INTRODUCTION
In December 2019, the World Health Organization declared an outbreak of febrile respiratory illness of an unknown etiology detected in Wuhan, China. Later, Chinese authorities confirmed a new type of coronavirus. Several strains of coronaviruses exist, but seven of them can infect humans. These are severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2, human coronavirus (HKU1, NL63, These are severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2, human coronavirus (HKU1, NL63, OC43, and 229E) with mild symptoms (Andersen et al., 2020; Corman et al., 2018; ICMRa, 2020). Since the disease outbreak, linked to the seafood market in Wuhan city, China (Cascella et al., 2020; ECDPC, 2020; ICMRa, 2020), the disease has spread to over 180 countries. The infection spreads by coughing and sneezing of an infected patient or through prolonged contact with an infected patient (ICMRa, 2020). Coronavirus has caused two large-scale epidemics in the last two decades, viz., SARS and MERS found mainly in bats (Drosten et al., 2003; Zaki et al., 2012; Zhou et al., 2020). Chan et al. (2020) carried out a bioinformatics-based analysis in 2019 on the latest novel coronavirus of 2019 (nCoV-2019) virus genome isolated from a Wuhan, China, patient cluster with atypical pneumonia and compared it with other associated coronavirus genomes. The genomes of nCoV-2019 showed 89% nucleotide identity with bat SARS-like-CoVZXC21 and 82% with human SARS-CoV. For this reason, the new virus was called SARS-CoV-2. The origin of the virus is still not clearly known. These genomic analyses suggested that SARS-CoV-2 probably evolved from a strain found in bats (Cascella et al., 2020).
Earlier, the most widely used standard test method for diagnosing the disease was real-time reverse–transcription polymerase chain reaction (qRT-–PCR). qRT-PCR cannot be used as a point-of-care testing because it requires long-time, highly qualified workers and specialized types of equipment. Coronavirus disease (COVID-19) is currently expanding rapidly in geographical terms with almost half the population is at risk in the world. Among the various available diagnostic solutions, rapid diagnostic tests (RDTs) are the most convenient and prompt. Therefore, there is a need to develop indigenous, cost-effective, and RDT kits that are more convenient and accurate. The immunochromatographic test kits appear to fulfill the requirement as these can be rapidly scaled up during an emergency. Researchers are employing their efforts in developing effective point-of-contact test kits and efficient laboratory techniques for molecular and serological diagnosis. These kits can be used in large populations for SARS-CoV-2 screening and can verify vaccine efficacy. The few approved SARS-CoV-2 lateral flow/rapid test (serological) kits from different countries are listed in Table 1.
Immunochromatographic test kits
Lateral flow immunochromatography assay is based on antigen–antibody reactions intended to detect small quantities of biological markers or target substances in the liquid sample without any need for specialized and expensive equipment. These tests are economic, simple, easy to use, and usually give results within 5–30 minutes. The analytics are identified by antigen–antibody label complexes using different labels, where gold nanoparticles (AuNPs) are used as a standard (Manta et al., 2015). The principle of the lateral flow immunochromatographic strip is shown in Figure 1. The immunochromatographic test strips work on the sandwich assay. The sandwich test kit is made up of three pads (sample, conjugate, and absorbent pads) and one nitrocellulose membrane (NCM) (Zhao et al., 2018). An antibody coupled with AuNPs was applied to the glass fiber membrane used as a conjugated pad. The test and control sample dots of antibodies were coated on the NCM. Natural untreated glass fiber membranes and cellulose absorbent pads were employed as sample and absorbent pads, respectively (Manta et al., 2015). In addition to speed, price, and ease of use, the rapid chromatographic immunoassay is more acceptable because of its simplicity and modality (Li et al., 2020). The immunochromatographic test kit consists of several components, viz., glass fiber sample pads, AuNPs-labeled conjugate pads, NCM, test lines, control lines, adsorbent pads, polyvinyl chloride support, and proteins (Zhao et al., 2018).
A recent study reported by Grifoni et al. (2020) found that unexposed human’s blood samples have 40%−60% of SARA-CoV-2-specific CD4-T cells, which leads to cross-reactivity. Therefore, to prevent cross-reactivity with prior human coronavirus (HCoV), MERS-CoV, SARA-CoV, and non-CoV infection, the selection of kit components and their masking is critical. The masking of a conjugate, absorbent pad, and AuNPs can be easily accomplished using various buffers and chemicals, as previously reported by different workers. However, the use for specific protein/reagents is a key for developing immunochromatographic test kits.
Key components of immunochromatographic test kits
Protein
Coronaviruses are single-stranded positive-polarity ribonucleic acid (RNA) genomes that code for 9,860 amino acids containing 29,891 nucleotides. The various biomarkers are membrane (M) protein, envelope (E) protein, nucleocapsid (N) protein, spike (S) protein, angiotensin-converting enzyme 2 (ACE2), and protein generated during thrombosis. The nucleocapsid (N) and spike (S1 + S2) proteins, around which most kits are developed, are indeed the two most important proteins. Figure 2 shows the various SARS-CoV-2 RNA proteins. In a research undertaken by Okba et al. (2020), human serum samples infected with HCoV, MERS-CoV, SARS-CoV, non-CoV, and SARS-CoV-2 found that S1 protein has a high specificity compared to N and S2 proteins. The N protein is also highly sensitive to antibody detection. They have also observed that SARS-CoV (coronavirus-2003) cannot be separated from SARS-CoV-2 (coronavirus-2019) in enzyme-linked immunosorbent assay (ELISA)enzyme-linked immunosorbent assayenzyme-linked immunosorbent assay by both of these proteins (Kilic et al., 2020; Okba et al., 2020).
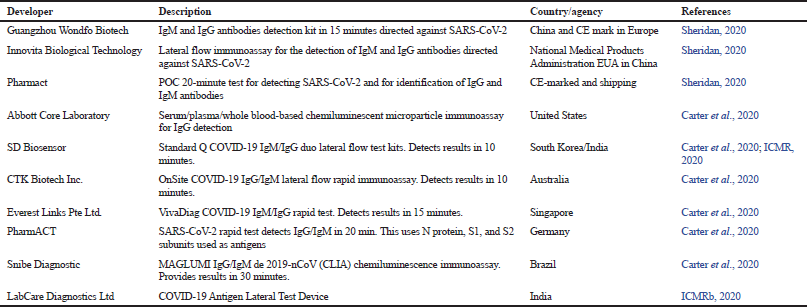 | Table 1. Few approved immunochromatographic-based rapid diagnostic test kits for SARS-CoV-2 in different countries. [Click here to view] |
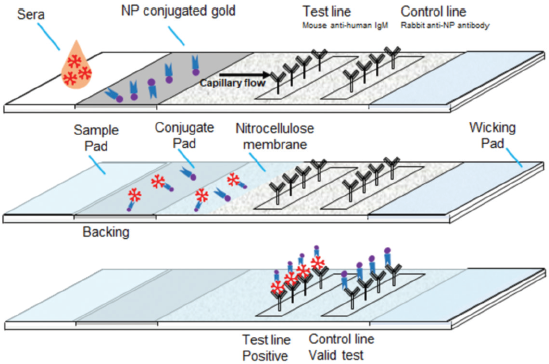 | Figure 1. The principle of the lateral flow immunochromatographic test strip sandwich assay (NP = Nanoparticles; IgM = Immunoglobulin M). [Click here to view] |
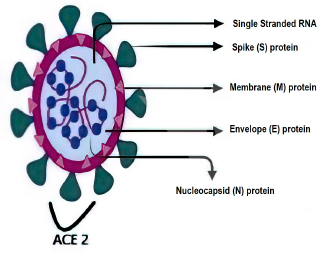 | Figure 2. The different proteins of SARS-CoV-2 RNA (RNA = Ribonucleic acid; S = Spike; N = Nucleocapsid; M = Membrane; E = Envelope; ACE2 = Angiotensin-converting enzyme 2). [Click here to view] |
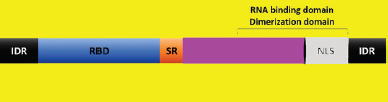 | Figure 3. N protein domains (RBD = RNA-binding domain; IDR = Intrinsically disordered region; SR = Serine–arginine-rich; NLS = Nuclear localization signal; RNA = Ribonucleic acid). [Click here to view] |
Demers-Mathieu et al. (2021) found that human immunoglobulin A (IgA), secretory immunoglobulin A (SIgA), secretory immunoglobulin M (SIgM), immunoglobulin M (IgM), and immunoglobulin G (IgG) antibodies can be induced by COVID-19 infection. The antibodies produced from SARS-CoV-2 infection in human milk react to N and S proteins and SIgM/IgM reactive to the nucleocapsid was found to be 1.2 times greater than the spike (S1 + S2) reactive protein. SIgA/IgA reactive to the nucleocapsid was 1.7 times lower to spike (S1 + S2). The detection of IgG reactive to the nucleocapsid was similar to spike protein. They do not disregard the risk of cross-reactivity either (Demers-Mathieu et al., 2020).
Therefore, particular proteins need to be chosen for the production of immune chromatographic test kits based on the abundance of SARS-CoV-2 reactive antibodies to a specific protein.
N protein (nucleocapsid phosphoprotein)
The nucleoprotein is a significant antigen of SARS-CoV with abundant antigenic sites. The N protein binds to RNA genome and interacts with the viral membrane. It also helps in RNA synthesis. Figure 3 shows a representation of the N protein structure. In comparison to other viral structural proteins, the low variable N protein number, as well as its period of existence, tests virus survival. N protein of SARS-CoV-1 (2003) and SARS-CoV-2 (COVID-19) are also very similar in their amino acid sequence. Therefore, tests developed using N protein detects both coronavirus (2003 & COVID-19) infections as they share 94% identity and 97% similarity to their amino acid sequence (Renkom Biotech, 2020).
Spike protein S1 & S2
The virus envelope membrane has three or four proteins, viz. membrane (M), envelope (E), and spike (S) (de Haan and Rottier, 2005). The S protein with the 1,160–1,422 amino acid type 1 glycoprotein chain is the largest of all membrane proteins. In some coronaviruses, during maturation, the S protein splits into S1 and S2 subunits and is non-covalently associated. S1 has a globular head and S2 has a stalk-like region (Cavanagh, 1995; de Groot et al., 1987; de Haan et al., 2006).
S1 subunit recognizes the host-receptor binding, while S2 subunit facilitates the fusion between the viral envelope and host cell membrane. S1 has two domains in the coronavirus; one is the N-terminal domain (S1-NTD) and the other is the C-terminal domain (S1-CTD). S1 receptor binding domain (RBD) binds to the ACE2. Figure 4 shows a representation of the S protein structure. One or both of these S1 domains bind to the receptors and function as the RBD. With ectodomain S1 of the spike glycoprotein, it is possible to differentiate COVID-19 from SARS-CoV 2003, as this protein is less similar than nucleocapsid with 70% identical and 81% similarities. For better test specificity, the best option is to select the S1 region of the spike protein. Considering these two domains of S1, S1-NTD has 66% identical and 79% similarities, while S1-CTD has 79% identical and 89% similarities (Renkom Biotech, 2020). The median incubation time is estimated to be 5.1 days. IgM antibodies specific for SARS-COV-2 can be detected 3–5 days after the onset of symptoms (Biopanda Reagents, 2020).
Approaches for the development of SARS-CoV-2 immunochromatographic test kit
For developing the SARS-CoV-2 immunochromatographic test kit, three alternative approaches and combinations can be used.
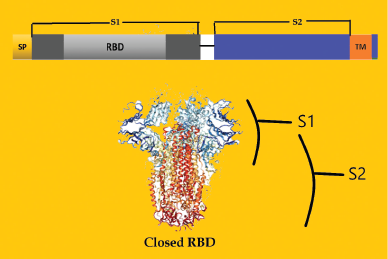 | Figure 4. S protein domains (RBD = Receptor-binding domain; SP = Signal peptide; SR = Serine–arginine-rich; TM = Transmembrane domain; S1= S1 subunit of Spike protein; S2 = S1 subunit of Spike protein). [Click here to view] |
Alternative I (the antibody detection kit)
This alternative is based on antibody detection. The first method detects specimen antibodies by binding to antigen-conjugated markers. The strips will be coated with IgG/IgM mAbs (monoclonal antibodies) and IgG/ IgM pAbs (polyclonal antibodies) of N and S proteins on the “T1 and T2” test line. Figure 5 depicts the kit developed from alternative 1 module. The “C” control line will be coated with an anti-chicken IgY antibody. The test strip contains recombinant SARS-CoV-2 antigen conjugated to colored gold nanoparticles. Free colloidal gold-labeled antibodies, mouse anti-human mIgM/mIgG, and chicken IgY are present in the release pad section. Diluted serum, plasma, or whole blood samples were applied to the release pad section. The mIgM/mIgG antibody binds to the coronavirus IgM/IgG antibodies and forms an IgM–mIgM / IgG–mIgG complex. The antibodies of the specimen move through the capillary action in the strips. If the sample contains coronavirus IgM/IgG antibodies, the test lines T1 and T2 will bind and develop the color through the IgM–IgM/IgG–IgG complex. If there are no coronaviruses present in the sample, the free mIgM/mIgG test line will not link to T1 and T2 and no color will develop. The free mIgM/mIgG then binds the chicken IgY antibody at control line C. This control line should be clear after sample testing as it indicates the proper functioning of the kit (Franzen, 2020; Gerbers, 2013; Manta et al., 2015). The Chinese CDC (center for disease control and prevention) used this kind of test kit widely during the COVID-19 outbreak in China (BioMedomics Inc., 2020). Various manufacturers also received CE + certification for the European market.
Alternative II (antigen detection kit)
The second alternative method involves the development of an antigen detection kit. This technique is considered to be more accurate than the antibody detection method. COVID-19 antigens are detectable in the nose and throat of the individual from the onset of symptoms. In this approach, N and S anti-human monoclonal or polyclonal antibody protein is conjugated with gold nanoparticles. After collecting the sample from the nose and throat, it is diluted with the diluent and applied to the sample pad section. The antigen present in the sample pad binds to the monoclonal or polyclonal antibody and forms an antigen–antibody complex at the conjugate pad section (Fig. 6). This mixture subsequently migrates along the membrane to the test area. Then, the complex will bind to the anti-mouse / rabbit SARS-CoV-2 antigen and develop color on the test line. If there is no coronavirus antigen in the sample, the free antigen will not link to the test line and no color will develop. In such a case, the free antigen will bind to the anti-chicken IgY antibody of the control line and the color of the control line ensures that the kit is working correctly.
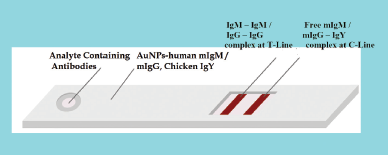 | Figure 5. The alternative 1 approach for IgG/IgM antibodies detection kit (IgG = Immunoglobulin G; IgM = Immunoglobulin M; IgY = Immunoglobulin Y; m = Anti-human; AuNPs = Gold nanoparticles; T = Test line; C = Control line). [Click here to view] |
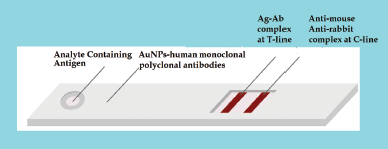 | Figure 6. The alternative 2 approaches for antigen detection kit (Ag = Antigen; Ab = Antibodies; AuNPs = Gold nanoparticles; T = Test line; C = Control line). [Click here to view] |
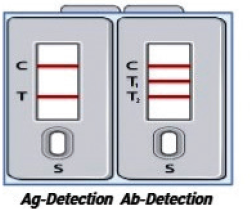 | Figure 7. The alternative 3 approaches for antigen and antibody detection kit in a single housing (Ag = Antigen; Ab = Antibodies; T = Test line for antigen detection; C = Control line; T1 = Test line 1 for immunoglobulin M detection; T2 = Test line 2 for immunoglobulin G detection; S = Sample port). [Click here to view] |
Alternate III (antigen and antibody detection kit in single housing)
For more precision, alternative III can be used. Both antibodies and antigen detection strips can be placed in a single house for comparative and simultaneous detection of antibodies and antigens from specimen samples. In alternative III, alternatives I and II are combined to form the test kits with two separate chambers. One section of test kits detects antibodies (Alternate I) and the other part detects antigens (alternate II) of the COVID-19. Figure 7 depicts the kit developed from alternative III module.
The test license for the manufacture of the COVID-19 kit is to be taken from the Central Drugs Standard Control Organization (CDSCO), Delhi. The test kit should be validated for sensitivity and specificity at the National Institute of Virology in Pune after it has been designed. The CDSCO is then approached for a commercial manufacturing license.
CONCLUSION
The urgent requirement in India is to meet the challenge of SARS-CoV-2. In India, there are currently no commercially available indigenous sources of reagents/protein for the production of rapid test kits. The reagents/proteins are imported for the kits designing. These imported reagents/proteins can cause false results because their selection and validation are not based upon risk analysis. Besides, pre-protein-coated uncut sheets can lead to false results in a selected population genotype. India needs to develop indigenous reagents/proteins that can have specificity for Indian pollution genotype. The use of pre-coated uncut sheet needs to be avoided. Utilizing imported reagents/proteins for antibodies detection kits, N protein reactive to IgA, secretory IgA (SIgA), secretory IgM (SIgM), and IgM can improve test sensitivity but can have poor specificity. N protein can also not distinguish between SARS-CoV and SARS-CoV-2 infection, but this consideration can be overlooked since SARS-CoV infection occurred 19 years ago and now there is a negligible likelihood of the presence of these antibodies. Using S1 protein is a better choice to increase the specificity of kits. On the contrary, N and S proteins have no role in IgG sensitivity and specificity. In addition, the combined antigen and antibody detection kit in a single housing would be a better choice for comparative screening of SARS-CoV-2.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
FUNDING
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
Not applicable.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med, 2020; 26(4):450–2. CrossRef
BioMedomics Inc. COVID-19 IgM/IgG rapid test. 2020. [Online] Available at:https://www.biomedomics.com/products/infectious-disease/covid-19-rt/ (Accessed 8 July 2020).
Biopanda Reagents. COVID-19 Rapid test CE. 2020. Available via https://www.biopanda.co.uk/php/products/rapid/infectious_diseases/covid19.php (Accessed 8 July 2020).
Carter LJ, Garner LV, Smoot JW, Li Y, Zhou Q, Saveson CJ, Sasso JM, Gregg AC, Soares DJ, Beskid TR, Jervey SR, Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent Sci, 2020; 6(5):591–605. CrossRef
Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, evaluation and treatment coronavirus (COVID-19). StatPearls Publishing, Treasure Island, FL, 2020.
Cavanagh D. The coronavirus surface glycoprotein. In: Siddell SG (ed.). The coronaviridae. Plenum Press, New York, NY, pp 73–113, 1995. CrossRef
Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect, 2020; 9(1):221–36. CrossRef
Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res, 2018; 100:163–88. CrossRef
de Groot RJ, Maduro J, Lenstra JA, Horzinek MC, van der Zeijst BA, Spaan WJ. cDNA cloning and sequence analysis of the gene encoding the peplomer protein of feline infectious peritonitis virus. J Gen Virol, 1987; 68:2639–46. CrossRef
de Haan CAM, Lintelo ET, Li Z, Raaben M, Wurdinger T, Bosch BJ, Rottier PJM. Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range. J Virol, 2006; 80(22):10909–18. CrossRef
de Haan CAM, Rottier PJM. Molecular interactions in the assembly of coronaviruses. In: Roy P (ed.). Virus structure and assembly, Elsevier Academic Press, San Diego, CA, vol 64, pp 165–230, 2005. CrossRef
Demers-Mathieu V, Do DM, Mathijssen GB, Sela DA, Seppo A, Jarvinen KM, Medo S. Difference in levels of SARS-CoV-2 S1 and S2 subunits- and nucleocapsid protein reactive SIgM/IgM, IgG and SIgA/IgA antibodies in human milk. J Perinatol, 2021; 44:850–9. CrossRef
Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguière AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Müller S, Rickerts V, Stürmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. Identification of a novel-coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003; 348(20):1967–76. CrossRef
ECDPC. Rapid risk assessment: outbreak of acute respiratory syndrome associated with a novel coronavirus, Wuhan, China. 2020. Available via https://www.ecdc.europa.eu/en/publications-data/risk-assessment-outbreak-acute-respiratorysyndrome-associated-novel-coronavirus (Accessed 08 July 2020) CrossRef
Franzen M. Covid-19 rapid test - Blue Paper. Alten Life Sci, 2020. Available via https://osf.io/jpukc (Accessed 8 July 2020).
Gerbers R. Development of enhanced lateral flow test devices for point-of-development of enhanced lateral flow test devices for point-of-care diagnostics care diagnostics. 2013. Available via https://digitalcommons.uri.edu/theses (Accessed 8 July 2020).
Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell, 2020; 181:1489–501. CrossRef
ICMRa. Revised strategy of COVID-19 testing in India. 2020. Available via https://www.mohfw.gov.in/pdf/ICMRrevisedtesting strategyforCOVID.pdf (Accessed 08 July 2020).
ICMRb. List of companies/vendors of rapid antigen test kits for covid19 validated/being validated by icmr dated. 2020. Available via https://www.icmr.gov.in/pdf/covid/kits/archive/List_of_rapid_antigen_kits_14082020.pdf (Accessed 20 November 2020).
Kilic T, Weissleder R, Lee H. Molecular and immunological diagnostic tests of COVID-19: current status and challenges. iScience, 2020; 23(8):101406. CrossRef
Manta P, Agarwal S, Singh, Bhamrah GSS. Formulation, development and sensitivity, specificity comparison of gold, platinum and silver nanoparticle-based HIV ½ and hCG IVD rapid test kit (Immune chromatographic test device). World J Pharm Pharm Sci, 2015; 4(05):1870–905.
Li M, Jin R, Peng Y, Wang C, Ren W, Lv F, Gong FF, Wang Q, Li J, Shen T, Sun H, Zhou L, Cui Y, Song H, Sun L. Generation of antibodies against COVID-19 virus for development of diagnostic tools. medRxiv, 2020. CrossRef
Okba N, Müller MA, Li W, Wang C, Geurtsvan Kessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken CBEM, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. Severe acute respiratory syndrome coronavirus 2−specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020; 26(7):1478–88. CrossRef
Renkom Biotech SL. Detection of the new coronavirus disease 2019 (covid-19) by rapid serodiagnostic assays. 2020. Available via https://www.rekombiotech.com/en/blog/detection-covid-19 (Accessed 8 July 2020).
Sheridan C. Coronavirus and the race to distribute reliable diagnostics. Nat Biotechnol, 2020; 38(4):382–4. CrossRef
Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med, 2012; 367(19):1814–20. CrossRef
Zhao Y, Yang M, Fu Q, Ouyang H, Wen W, Song Y, Zhu C, Lin Y, Du D. A nanozyme- and ambient light-based smartphone platform for simultaneous detection of dual biomarkers from exposure to organophosphorus pesticides. Anal Chem, 2018; 90(12):7391–8. CrossRef
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 2020; 579(7798):270–3. CrossRef