INTRODUCTION
Bacteria are becoming increasingly resistant to conventional antibiotics, resulting in the therapeutic failure of many antibiotics, even newly introduced agents (Cornaglia, 2009; Tan, 2008). The emerging crisis of antibiotic resistance throughout the world is largely attributed to poor patient compliance to treatment guidelines, overprescription, and misuse of anti-infective agents in medical institutions and antibiotic abuse in agriculture (Palumbi, 2001). Additionally, several pharmaceutical companies abandoned the market of antibiotics (Alanis, 2005). Several approaches have been proposed to tackle the development of antibiotic resistance, such as the introduction of combined therapies targeting multiple bacterial targets, the evolvement of bacteriophage therapies, microbiota transplantation, and targeting of key resistance players, such as inhibition of drug-modifying enzymes and drug efflux pumps (Kaur, 2016). However, these approaches resulted in limited success, typically due to cost, safety issues, and/or a narrow spectrum of efficacy. Therefore, increasing demand for new types of antimicrobial therapies to manage infectious diseases and bacterial resistance is reshaping drug discovery research and development.
Antimicrobial peptides (AMPs) are broadly expressed in nature, and they have been found in most living species as a part of the innate immune response against various pathogenic bacteria, viruses, fungi, and parasites and hence frequently referred to as host defense peptides (HDPs) (Bahar and Ren, 2013; Diamond et al., 2009). AMPs are short (6 to 100 amino acids) peptides, exhibiting a net positive charge and adopting amphipathic structures (Giuliani et al., 2007; Hartmann et al., 2010). HDPs can be categorized depending on their adopted structures into four subclasses: α-helical, ?exible, β-sheet, and loop peptides (Lee et al., 2015). The most predominant subgroup of AMPs is amphipathic α-helices such as maculatins, citropin, and melittin (Zhang et al., 2016). AMPs offer promising activities to tackle infections produced by pathogens resistant to conventional antibiotics. These molecules display broad-spectrum activities against pathogenic microorganisms by interacting with and disrupting bacterial cell membranes. Moreover, they can interfere with macromolecule synthesis like DNA and proteins (Kumar et al., 2018).
Mounting research is currently directed towards AMPs design, aiming for the development of lead entities in antimicrobial drug discovery and overcoming limitations associated with naturally existing peptides, particularly toxicity, and spectrum of activity. Additionally, several combinations of selected AMP and conventional antibiotics have been proposed, showing synergistic effects as well as reduced drug toxicity (Almaaytah et al., 2018). Mounting evidence suggests that AMPs influence the integrity of the bacterial membrane, thereby enhancing the permeability and sensitivity of bacterial strains towards antibiotics in combinations (Guilhelmelli et al., 2013; Zhang et al., 2014).
Brevinin-1E (FLPLLAGLAANFLPKIFCKITRKC) is a membrane-active agent that is isolated from the granular glands in the skins of the European frog Rana species (Zohrab et al., 2019). Brevinin-1E is characterized by a broad range of activities against various bacterial strains. Also, it displays strong hemolytic activity against erythrocytes which hinders its application as a therapeutic antimicrobial agent (Rinaldi, 2002). On the other hand, bombolitin IV (INIKDILAKLVKVLGHV) is expressed in the Megabombus pennsylvanicus species, where it is secreted into the bumblebee venom (Roberson et al., 2019). Similarly, it possesses a broad-spectrum antimicrobial that is accompanied by hemolytic effects that limit its use.
In this research, we designed a novel peptide that comprises merged active fragments of the brevinin-1E and bombolitin IV peptides to create a hybrid peptide named the SAMA peptide possessing increased antimicrobial effects and improved safety profile in comparison with the parent peptides. Our peptide SAMA displayed promising antimicrobial and antibiofilm activities when tested on a broad range of selected bacterial strains and importantly exhibited lower hemolytic activity compared to the parent peptides.
MATERIALS AND METHODS
Cell lines
Bacterial strains were obtained from the American Type Tissue Culture Collection (ATCC) and these involve two Gram-positive bacteria, methicillin-susceptible Staphylococcus aureus (ATCC 29215) and methicillin-resistant S. aureus (ATCC BAA-41). Additionally, two Gram-negative strains involving susceptible Escherichia coli (ATCC 25922) and the drug-resistant strain E. coli (ATCC BAA 2452) were also obtained. Adenocarcinoma human alveolar basal epithelial cells (A549) were also purchased from the ATCC (ATCC CCL-185).
Antibiotics
Levofloxacin, chloramphenicol, and doxycycline were purchased from Sigma-Aldrich, China. Ampicillin was obtained from Sigma-Aldrich, Canada. Rifampicin was obtained from Bio Basic, Canada. Stock solutions were made as per the manufacturer’s instructions and stored at the recommended storage temperature for each antibiotic.
Synthesis and purification of SAMA
The SAMA peptide (NH2-GRLIDKIARKLVKKIQRFARKFF-COOH) was obtained from GL Biochem (Shanghai, China). The solid-phase methods and Fmoc chemistry were used to synthesize the peptide, followed by reverse-phase high performance liquid chromatography for peptide purification by 95.5%. Electrospray ionization mass spectrometry was used to verify the identity of the peptide.
Bioinformatic analysis
The estimated helicity of SAMA was predicted using the Network Protein Sequence Analysis software. On the other hand, the HeliQuest software was used to calculate SAMA hydrophobicity (H) and hydrophobic moment (μH). However, molecular weight, water solubility, isoelectric point, and net charge at neutral pH for the hybrid peptide were predicted using Innovagen’s peptide calculator. Besides, Antimicrobial Peptide Calculator and Predictor tool in Antimicrobial Peptide Database (APD3) was used for the estimation of the protein-binding potential (Boman index) of the hybrid peptide. Model reliability confirmation and 3D structure prediction were carried out using the I-TASSER software. Server webpages were accessed in the period of 02/2020–09/2020.
Bacterial susceptibility assay
The antibacterial activity of the SAMA peptide was assessed by measuring the minimum inhibitory concentration (MIC) and the minimum bactericidal concentration (MBC) using the broth microdilution method outlined by the Clinical and Laboratory Standards Institute guidelines (Brown et al., 2005). In brief, different strains of bacteria at a 106 CFU/ml concentration were prepared by diluting overnight cell culture using a fresh Mueller Hinton Broth. 50 μl of diluted bacterial suspension was mixed with 50 μl of different peptide concentrations in 96-well microtiter plates. The growth of bacteria was assessed by determining optical density at λ = 600 nm. MBC was determined by sampling of 10 μl from each well, spreading into Mueller Hinton Agar (Oxoid Ltd., Basingstoke, UK), and incubating at 37°C for 24 hours before reading the MBCs. The MBC was determined as the lowest concentration of the peptide that is needed to diminish the number of survival bacterial cells to <0.1% after 24 hours of incubation.
Antibiofilm activity
Attenuation of biofilm formation was examined as described previously (Luca et al., 2013), using the Calgary Biofilm Device. Briefly, 150 μl of the bacterial inoculum was added to each well of the 96-microtiter plate. Lids containing pegs were placed into the 96-microtiter plate on which biofilm-forming cells can adhere and grow, followed by orbital shaking at 125 rpm and incubation at 37°C for 20 hours. Plates were next rinsed three times with 200 μl of phosphate-buffered saline to eliminate planktonic cells. Next, pegs-lids were moved into a “challenge 96-well microtiter plate” containing peptide solutions (200 μl) at different concentrations, incubated at 37°C for 2 hours, and then transferred into a recovery plate. The growth of recovered bacteria was examined by determining the absorbance of the plate at wavelength of 595 define the minimum biofilm eradication concentration (MBEC).
Synergistic checkerboard assay
Synergistic effects of combinations of SAMA and selected antibiotics (levofloxacin, chloramphenicol, doxycycline, ampicillin, and rifampicin) were examined against four bacterial strains according to previous studies (Dundar and Otkun, 2010; Orhan et al., 2005). Briefly, different antibiotic and peptide concentrations (25 μl each) were added to 50 μl of diluted bacterial suspension in 96-well plates. After incubation for 18 hours at 37°C, absorbance was measured at 600 nm. The fractional inhibitory concentrations (FICs) index was calculated according to the following equation:
When the FIC index was ≤0.5, the combination was considered synergistic. However, it was considered additive when 0.5 ≤ FICs ≤ 1, indifferent when 1 < FICs ≤ 4, and antagonistic when 4 < FICs.
Hemolytic assay
2 ml of human blood (Sigma-Aldrich, St. Louis, MO) was added to 48 ml of phosphate-buffered saline to reach a final concentration of 4% red blood cell (RBC) and centrifuged for 5 minutes at 2,000 rpm. The process was repeated twice by aspirating the supernatant and replacing it with a fresh buffer. 2 ml of different peptide solution concentrations were then mixed with 2 ml of RBC suspension and then incubated for 60 minutes at 37°C. Triton X-100 (Santa Cruz Biotechnology, Dallas, TX) was used as a positive control. After incubation and gentle vortexing, 1 ml was transferred from each sample and then centrifuged at 2,000 rpm for 5 minutes and then 200 μl was transferred from each supernatant into a 96-well plate, where absorbance was measured at 450 nm. Hemolysis percentage was measured using the following equation:
where “A” is the absorbance value of the tested solutions, “An” is the absorbance value of the negative control, and “Ap” is the absorbance value of the positive control.
MTT assay
As described previously (Nosrati et al., 2020), briefly, adenocarcinoma human alveolar basal epithelial cells (A549) were cultured in the F-12K Medium provided with 10% of fetal bovine serum and 1% of a penicillin-streptomycin solution at 37°C. Cells were seeded at a concentration of 7 × 103 cells/well in a 96-well plate and incubated overnight at 37°C under 5% CO2 with different concentrations of the peptide (100, 80, 60, 40, 20, 10, 5, and 1 μM). Then, 10 μl MTT solution (5 mg/ml in serum-free media) was added to each well and then the plates were incubated again at 37°C, 5% CO2, and humidity of 95% for 4 hours. Media were then aspired and 200 μl of dimethyl sulfoxide per well was added. Plates were shaken for 1 minute and absorbance was measured at wavelength 590 nm. The viability percentage was determined according to the following equation:
The IC50 was statistically estimated using the GraphPad Prism software (V8.4).
RESULTS
Peptide design and characterization
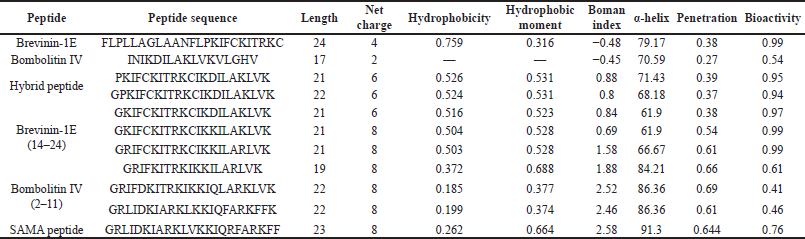
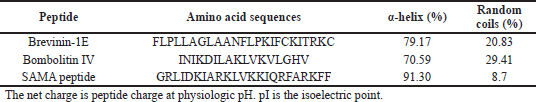
The SAMA peptide comprises 23 amino acids. The design started with the hybridization of the peptide region (14–24)of brevinin-1E to form the N-terminus of our peptide and the peptide region (2–11) of bombolitin IV to form its C-terminus. The in silico characterization of the hybrid peptide suggested the need for further sequence modification to optimize the physicochemical parameters essential for antimicrobial activity, such as helicity, hydrophobicity, and net charge. Therefore, a series of amino acid substitutions and additions were performed to optimize the parameters essential for antimicrobial activity as predicted by in silico characterization of derivatives (Supplementary Tables 1 and 2),until a final sequence of the SAMA peptide was obtained with satisfactory physicochemical characters (Tables 1 and 2).
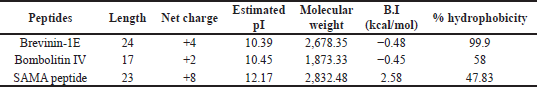
The SAMA peptide displayed features essential for beneficial antimicrobial activity: higher helicity than parent peptides, a net charge of +8 at physiological pH, and intermediate level of hydrophobicity. The helical structure as predicted by the I-TASSER software (Zhang, 2008) of the SAMA peptide is depicted in Figure 1.
 | Supplementary Table 1. Predicted physicochemical parameters of parents’ peptides and modified hybrid peptide derivatives. [Click here to view] |
 | Table 1. Sequences and estimated α-helical percentage of the parent peptides (brevinin-1E and bombolitin IV) and the SAMA peptide. [Click here to view] |
 | Table 2. In silico characterization of the SAMA peptide compared with parent peptides. [Click here to view] |
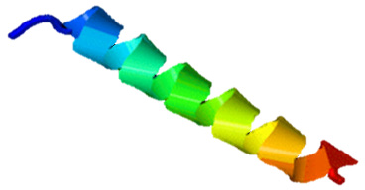 | Figure 1. Predicted 3D structure of the SAMA hybrid peptide. The structure was created by the I-TASSER software. [Click here to view] |
Antimicrobial activity of SAMA
To evaluate the activity and the spectrum of SAMA’s antimicrobial effects, four different strains of bacteria were used: a sensitive strain of Gram-positive S. aureus (ATCC 29215) and a multidrug-resistant S. aureus (ATCC BAA-41), along with sensitive and multi-drug-resistant strains of Gram-negative E. coli (ATCC 25922 and ATCC 2452, respectively). The SAMA peptide exhibits significant antimicrobial effects against Gram-positivestrains, with MIC and MBC values of 5–15 μM (Table 3). Additionally, SAMA was more potent against Gram-negative strains with MIC and MBC values of 5 μM. Together, these results demonstrated a broad-spectrum bactericidal activity of the SAMA peptide.
Antibiofilm activity of SAMA
Biofilm formation is among the key bacterial virulent factors through which bacterial strains acquire their resistance to a broad array of antibiotics. Therefore, we investigated the potential attenuation of biofilm formation in bacteria upon SAMA treatment to determine the MBEC. Indeed, the recovery and growth of bacteria from SAMA-treated S. aureus (ATCC 29215) and S. aureus (ATCC BAA-41) biofilms were attenuated even after 8 hours of incubation in recovery media. While a peptide concentration up to 40 μM was sufficient to attenuate biofilm formation in Gram-positive strains (Table 4), biofilm-forming Gram-negative strains exhibited more resistance to SAMA activity, and concentrations up to 120 μM were not sufficient to attenuate biofilm formation.
Synergistic antimicrobial activity of SAMA
Given that modern therapeutics often relies on combined regimens that allow the use of lower doses, maximize the effects, and minimize drug toxicities, we next investigated potential additive or synergistic effects of SAMA when combined with selected antibiotics using the checkerboard microtiter plate assay. We determined MIC values of the antibiotics alone and then MICs of SAMA-antibiotic combinations. As summarized in Table 5, the calculated MICs of both SAMA and the selected drugs reduced dramatically in combinations with various percentages, and many combinations displayed synergistic rather than additive effects on the planktonic cells of the studied strains.
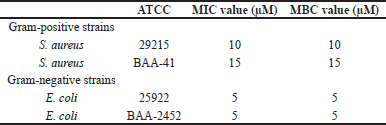 | Table 3. MIC and MBC of the SAMA peptide against different bacterial strains. [Click here to view] |
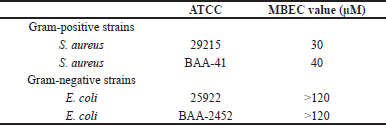 | Table 4. MBEC of the SAMA peptide on different strains. [Click here to view] |
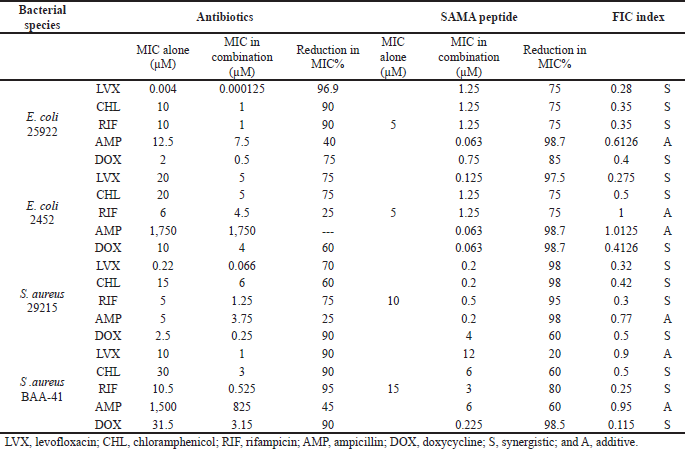 | Table 5. MICs of SAMA and antibiotics and the calculated FIC. [Click here to view] |
While the levofloxacin combination with SAMA displayed synergistic effects, with a more substantial reduction in MIC values against Gram-negative than Gram-positive strains, this combination exhibited additive effects against the methicillin-resistant Gram-positive strain, with an FIC score of 0.9. On the other hand, the chloramphenicol-SAMA combination displayed synergistic effects on all tested bacterial strains, with MICs reduction percentages ranging from 60% to 90%. Similarly, the doxycycline-SAMA combination displayed synergistic effects on tested strains, with various percentages of reductions in MICs. Importantly, both rifampicin and ampicillin showed neither additive nor synergistic effects on multi-drug-resistant E. coli strain, while their combinations with SAMA displayed synergistic and additive effects on the other strains, respectively. Together, these data demonstrated that the beneficial antimicrobial activity of SAMA could be further exploited to lower drug resistance rates commonly reported with other antibiotics.
Toxicity evaluation of SAMA
Many cationic peptides are known for their hemolytic toxicity on RBCs. Therefore, we next examined the safety profiles of the SAMA peptide at its reported MIC and MBC. The in vitro hemolytic assay showed minimal hemolytic activity of SAMA at concentrations up to 10 μM (Table 6). Incubation of RBCs with higher concentrations of SAMA resulted in increased hemolytic toxicity that approaches 35% with concentrations beyond 65 μM. These results suggested a low toxicity profile of SAMA at therapeutic concentrations required for antimicrobial activity.
 | Table 6. SAMA hemolytic effects on human RBCs upon exposure time of 60 minutes at different concentrations. [Click here to view] |
Potential anticancer activity of SAMA
Several AMP exhibited anticancer activity against various cancer cell lines. In this direction, we examined the potential anticancer activity of the SAMA peptide by examining its effects on the viability of adenocarcinomic human alveolar basal epithelial cells (A549). As shown in Figure 2, SAMA displayed potent antiproliferative effects on A549 cells, with an IC50 around 4.5 μM.
DISCUSSION
Antimicrobial resistance exemplifies one of the most challenging health problems nowadays. Therefore, it becomes imperative to direct efforts towards the development of antimicrobial therapies with improved safety and efficacy to tackle infections produced by drug-resistant strains (Aslam et al., 2018). AMPs have emerged as promising treatment options (Jenssen et al., 2006), and mounting research is currently directed to designing and developing novel peptides for managing bacterial infections. Here, we designed and characterized a novel peptide derived from the sequences of two natural AMPs, which demonstrated promising antimicrobial and anticancer activities.
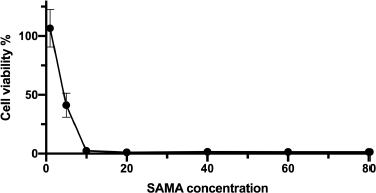 | Figure 2. Growth curve of A549 cells after treatment with different concentrations (μM) of the SAMA peptide. Values represent the means (±SD) of six different experiments. [Click here to view] |
Hybridization is one of the peptidomimetics strategies used to reduce toxicity and enhance the activity of antimicrobials of AMPs (Wang et al., 2015). This approach relies on combining the bioactive fragments of natural AMPs. The resulting hybrid peptide can be further modified with selected point mutations for improved physicochemical properties and enhanced biological activity. In this direction, we designed a hybrid peptide (PKIFCKITRKCIKDILAKLVK) comprising the bioactive fragments obtained from 14 to 24 of brevinin-1E and from 2 to 11of bombolitin IV. Accumulated evidence suggests that the antimicrobial activity for peptides is predominantly a function of helicity content, hydrophobicity, and the net charge, wherein higher helicity, lesser than 50% hydrophobicity, and a cationic net charge (~ +7) are features of a biologically active peptide. Therefore, the sequence of the hybrid peptide was modified, and derivatives were in silico characterized (Supplementary Tables 1 and 2) to result in the SAMA peptide with a sequence of GRLIDKIARKLVKKIQRFARKFF. SAMA comprises 23 amino acids with an enhanced helicity percentage of 91.3% (Table 1; Fig. 1). The modified SAMA peptide contains nine positively charged residues including five lysine and four arginine residues, with one aspartic acid residue contributing one negative charge, bringing the net charge to +8, which is sufficient for the initial electrostatic interaction with the negatively charged phospholipids on the bacterial membranes. SAMA also contains 11 hydrophobic residues (1 valine, 2 alanine, 2 leucine, 3 phenylalanine, and three isoleucine) with a total hydrophobic percentage of 47.83% with a hydrophobic momentum of 72% reflecting the amphipathicity of the peptide, which facilitated the antimicrobial activity, with minimal hemolytic toxicity. Several studies indicated that less than 50% hydrophobicity had been determined as an optimum value for AMPs to bind and disorder the integrity of bacterial membranes (Yin et al., 2012), whereas a higher level of hydrophobicity results in a decrease in the antimicrobial specificity (Chen et al., 2007). A high Boman index value indicates that SAMA would hit multi-intracellular targets in bacteria, owing to a high propensity for interaction with a wide range of proteins.
Upon in vitro functional characterization, the SAMA peptide effectively attenuated the growth of Gram-negative bacteria at an MIC of 5 μM. With a lesser potency, it inhibited the growth of Gram-positive bacteria within the range of 10–15 μM. The difference in the composition of the cell wall seems to affect the permeability of the SAMA peptide. Therefore, the interaction of the SAMA peptide with teichoic acids in Gram-positive bacteria may act as a trap for it on its way to the cytoplasmic membrane, resulting in a decreased peptide concentration on the inner membrane (Malanovic and Lohner, 2016). In addition, the MBC values of the SAMA peptide were similar to the MIC values, suggesting a bactericidal antimicrobial activity. In addition to microbial growth inhibition, we examined the antibiofilm activity of the SAMA peptide, which was solely efficient against Gram-positive strains, but not Gram-negative ones. These findings could be explained by the unique biochemistry of the E. coli outer membranes. It has been reported that E. coli secretes an anionic polysaccharide identified as alginate, which in turn can interact with cationic AMPs, resulting in the capture and the neutralization of these peptides and subsequent protection of E. coli in the biofilm from the bactericidal actions of positively charged AMPs (Yasir et al., 2018). Moreover, Gram-negative bacteria such as E. coli have a protective genetic system to add aminoarabinose to lipid A that results in decreasing the outer membrane net negative charge. Indeed, it was shown that modification of lipid A attenuated the electrostatic attraction of cationic AMPs toward the Gram-negative bacteria (Band and Weiss, 2015).
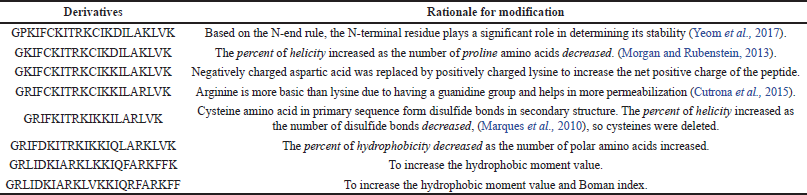 | Supplementary Table 2. Amino acid sequences of the modified hybrid peptide and the reason for the modification. [Click here to view] |
Different antibiotic combinations were evaluated to reduce the toxicity and improve the antibacterial activity of both the SAMA peptide and individual conventional antibiotics. All tested antibiotics are active intracellularly except ampicillin, which inhibits bacterial growth by targeting the bacterial cell wall. Therefore, the combination of the SAMA peptide with ampicillin displays no synergistic effects, because of the different sites of actions of the two peptides. Other combinations, however, showed synergistic or at least additive effects. Based on the mechanism of action that is suggested for the SAMA peptide, it may affect the integrity of the bacterial membrane through pore formation resulting in enhancing the permeability of bacterial cells and increasing influx and uptake of antibiotics into the cells to accomplish the destruction process. As a result, more intracellular accumulation of antibiotics is expected and hence the increased sensitivity of the bacterial strains, which are normally resistant to conventional antibiotics.
Parallel to functional characterization, the SAMA peptide toxicity was investigated for potential hemolytic activity against RBCs. Hemolysis is a common side effect for cationic peptides and is usually linked to peptide hydrophobicity and amphipathicity. At therapeutic concentrations, SAMA displayed minimal hemolytic activity of lesser than 10% and demonstrated a safer profile compared to the parent peptides (brevinin-1E and bombolitin IV), which exhibited significant hemolytic effects at even lower concentrations (Son et al., 2003; Waghu et al., 2016). This could be attributed to the more abundant polar amino acids in the SAMA N-terminus that could contribute to breaking the nonpolar side of the helix upon binding the RBC membrane, the feature that is deemed essential for peptide-mediated hemolytic activity.
Several studies have demonstrated that AMPs possess promising anticancer activity against several cancer cell types. Importantly, the antiproliferative activity was selective for cancer cells over the normal cell (Papo and Shai, 2005). Here, the anticancer activity of the SAMA peptide was examined against adenocarcinomic human alveolar basal epithelial cells. SAMA proved to be a potential anticancer agent against A549 cells (Fig. 2). This could be explained by the mechanism suggested for melittin and the defensins against cancer cells (Hanaoka et al., 2016; Moreno and Giralt, 2015). These peptides bind to the cancer cell membrane, destabilizing it, causing their necrotic death. Mounting evidence suggests that these peptides would display selectivity toward cancerous cells, as the cytoplasmic membrane of cancer cells contains more abundant negatively charged phosphatidylserine in addition to O-glycosylated mucins (Deslouches and Di, 2017). Another explanation is the presence of a high percentage of microvilli on cancerous cells relative to the normal cells and, hence, increased surface area on cancerous cells and increased accumulation and binding of the peptide on cell surface (Papo and Shai, 2005).
CONCLUSION
In summary, we have designed and characterized a modified hybrid peptide using the sequences of brevinin-1E and bombolitin IV. SAMA exhibited improved antimicrobial effects on planktonic Gram-negative and Gram-positive bacterial cells with a promising safety profile. SAMA displayed a low hemolytic effect against mammalian erythrocytes. Furthermore, lower MICs and hence improved safety for SAMA were achieved upon the combination with five kinds of conventional antibiotics. In addition, SAMA exhibited antibiofilm activities, albeit at higher concentrations, extending the use of peptides for nonautoclavable inanimate surfaces.
ACKNOWLEDGMENTS
This work was funded by the Deanship of Scientific Research at Jordan University of Science and Technology (Grant no. 20190066).
AUTHORS’ CONTRIBUTIONS
M. Alsaggar and Y. Al Tall conceptualized the study. A. Al-Tarawneh and M. Al-Hazabreh conducted the methodology. M. Alsaggar and A. Al-Tarawneh carried out the analysis and investigation. M. Alsaggar and Y Al Tall wrote and edited the manuscript. M. Alsaggar acquired the funding. All authors have read and agreed to the published version of the manuscript.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Alanis AJ. Resistance to antibiotics: are we in the post-antibiotic era? Arch Med Res, 2005; 36(6):697–705. CrossRef
Almaaytah A, Qaoud MT, Abualhaijaa A, Al-Balas Q, Alzoubi KH. Hybridization and antibiotic synergism as a tool for reducing the cytotoxicity of antimicrobial peptides. Infect Drug Resist, 2018; 11:835. CrossRef
Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, Nisar MA, Alvi RF, Aslam MA, Qamar MU, Salamat MKF, Baloch Z. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist, 2018; 11:1645–58. CrossRef
Bahar A, Ren D. Antimicrobial peptides. Pharmaceuticals, 2013; 6(12):1543–75. CrossRef
Band VI, Weiss DS. Mechanisms of antimicrobial peptide resistance in Gram-negative bacteria. Antibiotics, 2015; 4(1):18–41. CrossRef
Brown DF, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, Wren MW. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J Antimicrob Chemother, 2005; 56(6):1000–18. CrossRef
Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob Agents Chemother, 2007; 51(4):1398–406. CrossRef
Cornaglia G. Fighting infections due to multidrug-resistant Gram-positive pathogens. Clin Microbiol Infect, 2009; 15(3):209–11. CrossRef
Cutrona KJ, Kaufman BA, Figueroa DM, Elmore DE. Elmore. Role of arginine and lysine in the antimicrobial mechanism of histone-derived antimicrobial peptides. FEBS Lett, 2015; 589(24 Pt B):3915–20. CrossRef
Deslouches B, Di YP. Antimicrobial peptides with selective antitumor mechanisms: prospect for anticancer applications. Oncotarget, 2017; 8(28):46635–51. CrossRef
Diamond G, Beckloff N, Weinberg A, Kisich KO. The roles of antimicrobial peptides in innate host defense. Curr Pharm Des, 2009; 15(21):2377–92. CrossRef
Dundar D, Otkun M. In-vitro efficacy of synergistic antibiotic combinations in multidrug resistant Pseudomonas aeruginosa strains. Yonsei Med J, 2010; 51(1):111–6. CrossRef
Giuliani A, Pirri G, Nicoletto S. Antimicrobial peptides: an overview of a promising class of therapeutics. Open Life Sci, 2007; 2(1):1–33. CrossRef
Guilhelmelli F, Vilela N, Albuquerque P, Derengowski L, Silva-Pereira I, Kyaw C. Antibiotic development challenges: the various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front Microbiol, 2013; 4:353. CrossRef
Hanaoka Y, Yamaguchi Y, Yamamoto H, Ishii M, Nagase T, Kurihara H, Akishita M, Ouchi Y. In vitro and in vivo anticancer activity of human β-defensin-3 and its mouse homolog. Anticancer Res, 2016; 36(11):5999–6004. CrossRef
Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob Agents Chemother, 2010; 54(8):3132–42. CrossRef
Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev, 2006; 19(3):491–511. CrossRef
Kaur I. Novel strategies to combat antimicrobial resistance. J Infect Dis Ther, 2016; 4(4):292. CrossRef
Kumar P, Kizhakkedathu JN, Straus SK. Antimicrobial peptides: diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules, 2018; 8(1):4. CrossRef
Lee HT, Lee CC, Yang JR, Lai JZ, Chang KY. A large-scale structural classification of antimicrobial peptides. BioMed Res Int, 2015; 2015:1–6. CrossRef
Luca V, Stringaro A, Colone M, Pini A, Mangoni ML. Esculentin (1-21), an amphibian skin membrane-active peptide with potent activity on both planktonic and biofilm cells of the bacterial pathogen Pseudomonas aeruginosa. Cell Mol Life Sci, 2013; 70(15):2773–86. CrossRef
Malanovic N, Lohner K. Antimicrobial peptides targeting Gram-positive bacteria. Pharmaceuticals, 2016; 9(3):59. CrossRef
Marques JR, Fonseca RR, Drury B, Melo A. Amino acid patterns around disulfide bonds. Int J Mol Sci, 2010; 11(11):4673–86. CrossRef
Moreno M, Giralt E. Three valuable peptides from bee and wasp venoms for therapeutic and biotechnological use: melittin, apamin and mastoparan. Toxins (Basel), 2015; 7(4):1126–50. CrossRef
Morgan AA, Rubenstein E. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One, 2013; 8(1):e53785. CrossRef
Nosrati H, Khodaei M, Banitalebi-Dehkordi M, Alizadeh M, Asadpour S, Sharifi E, Ai J, Soleimannejad M. Preparation and characterization of poly(ethylene oxide)/zinc oxide nanofibrous scaffold for chronic wound healing applications. Polym Med, 2020; 50(1):41–51. CrossRef
Orhan G, Bayram A, Zer Y, Balci I. Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis. J Clin Microbiol, 2005; 43(1):140–3. CrossRef
Palumbi SR. Humans as the world’s greatest evolutionary force. Science, 2001; 293(5536):1786–90. CrossRef
Papo N, Shai Y. Host defense peptides as new weapons in cancer treatment. Cell Mol Life Sci, 2005; 62(7–8):784–90. CrossRef
Rinaldi AC. Antimicrobial peptides from amphibian skin: an expanding scenario: commentary. Curr Opin Chem Biol, 2002; 6(6):799–804. CrossRef
Roberson MG, Smith DK, White SM, Wallace IS, Tucker MJ. Interspecies bombolitins exhibit structural diversity upon membrane binding, leading to cell specificity. Biophys J, 2019; 116(6):1064–74. CrossRef
Son WS, Kim JS, Kim HE, Park SH, Lee BJ. Structural studies on the antimicrobial peptide Brevinin 1E by spectroscopic methods. Spectroscopy, 2003; 17:650369. CrossRef
Tan TT. “Future” threat of Gram-negative resistance in Singapore. Ann Acad Med Singap, 2008; 37(10):884–90.
Waghu FH, Barai RS, Gurung P, Idicula-Thomas S. CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res, 2016; 44(D1):D1094–7. CrossRef
Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res, 2015; 44(D1):D1087–93. CrossRef
Yasir M, Willcox MD, Dutta D. Dutta. Action of antimicrobial peptides against bacterial biofilms. Materials (Basel), 2018; 11(12):2468. CrossRef
Yeom J, Ju S, Choi Y, Paek E, Lee C. Comprehensive analysis of human protein N-termini enables assessment of various protein forms. Sci Rep, 2017; 7(1):6599. CrossRef
Yin LM, Edwards MA, Li J, Yip CM, Deber CM. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J Biol Chem, 2012; 287(10):7738–45. CrossRef
Zhang SK, Song JW, Gong F, Li SB, Chang HY, Xie HM, Gao HW, Tan YX, Ji SP. Design of an α-helical antimicrobial peptide with improved cell-selective and potent anti-biofilm activity. Sci Rep, 2016; 6(1):27394. CrossRef
Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics, 2008; 9(1):40. CrossRef
Zhang Y, Liu Y, Sun Y, Liu Q, Wang X, Li Z, Hao J. In vitro synergistic activities of antimicrobial peptide brevinin-2CE with five kinds of antibiotics against multidrug-resistant clinical isolates. Curr Microbiol, 2014; 68(6):685–92. CrossRef
Zohrab F, Askarian S, Jalili A, Oskuee RK. Biological Properties, current applications and potential therapeautic applications of brevinin peptide superfamily. Int J Pept Res Ther, 2019; 25(1):39–48. CrossRef