INTRODUCTION
Human epidermal growth factor (hEGF) is a protein with 53 amino acid residues with 6.2 kDa of molecular weight. This polypeptide has three intramolecular disulfide bonds and heat resistance properties (Eissazadeh et al., 2017; Sriwidodo et al., 2019; Tang et al., 2016). hEGF plays a role in stimulating cell proliferation, differentiation, and migration in the wound-healing process. This has led to the high demand for hEGF in the clinical field, thus encouraging efforts to increase hEGF production through recombinant DNA technology (Ma et al., 2016; Zheng et al., 2016).
E. coli was one of the most widely used hosts for recombinant protein expression (Hayat et al., 2018). It has a high rate of protein expression and rapid growth, is inexpensive, and produces culture in the high-density cell (Kaur et al., 2018; Kim et al., 2017; Maksum et al., 2019). There were previous studies on the extracellular expression of hEGF in E. coli using various signal peptides (Indriyani et al., 2019; Maksum et al., 2017; Melati et al., 2019; Sriwidodo et al., 2017). Nevertheless, the extracellular expression system has a lower expression rate than the intracellular system (Su et al., 2006). To overcome this problem, the intracellular expression approach was currently used to increase hEGF expression.
Intracellular protein expression in E. coli especially small protein such as hEGF mostly did not work well because the small protein was easily degraded by proteases from the host cell. This causes a significant loss in yields and made it difficult in the purification process. Alternatively, the protein was fused with a fusion partner so the target protein could avoid degradation by proteases (Sun et al., 2005; Zhang et al., 2015). Several fusion tags were commonly used in fusion protein expression systems such as glutathione S-transferase, small ubiquitin-related modifier, thioredoxin, and maltose-binding protein. However, there were some difficulties in fusion tag removal such as unspecific cleavage site, the steric hindrance at the cleavage site, and the expensive cost of specific protease (Costa et al., 2014; Zheng et al., 2016).
The intein is a protein segment that can cleavage itself from protein precursors where it fuses two segments of flanking proteins or amino acids (Li, 2011). The intein-mediated expression system has been developed as a powerful tool for protein expression that utilized its ability to self-cleavage. Target protein synthesized in the fusion of intein form which has been genetically designed to do controllable cleavage from peptide bond at either N- or C-terminal of the intein. After protein cleavage by intein, the target protein was produced with the desired terminal amino acid (Jiang et al., 2015). Ssp DnaB mini-intein derived from Synechocystis sp. was used in the commercial plasmid, such as IMPACTTM system (Ding et al., 2003). The release of the target protein can be induced simply by changing pH, giving the advantage of cleaving peptide bonds without using specific protease (Sun et al., 2005).
The expression of recombinant protein in E. coli frequently leads to the formation of inclusion bodies. Inclusion bodies are protein aggregate that consists of partially folded and misfolded protein that interacts via hydrophobic interaction. Their formation is often undesirable because they generally lack biological activity (Gomes et al., 2016; Silaban et al., 2019; Singh and Panda, 2005). This approach can give the advantage by refolding the inactive protein in inclusion bodies into its active form (Upadhyay et al., 2016). Moreover, inclusion bodies are produced in high amount and easy to separate because they were denser than the other cellular components (Singh and Panda, 2005; Singh et al., 2015). To recover active protein from inclusion bodies, inclusion bodies were solubilized in presence of chaotropic agents such as urea and guanidine chloride and afterward solubilized protein was refolded by decreasing the concentration of chaotropic agents (Singh et al., 2015; Vallejo and Rinas, 2004).
In this report, we construct fusion protein CBD-Ssp DnaB-hEGF by fused hEGF gene with the CBD-Ssp DnaB gene in pD861 plasmid. E. coli BL21(DE3) was transformed with recombinant plasmid. CBD-Ssp DnaB-hEGF was successfully expressed as inclusion bodies. Inclusion bodies were isolated, solubilized, and refolded. Ssp DnaB mini-intein splicing activity could be achieved by gradually shifting pH from 8.5 to 6.0. After Ssp DnaB mini-intein cleavage, hEGF protein was obtained with a concentration of 0.163 mg/ml. The result demonstrated that hEGF protein could be recovered by inclusion bodies obtained from intracellular expression in E. coli.
MATERIALS AND METHODS
Bacterial strain, plasmid, and materials
E. coli BL21(DE3) was taken from our laboratory stock. Plasmid pD861-CBD-Ssp DnaB-hEGF was constructed by fused CBD-Ssp DnaB gene [National Center for Biotechnology Information (NCBI) access code HG792069.1] and hEGF gene (NCBI access code GQ214314.1) into plasmid pD861 and synthesized by ATUM, Newark, CA. Citric acid, sodium chloride, urea, Tris base, β-mercaptoethanol, and calcium chloride were purchased from Merck, Kenilworth, NJ. Yeast extract was purchased from Oxoid, United Kingdom. Agar bacto, ethylenediaminetetraacetic acid (EDTA), tryptone, and glycerol were purchased from 1st base, Singapore. L-rhamnose, tricine, Triton X-100, kanamycin sulfate, and N,N,N’,N’-tetramethylethylenediamine (TEMED) were purchased from Sigma-Aldrich, Singapore.
Expression of recombinant CBD-Ssp DnaB-hEGF fusion protein
To express CBD-Ssp DnaB-hEGF, CaCl2 competent E. coli BL21(DE3) cells were transformed with pD861-CBD-Ssp DnaB-hEGF plasmid using the heat shock method. Transformed cells were grown in LB agar plates supplemented with 50 μg/ml kanamycin at 37°C for 16–18 hours. Single colonies were replicated and the selected replica was grown in 5 ml LB medium supplemented with 50 μg/ml kanamycin at 37°C for 16–18 hours. A total of 1 ml of starter culture was transferred into 100 ml LB medium and incubated at 37°C with shaking at 200 rpm until OD600 0.6 was reached and then was induced by adding L-rhamnose until the final concentration of 4 mM and incubated for 5 hours. The culture was centrifuged at 6,000 g for 20 minutes at 4°C. The cell pellet was stored at −20°C for further analysis.
Inclusion bodies preparation
Cell pellet (approximately 2 g wet weight) were resuspended in 1:4 ratio with lysis buffer (20 mM Tris-Cl, 1 mM EDTA, and pH 8.5); then, the suspension was sonicated for 1 minute (1 second on and 1 second off pulse). Sonication was performed on ice for 10 cycles with 1 minute gap for each cycle. The lysed cell suspension was centrifuged at 12,000 g for 20 minutes at 4°C. The pellet obtained was washed twice using washing buffer I (20 mM Tris-Cl, 1 mM EDTA, 1% Triton X-100, 1 M urea, and pH 8.5), once using washing buffer II (20 mM Tris-Cl, 1 mM EDTA, 1% Triton X-100, and pH 8.5), and twice using washing buffer III (20 mM Tris-Cl and pH 8.5). Purified inclusion bodies were resuspended in 20 mM Tris-Cl and pH 8.5 and used for solubilization.
Optimization of urea concentration for solubilization of inclusion bodies
Purified inclusion bodies’ suspension was solubilized with 20 mM Tris-Cl pH 8.5 containing different urea concentrations to optimize solubilization conditions for inclusion bodies of CBD-Ssp DnaB-hEGF fusion protein. Inclusion bodies’ suspension was solubilized with 1:10 solubilizing buffer (20 mM Tris-Cl and pH 8.5) with different concentrations of urea (0–8 M urea). The mixture was incubated for 2 hours at room temperature with shaking at 150 rpm and after that centrifuged at 13,000 rpm for 20 minutes at 4°C. Solubilized inclusion bodies were characterized by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by ImageJ software.
Solubilization and refolding of solubilized CBD-Ssp DnaB-hEGF
Purified inclusion bodies were solubilized with 25 ml solubilizing buffer (10 mM Tris-Cl, 5 mM β-mercaptoethanol, 8 M urea, and pH 8.5) (Zhang et al., 2015) for 2 hours at room temperature with shaking at 150 rpm and centrifuged at 10,000 g for 30 minutes at 4°C. Solubilized inclusion bodies were refolded using the dialysis method according to Zhang et al. (2015) with some modification. Solubilized inclusion bodies (10 ml) was dialyzed with 1 L refolding buffer I (10 mM Tris-Cl, 0.5 mM EDTA, 50 mM NaCl, 5 mM β-mercaptoethanol, 2 M urea, and pH 8.5) for 24 hours at 4°C, followed by 1 L refolding buffer II (10 mM Tris-Cl, 0.5 mM EDTA, 50 mM NaCl, and pH 8.5). Refolded protein was cleaved by incubating it in buffer with different pH (8.0, 7.5, 7.0, 6.5, 6.0, and 5.6) for 24 hours at 4°C, respectively. The refolded protein and cleaved protein were characterized by SDS-PAGE and analyzed by ImageJ software.
RESULTS  AND DISCUSSION
Construction CBD-Ssp DnaB-hEGF gene in recombinant plasmid
CBD-Ssp DnaB-hEGF gene was constructed from the sequence of the hEGF (GeneBank GQ214314.1) and CBD-Ssp DnaB (GeneBank HG792069.1). The first methionine from hEGF was eliminated and fused to asparagine at the end of the C-terminal of CBD-Ssp DnaB (Fig. 1). Asparagine residue on C-terminal of Ssp DnaB mini-intein was directly fused to alanine residue on N-terminal of hEGF. The expression rate of heterologous protein on E. coli was closely related to the codon usage bias phenomenon (Gomes et al., 2016). Bias codon could be avoided by optimizing the CBD-Ssp DnaB-hEGF gene to have high CAI value and GC content in the range of 30%–70% (Parret et al., 2016; Silaban et al., 2014; Sriwidodo et al., 2017). CBD-Ssp DnaB-hEGF gene was optimized by using E. coli preference codon to achieve better protein expression. The optimized CBD-Ssp DnaB-hEGF gene had a length of 843 bp. CAI value of gene was 1.00 with GC content of 51.89%. CBD-Ssp DnaB-hEGF gene was inserted into plasmid pD861 and synthesized by ATUM (Fig. 2). The promoter was a key component in protein expression because it plays a role in regulating the expression of the gene of interest. L-rhamnose promoters provide more tight regulation which will prevent leakage of protein (Kaur et al., 2018). Therefore, it is assumed to be able to increase protein acquisition after the addition of an inducer. The use of plasmid with high copy numbers and a strong ribosomal binding site can also increase the rate of protein expression (Hu et al., 2012; Marschall et al., 2016).
Expression of recombinant CBD-Ssp DnaB-hEGF and inclusion bodies preparation
Recombinant CBD-Ssp DnaB-hEGF was successfully expressed as 31.2 kDa protein in E. coli. CBD-Ssp DnaB-hEGF was majorly expressed in inclusion bodies form compared to soluble form (Fig. 3a). Induction at mid-log phase (OD600 0.6) provides a high expression rate due to E. coli rapid growth which leads to a higher protein expression rate (Fazaeli et al., 2019; Kaur et al., 2018). In a previous study, the L-rhamnose promoter was successfully expressing recombinant protein extracellularly using signal peptide (Indriyani et al., 2019; Maksum et al., 2019). However, a recent study showed that the L-rhamnose promoter can also be utilized for intracellular expression of CBD-Ssp DnaB-hEGF as well. Inclusion bodies were washed by detergent to remove contaminants such as cell membrane and cellular protein following aggregate with inclusion bodies. Triton-X100 was used to wash cell membranes and also the addition of a low concentration of urea could help to remove cellular protein (Qi et al., 2015; Singh et al., 2015). Purified inclusion bodies were obtained after several washing steps (Fig. 3b) and it was used for solubilization and refolding.
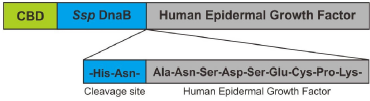 | Figure 1. Scheme of CBD-Ssp DnaB-hEGF fusion protein assembly (modified from Király et al., 2006 ). [Click here to view] |
 | Figure 2. Construction of plasmid pD861-CBD-Ssp DnaB-hEGF. The plasmid’s characteristics were rhamnose promoter, kanamycin selection marker, Ori pUC, and strong RBS. [Click here to view] |
 | Figure 3. Expression of CBD-Ssp DnaB-hEGF and inclusion bodies preparation. (a) Tricine SDS-PAGE analysis of CBD-Ssp DnaB-hEGF expression as inclusion bodies. Lane 1: soluble fraction, lane 2: insoluble fraction, and lane M: protein marker. (b) Tricine SDS-PAGE analysis of purified inclusion bodies after several washing steps. Lane 1: purified IB and lane M: protein marker (size of protein in kilodalton is shown on the left). [Click here to view] |
Optimization of urea concentration for solubilizing inclusion bodies
Different concentration of urea (0–8 M) was used to solubilize CBD-Ssp DnaB-hEGF from inclusion bodies. Urea would help to disrupt hydrophobic interaction between proteins in inclusion bodies resulting in solubilized CBD-Ssp DnaB-hEGF (Patra et al., 2000; Singh and Panda, 2005). CBD-Ssp DnaB-hEGF was slightly solubilized from inclusion bodies in the presence of 2 M urea and conversely showed better solubility in the presence of 6–8 M urea. CBD-Ssp DnaB-hEGF solubility tended to increase when the concentration of urea was increased (Fig. 4a). The highest amount of solubilized CBD-Ssp DnaB-hEGF was achieved by using a solubilization buffer containing 8 M urea (Fig. 4b). This result showed that the solubilization buffer containing 8 M urea could give better solubilization of CBD-Ssp DnaB-hEGF from inclusion bodies. The use of higher concentration of urea will give more solubilized protein that would be in denatured state, so refolding was necessary to convert protein into its native conformation by removal of denaturant through dialysis (Yamaguchi et al., 2013).
Refolding and simultaneous cleavage of solubilized CBD-Ssp DnaB-hEGF
Solubilized CBD-Ssp DnaB-hEGF was refolded by lowering the urea concentration using a stepwise dialysis method. The dialysis method could prevent protein loss because of the uncontrolled cleavage of Ssp DnaB mini-intein (Zhang et al., 2015). The concentration of urea was lowered from 8 to 2 M then until approximately reaching 0 M. No aggregate was observed after dialysis; this showed that the refolding process was successful. Solubilized CBD-Ssp DnaB-hEGF started to refold as the urea concentration was decreasing (Singh et al., 2015; Yamaguchi and Miyazaki, 2014). On the other side, a proper disulfide bond was formed from reduced disulfide bond through disulfide shuffling while stepwise dialysis was performed (Yamaguchi et al., 2013). Even stepwise dialysis was taking more time and “slow,” this method could give good results. There was no aggregation observed after dialysis was performed.
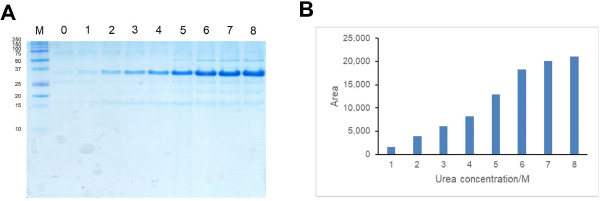 | Figure 4. Optimization of solubilization condition of inclusion bodies in different concentrations of urea. (a) Tricine SDS-PAGE analysis of CBD-Ssp DnaB-hEGF inclusion bodies solubilized in different concentrations of urea. Lanes 0–8: CBD-Ssp DnaB-hEGF inclusion bodies solubilized in urea with a concentration of 0–8 M. Lane M: protein marker (size of protein in kilodalton is shown on the left). (b) Solubilized CBD-Ssp DnaB-hEGF quantification by using ImageJ software. [Click here to view] |
 | Figure 5. Refolding and cleavage of solubilized CBD-Ssp DnaB-hEGF using dialysis method. (a) Refolded CBD-Ssp DnaB-hEGF. Lane 1: refolded CBD-Ssp DnaB-hEGF. Lane M: protein marker. (b) Optimization of pH condition for induced splicing of refolded CBD-Ssp DnaB-hEGF. Lane 1–5: induced splicing by lowering the pH to 8.0; 7.5; 7.0; 6.5; 6.0. Lane 6: induced splicing by unmodified cleavage buffer pH 6.0. Lane M: protein marker. (c) Supernatant of cleavage of refolded CBD-Ssp DnaB-hEGF. Lane 1: refolded CBD-Ssp DnaB-hEGF. Lane 2: cleavage result of CBD-Ssp DnaB-hEGF. Lane M: protein marker (size of protein in kilodalton is shown on the left). [Click here to view] |
CBD-Ssp DnaB-hEGF shows Ssp DnaB splicing activity which is characterized by a band at 25 kDa; this band belonged to CBD-Ssp DnaB (Fig. 5a). The pH condition for splicing was investigated (pH 8.0-6.0) to ensure the pH optimum for splicing. Protein splicing occurred even at pH 8.0 and so did at the lower pH until pH 6.0 (Fig. 5b). This showed that gradually lowering pH until 6.0 could induce Ssp DnaB splicing activity. hEGF was released from CBD-Ssp DnaB-hEGF, which showed by a band at 6.2 kDa (Fig. 5c). Splicing activity of Ssp DnaB intein involved charge relay between amino acid residues and was ended by asparagine cyclization at C-terminal of intein (Ding et al., 2003; Topilina and Mills, 2014). Uncontrolled splicing activity was caused by Ssp DnaB intein which was sensitive to pH changes (Chen et al., 2002) and the cleavage was observed even at pH 8.0. By gradually decreasing pH from 8.0 to 6.0, hEGF was obtained. hEGF was obtained with a concentration of 0.163 mg/ml which was quantified by the densitometry method.
CONCLUSION
In summary, CBD-Ssp DnaB-hEGF was successfully expressed as inclusion bodies in E. coli BL21(DE3). Refolded hEGF was recovered from inclusion bodies after isolation, solubilization, and refolding steps with a concentration of 0.163 mg/ml. The solubilization and refolding method presented in this study could be used for the recovery of refolded hEGF from CBD-Ssp DnaB-hEGF inclusion bodies. The result shows a great opportunity to produce large amounts of hEGF from bacterial inclusion bodies. This finding leads to the optimization and development method in the future for maximizing the recovery of refolded hEGF. So, it could be applicable for a larger scale, especially at the industrial scale.
ACKNOWLEDGMENTS
This work was supported by grant PD No. 1827/UN6.3.1/LT/2020 from the Directorate General of Research and Development Strengthening, Ministry of Technology Research and Higher Education, Republic of Indonesia.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
Not applicable.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Chen XH, Xu MQ, Ding Y, Ferrandon S, Rao ZH. Purification and initial crystallization studies of a DnaB intein from Synechocystis sp PCC 6803. Acta Crystallogr Sect D-Biol Crystallogr, 2002; 58:1201–3. CrossRef
Costa S, Almeida A, Castro A, Domingues L. Fusion tags for protein solubility, purification, and immunogenicity in Escherichia coli: the novel Fh8 system. Front Microbiol, 2014; 5:1–20. CrossRef
Ding Y, Xu MQ, Ghosh I, Chen X, Ferrandon S, Lesage G, Rao Z. Crystal structure of a mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing. J Biol Chem, 2003; 278(40):39133–42. CrossRef
Eissazadeh S, Moeini H, Dezfouli MG, Heidary S, Nelofer R, Abdullah MP. Production of recombinant human epidermal growth factor in Pichia pastoris. Braz J Microbiol, 2017; 48(2):286–93. CrossRef
Fazaeli A, Golestani A, Lakzaei M, Varaei SSR, Aminian M. Expression optimization, purification, and functional characterization of cholesterol oxidase from Chromobacterium sp. DS1. PLoS One, 2019; 14(2):1–15. CrossRef
Gomes A, Byregowda S, Veeregowda B, Vinayagamurthy B. An overview of heterologous expression host systems for the production of recombinant proteins. Adv Anim Vet Sci, 2016; 4:346–56. CrossRef
Hayat SMG, Farahani N, Golichenari B, Sahebkar A. Recombinant protein expression in Escherichia coli (E. coli): what we need to know. Curr Pharm Des, 2018; 24(6):718–25. CrossRef
Hu Y, Feng L, Li Y, Zhang Y, Lu P, Rayner S, Chen S. Ribosomal binding site switching: an effective strategy for high-throughput cloning constructions. PLoS One, 2012; 7(11):e50142. CrossRef
Indriyani A, Anggraeni NI, Sriwidodo, Maksum IP. Optimization extracellular secretion of recombinant human epidermal growth factor (hEGF) in Escherichia coli BL21(DE3) pD881-OmpA-hEGF by using Response Surface Method (RSM). Int J Res Pharm Sci, 2019; 10(3):1824–31. CrossRef
Jiang A, Jin W, Zhao F, Tang Y, Sun Z, Liu J-N. Split Ssp DnaB mini-intein-mediated production of recombinant human glucagon-like peptide-1/7-36. Biotechnol Appl Biochem, 2015; 62(3):309–15. CrossRef
Kaur J, Kumar A, Kaur, J. Strategies for optimization of heterologous protein expression in E. coli: roadblocks and reinforcements. Int J Biol Macromol, 2018; 106:803–22. CrossRef
Kim S, Jeong H, Kim EY, Kim JF, Lee SY, Yoon, SH. Genomic and transcriptomic landscape of Escherichia coli BL21(DE3). Nucleic Acids Res, 2017; 45(9):5285–93. CrossRef
Kiraly O, Guan L, Sepessy E, Tóth M, Kukor Z, Sahin-Tóth M. Expression of human cationic trypsinogen with an autenthic N terminus using intein-mediated splicing in aminopeptidase P (pepP) deficient Escherichia coli. Protein Expr Purif, 2006; 48:104–11. CrossRef
Li Y. Self-cleaving fusion tags for recombinant protein production. Biotechnol Lett, 2011; 33(5):869–81. CrossRef
Ma Y, Yu J, Lin J, Wu S, Li S, Wang J. High efficient expression, purification, and functional characterization of native human epidermal growth factor in Escherichia coli. Biomed Res Int, 2016; 2016. CrossRef
Maksum I, Lestari A, Fauzia R, Rachman S, Soedjanaatmadja U. Escherichia coli BL21(DE3) expression system using TorA signal peptide for Recombinant Human Albumin (rHA) secretion. Int J Res Pharm Sci, 2019; 10:3319–24. CrossRef
Maksum IP, Utama E, Sriwidodo Subroto T. Extracellular secretion of recombinant human epidermal growth factor by using trimethylamine N-Oxide reductase a (TorA) signal peptide in Escherichia coli BL21(DE3). J Pharm Sci Res, 2017; 9(6):1007–16.
Marschall L, Sagmeister P, Herwig C. Tunable recombinant protein expression in E. coli: enabler for continuous processing? Appl Microbiol Biotechnol, 2016; 100(13):5719–28. CrossRef
Melati R, Indriyani A, Gaffar S, Sriwidodo, Maksum IP. Comparison of extracellular secretion of recombinant human epidermal growth factor using TorA and PelB signal peptides in Escherichia coli BL21 (DE3). Asian J Pharm Clin Res, 2019; 12(11):81–4. CrossRef
Parret AH, Besir H, Meijers R. Critical reflections on synthetic gene design for recombinant protein expression. Curr Opin Struct Biol, 2016; 38:155–62. CrossRef
Patra AK, Mukhopadhyay R, Mukhija R, Krishnan A, Garg LC, Panda AK. Optimization of inclusion body solubilization and renaturation of recombinant human growth hormone from Escherichia coli. Protein Expr Purif, 2000; 18(2):182–92. CrossRef
Qi X, Sun Y, Xiong S. A single freeze-thawing cycle for highly efficient solubilization of inclusion body proteins and its refolding into bioactive form. Microb Cell Fact, 2015; 14(1):1–12. CrossRef
Silaban S, Gaffar S, Simorangkir M, Maksum IP, Subroto T. Effect of IPTG concentration on recombinant human prethrombin-2 expression in Escherichia coli BL21(DE3) ArcticExpress. IOP Conf Ser Earth Environ Sci, 2018; 217(1):1–6. CrossRef
Silaban S, Maksum I, Gaffar S, Hasan K, Enus S, Subroto T, Soemitro S. Codon optimization and chaperone assisted solubilization of recombinant human prethrombin-2 expressed in Escherichia coli. Microbiol Indones, 2014; 8(4):170–5. CrossRef
Singh A, Upadhyay V, Upadhyay AK, Singh SM, Panda AK. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb Cell Fact, 2015; 14(1):1–10. CrossRef
Singh SM, Panda AK. Solubilization and refolding of bacterial inclusion body proteins. J Biosci Bioeng, 2005; 99(4):303–10. CrossRef
Sriwidodo S, Maksum I, Riswanto N, Rostinawati T, Subroto T. Extracellular secretion recombinant of human epidermal growth factor (hEGF) using pectate lyase B (PelB) signal peptide in Escherichia coli BL21(DE3). Int J Res Pharm Sci, 2017; 8:33–40.
Sriwidodo S, Subroto T, Maksum IP, Wathoni N, Rostinawati T, Ulya H, Putri IU. Optimization of secreted recombinant human epidermal growth factor production using pectate lyase B from Escherichia Coli BL21(DE3) by central composite design and its production in high cell density culture. J Pharm Bioallied Sci, 2019; 11(Suppl 4):S562–6. CrossRef
Su Z, Huang Y, Zhou Q, Wu Z, Wu X, Zheng Q, Ding C, Li X. High-level expression and purification of human epidermal growth factor with SUMO fusion in Escherichia coli. Protein Pept Lett, 2006; 13(8):785–92. CrossRef
Sun Z, Chen J, Yao H, Liu L, Wang J, Zhang J, Liu JN. Use of Ssp dnaB derived mini-intein as a fusion partner for production of recombinant human brain natriuretic peptide in Escherichia coli. Protein Expr Purif, 2005; 43(1):26–32. CrossRef
Tang X, Liu H, Yang S, Li Z, Zhong J, Fang, R. Epidermal growth factor and intestinal barrier function. Mediators Inflamm, 2016;2016:27–30. CrossRef
Topilina NI, Mills KV. Recent advances in in vivo applications of intein-mediated protein splicing. Mob DNA, 2014; 5(1):1–14. CrossRef
Upadhyay V, Singh A, Panda AK. Purification of recombinant ovalbumin from inclusion bodies of Escherichia coli. Protein Expr Purif, 2016; 117:52–8. CrossRef
Vallejo LF, Rinas U. Strategies for the recovery of active proteins through refolding of bacterial inclusion body proteins. Microb Cell Fact, 2004; 3:1–12. CrossRef
Yamaguchi H, Miyazaki M. Refolding techniques for recovering biologically active recombinant proteins from inclusion bodies. Biomolecules, 2014; 4(1):235–51. CrossRef
Yamaguchi S, Yamamoto E, Mannen T, Nagamune T. Protein refolding using chemical refolding additives. Biotechnol J, 2013; 8(1):17–31. CrossRef
Zhang Y, Zhang K, Wan Y, Zi J, Wang Y, Wang J, Wang L, Xue X. A pH-induced, intein-mediated expression and purification of recombinant human epidermal growth factor in Escherichia coli. Biotechnol Prog, 2015; 31(3):758–64. CrossRef
Zheng X, Wu X, Fu X, Dai D, Wang F. Expression and purification of human epidermal growth factor (hEGF) fused with GB1. Biotechnol Biotechnol Equip, 2016; 30(4):813–8. CrossRef