On the other hand, a high prevalence of Pseudomonas infections was found in the 35–50 years age group. The incidence of MDR P. aeruginosa strains was more elevated in elderly males than elderly female patients (Mohanasoundaram, 2011)
Prior antibiotic treatment and the placement of a urinary catheter were the two most common risk factors for nosocomial MDR-PSA infection. Similar to our findings, previous trials showed that P. aeruginosa had a worse prognosis secondary to inappropriate therapy, reaching 53.8%, according to Morata et al. (2012). The use of multiple catheters, catheter insertion in emergency conditions, and prolonged use of catheters are the leading causes of nosocomial infections caused by several highly resistant nosocomial pathogens, including P. aeruginosa (Özdemir and Dizbay, 2015).
Our recommendations include cleaning our hospital ICUs thoroughly, implementing comprehensive antimicrobial stewardship plans with particular attention to carbapenem use, and using routine infection control methods to protect patients. The standardization of the phenotypic method used to detect MBL manufacturers necessitates more investigations. Despite the high cost of molecular methods used for the recognition of carbapenemase-producing isolates, they could be applied due to their high sensitivity and specificity as the presence of hidden MBL genes that are hard to be detected by phenotypic tests. This can lead to disseminating these MBL genes within workers who could act as transporters for MBL genes in the future.
CONCLUSION
Pseudomonas aeruginosa infections represented 18% of nosocomial infections in studied patients in different ICUs in Al-Azhar University Hospital. P. aeruginosa isolates were resistant to imipenem (36.5%) and meropenem (39.2%). The high prevalence of antibiotic resistance observed in our study was significant, leading to the emergence of MDR, extensive drug resistant, and pan-drug resistant isolates with worrisome percentages. Phenotypic methods for detecting carbapenemases and MBLs production in P. aeruginosa isolates could not identify the hidden MBL genes. So, molecular confirmation by PCR and analysis of different carbapenemase producers is required for all isolates.
ACKNOWLEDGMENT
The authors would like to express their gratitude to the Molecular Biology Research Unit at Assiut University in Egypt for providing the required resources for this study. This study was completely funded by the authors themselves.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FUNDING
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Al-Agamy MH, Shibl AM, Zaki SA, Tawfik AF. Antimicrobial resistance pattern and prevalence of metallo–lactamases in Pseudomonas aeruginosa from Saudi Arabia. Afr J Microbiol Res, 2011; 5:5528–33. CrossRef
Aminizadeh Z, Kashi MS. Prevalence of multidrug resistance and pandrug resistance among multiple Gram-negative species: experience in one teaching hospital, Tehran, Iran. Int Res J Microbiol, 2011; 2:90–5.
Arora D, Jindal N, Kumar R, Romit. Emerging antibiotic resistance in Pseudomonas-A challenge. Int J Pharm Pharm Sci, 2011; 3:82–4.
Ashour HM, El-Sharif A. Species distribution and antimicrobial susceptibility of Gram-negative aerobic bacteria in hospitalized cancer patients. J Transl Med, 2009; 7:14:1–13. CrossRef
Bashir D, Thokar MA, Fomda BA, Bashir G, Zahoor D, Ahmad S, Toboli AS. Detection of metallo-beta-lactamase (MΒL) producing Pseudomonas aeruginosa at a tertiary care hospital in Kashmir. Afr J Microbiol Res, 2011; 5:164–72.
Bendouah Z, Barbeau J, Hamad WA, Desrosiers M. Biofilm formation by Staphylococcus aureus and Pseudomonas aeruginosa is associated with an unfavorable evolution after surgery for chronic sinusitis and nasal polyposis. Otolaryngol Head Neck Surg, 2006; 134:991–6. CrossRef
Delissalde F, Amábile-Cuevas CF. Comparison of antibiotic susceptibility and plasmid content, between biofilm producing and non-producing clinical isolates of Pseudomonas aeruginosa. Int J Antimicrob Agents, 2004; 24:405–8. CrossRef
Fallah F, Borhan RS, Hashemi A. Detection of bla (IMP) and bla (VIM) metallo-β-lactamases genes among Pseudomonas aeruginosa strains. Int J Burn Trauma, 2013; 3:122–4.
Fazlul M, Zaini M, Rashid M, Nazmul M. Antibiotic susceptibility profiles of clinical isolates of Pseudomonas aeruginosa from Selayang Hospital, Malaysia. Biomed Res, 2011; 22:263–6.
Franco MRG, Caiaffa-Filho HH, Burattini MN, Rossi F. Metallo-beta-lactamases among imipenem-resistant Pseudomonas aeruginosa in a Brazilian University Hospital. Clinics, 2010; 65:825–9. CrossRef
Gad GF, El-Domany RA, Zaki S, Ashour HM. Characterization of Pseudomonas aeruginosa isolated from clinical and environmental samples in Minia, Egypt: prevalence, antibiogram and resistance mechanisms. J Antimicrob Chemoth, 2007; 60:1010–7. CrossRef
Govan JW. Pseudomonas and non-fermenters. In: Greenwood D, Slack R, Peutherer J, Barer M (eds.). Medical microbiology. A guide to microbial infections: pathogenesis, immunity, laboratory diagnosis and control. 7th edition, Churchill Livingstone-Elsevier, London, UK, pp 293–9, 2007.
Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharm Out Res, 2010; 10:3717–22. CrossRef
Jain A, Agarwal A. Biofilm production, a marker of pathogenic potential of colonizing and commensal staphylococci. J Microbiol Methods, 2009; 76:88–92. CrossRef
Kotsakis SD, Papagiannitsis CC, Tzelepi, E, Legakis NJ, Miriagou V, Tzouvelekis LS. GES-13, a β-lactam variant possessing Lys-104 and Asn-170 in Pseudomonas aeruginosa. Antimicrob Agents Chemother, 2010; 54:1331–3. CrossRef
Lagatolla C, Tonin EA, Monti-Bragadin C, Dolzani L, Gombac F, Bearzi C, Rossolini GM. Endemic carbapenem-resistant Pseudomonas aeruginosa with acquired metallo-β-lactamase determinants in European hospital. Emerg Infect Dis, 2004; 10:535–8. CrossRef
Landman D, Quale JM, Mayorga D, Adedeji A, Vangala K, Ravishankar J, Brooks S. Citywide clonal outbreak of multiresistant Acinetobacter baumannii and Pseudomonas aeruginosa in Brooklyn, NY: the preantibiotic era has returned. Arch Intern Med, 2002; 162:1515–20. CrossRef
Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev, 2009; 22:582–610. CrossRef
Mahmoud AB, Zahran WA, Hindawi GR, Labib AZ, Galal R. Prevalence of multidrug-resistant Pseudomonas aeruginosa in patients with nosocomial infections at a university hospital in Egypt, with special reference to typing methods. J Virol Microbiol, 2013; 13:165–59. CrossRef
Manjunath G, Prakash R, Vamseedhar Annam KS. Changing trends in the spectrum of antimicrobial drug resistance pattern of uropathogens isolated from hospitals and community patients with urinary tract infections in Tumkur and Bangalore. Int J Biol Med Res, 2011; 2:504–7.
Mansour S, Eldaly O, Jiman Fatani A, Mohamed M, Ibrahim E. Epidemiological characterization of P. aeruginosa isolates of intensive care units in Egypt and Saudi Arabia. East Mediter Health J, 2013; 19:71–80. CrossRef
Mendes RE, Castanheira M, Pignatari ACC, Gales AC. Metallo-beta-lactamases. J Bras Patol Med Lab, 2006; 42:103–13. CrossRef
Mohanasoundaram K. The antimicrobial resistance pattern in the clinical isolates of Pseudomonas aeruginosa in a tertiary care hospital; 2008–2010 (A 3 year study). Clinic Diagnost Res J, 2011; 5:491–4.
Morata L, Cobos-Trigueros N, Martínez JA, Soriano Á, Almela M, Marco F, Mensa J. Influence of multidrug resistance and appropriate empirical therapy on the 30-day mortality rate of Pseudomonas aeruginosa bacteremia. Antimicrob Agents Chemother, 2012; 56:4833–7. CrossRef
Özdemir K, Dizbay M. Nosocomial infection and risk factors in elderly patients in intensive care units. J Microbiol Infect Dis, 2015; 5:38–43. CrossRef
Paranjothi S, Dheepa R. Screening for multidrug resistance bacteria Pseudomonas aeruginosa in hospitalized patients in Hosur, Krishnagiri (dt). Int J Pharm Bio Sci, 2010; 1:10–9.
Picao RC, Poirel L, Gales AC, Nordmann P. Diversity of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolates causing bloodstream infections in Brazil. Antimicrob Agents Chemother, 2009; 53:3908–13. CrossRef
Rodrigues ACS, Chang MR, Nóbrega GD, Rodrigues MS, Carvalho NCP, Gomes BG, Asensi MD. Metallo-β-lactamase and genetic diversity of Pseudomonas aeruginosa in intensive care units in Campo Grande, MS, Brazil. Braz J Infect Dis, 2011; 15:195–9. CrossRef
Ruppé É, Woerther PL, Barbier F. Mechanisms of antimicrobial resistance in Gram-negative Bacilli. Ann Intensive Care, 2015; 5:21–36. CrossRef
Sader HS, Castanheira M, Mendes RE, Toleman M, Walsh TR, Jones RN. Dissemination and diversity of metallo-β-lactamases in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Int J Antimicrob Agents, 2005; 25:57–61. CrossRef
Sarhangi M, Motamedifar M, Sarvari J. Dissemination of Pseudomonas aeruginosa producing blaIMP1, blaVIM2, blaSIM1, blaSPM1 in Shiraz, Iran. Jundishapur J Microbiol, 2013; 6:e6920–5. CrossRef
Sekiguchi JI, Asagi T, Miyoshi-Akiyama T, Fujino T, Kobayashi I, Morita K, Kirikae T. Multidrug-resistant Pseudomonas aeruginosa strain that caused an outbreak in a neurosurgery ward and its aac (6′)-Iae gene cassette encoding a novel aminoglycoside acetyltransferase. Antimicrob Agents Chemother, 2005; 49:3734–42. CrossRef
Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, Arakawa Y. PCR typing of genetic determinants for metallo-β-lactamases and integrases carried by Gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J Clin Microbiol, 2003; 41:5407–13. CrossRef
Stepanovic S, Vukovic D, Hola V, Di Bonaventura G, Djukic S, Cirkovic I, Ruzicka F. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS, 2007; 115(8):891–9. CrossRef
Stewart PS, Mukherjee PK, Ghannoum MA. Biofilm antimicrobial resistance. In: Ghannoum M, GA O’Toole (eds.). Microbial biofilms. 1st edition, ASM Press, Washington, DC, pp 250–68, 2004. CrossRef
Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev, 2005; 18:306–25. CrossRef
Wassef M, Mahallawy H, Zafer M, Ghaith D, Hamid R. Lab based surveillance of multidrug resistant Pseudomonas aeruginosa in Cairo University Hospitals. Egypt J Microbiol Exp, 2015; 2:39–43. CrossRef
Wayne P. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne, PA, vol 31, pp 100–121, 2011.
Yang W, Shi L, Jia WZ, Yin X, Su JY, Kou Y, Yi X, Shinoda S, Miyoshi SI. Evaluation of the biofilm-forming ability and genetic typing for clinical isolates of Pseudomonas aeruginosa by enterobacterial repetitive intergenic consensus-based PCR. Microbiol Immunol, 2005; 49:1057–61. CrossRef