INTRODUCTION
The incidence of dengue and cancer has increased sharply over the past decade, and an estimate of 390 million dengue virus infections per year is indicated by a model (Bhatt et al., 2013). Until now, there have been no specific and effective treatments for dengue virus infection and cancer diseases. Interest in the study of the physiological activity of algae and their derivatives has been growing lately, leading to the exploration of millions of novel compounds with biological properties from various marine microsymbiont organisms (Yasuhara-Bell and Lu, 2010). Indonesia has a coastline of 81,000 km, second only to Canada, and comprises 17,000 islands. Such conditions make it an appropriate place for algae cultivation (Wouthuyzen et al., 2016).
Marine algae are classified into three groups, namely, Chlorophyceae, Rhodophyceae, and Phaeophyceae. Sargassum sp. which belongs to the group Phaeophyceae (brown algae) is widely cultivated in Indonesian waters. Based on the literature review, Sargassum sp. is often utilized as a producer of carrageenan (Manuel et al., 2015). Even though the proteins of the exophytic bacteria derived from Sargassum sp. repeatedly showed a unique biological activity such as antibacterial, antifungal, anticancer, and antidengue (Ahmad et al. 2019; Koishi et al., 2012; Rodrigues et al., 2009; Sugrani et al., 2019a), only a few studies have utilized the exophytic bacteria derived from Sargassum sp. Furthermore, Schmidt et al. (2010) demonstrated and reported that many peptides could inhibit dengue virus replication and infection. In recent years, some studies also released and reported that many types of peptides were used as antidengue agents (Chew et al., 2017; Hrobowski et al., 2005). Most recently, our group research also reported that many protein hydrolysates from different sources of microsymbionts marine algae could inhibit the pathogenic bacteria and cancer cells line (HeLa) (Asmi et al., 2020; Sugrani et al., 2020).
In our previous study, we examined the potential for the antidengue activity of proteins from the exophytic bacteria Enterobacter agglomerans SB 5 (1) derived from Sargassum sp. Potential protein fraction on antidengue activity was demonstrated in the 60%–80% saturation of ammonium sulfate fraction with the inhibition percentage and also CC50 value of 70% and 260.37 μg/ml, respectively (Ahmad et al., 2019). Nevertheless, the bioactivity results of antidengue and cytotoxic activity studies have not exhibited the maximum results. According to Wang and Zhang (2016), protein activity will increase from protein hydrolysis. Besides hydrolysis, low molecular weights are also related to the activity of protein hydrolysate. In this study, the protein of exophytic bacteria derived from Sargassum sp. would be fractioned, identified, and characterized by its bioactive protein hydrolysate with low molecular weight and its potential as a new antidengue and anticancer agents.
MATERIALS AND METHOD
Materials
This study used the exophytic bacteria E. agglomerans SB 5(1) from Sargassum sp. obtained from Biochemistry Laboratory, Hasanuddin University, Indonesia, egg of Artemia salina, Leach., HeLa cells, Vero cells, dengue virus serotype-2 (DENV-2), Viral ToxGlo™ Assay kit, Dulbecco's modified eagle medium (DMEM) media, 1% (w/v) L-glutamine, Bovine Serum Albumin (BSA), Lowry reagents, ammonium sulfate, cellophane bags, and sterile seawater.
Preparation of protein fraction
The cell cultures of the exophytic bacteria E. agglomerans SB 5 (1) were prepared according to the method described by Ahmad et al. (2019), with a few modifications. Tris–HCl buffer (20 mM, pH 8.2) was added to 30 g of cell pellet and then homogenized. It was then centrifuged at 4,000 rpm, at 4°C for 45 minutes. The supernatant was collected, then fractionated 20% by ammonium sulfate, stirred overnight at 4°C, and then centrifuged at 10,000 rpm at 4°C for 60 minutes to obtain the deposition of protein. This protein fraction was stored in the refrigerator for enzymatic hydrolysis (Uluko et al., 2015).
Determination of protein content
The determination of protein content by the Lowry method was measured by a UV–Vis spectrophotometer with BSA as standard (Ahmad et al., 2019).
Preparation of protein hydrolysates
Protein fractions were dissolved 3% (v/v) in 20 mM Tris–HCl buffer at pH 7, and protein suspensions were hydrolyzed by pepsin enzyme (EC 3.4.23.1) using a 6% w/w enzyme to substrate ratio, at the temperature of 37°C, pH 2 for 2 hours, and the enzymatic hydrolysis ceased by heating at 95°C for 10 minutes (Sugrani et al., 2020). The protein hydrolysate samples were placed at −20°C prior to use. The degree of hydrolysis (DH) was calculated using equation (1). Pepsin hydrolysates were separated using membrane molecular weight cut-off (MWCO) membranes (3, 5, and 10 kDa). The fraction was frozen at −20°C for further analysis (Uluko et al., 2015).
DH (%) = [(h−h0)/Htot] × 100 (1)
where “h” is the number of peptide bonds cleaved during hydrolysis, “h0” is the content of free amino groups of substrates, and “Htot” is the total number of peptide bonds in the protein substrate (8.6 mEq/g protein) (Cian et al., 2012).
Ultrafiltration
The separation of MWCO, which used a Vivaspin 20 Fisher Scientific, was the ultrafiltrates lyophilized hydrolysate process. The fractions were collected from the molecular weight (MW) range of >10 kDa, 5–10 kDa, 3–5 kDa, and <3 kDa and stored at −20°C (Guo et al., 2008). These fractions were Peptides Enterobacter from Algae (PEA): PEA1a, PEA1b, PEA1c, and PEA1d.
SDS-PAGE electrophoresis
The purified level and molecular weight of the peptide were determined using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 7% and 10% SDS, using a Mini-PROTEAN 3 cells apparatus (Laboratories of Bio-Rad, Irvine, CA). About 3 μg/μl of peptide and marker was dissolved in the sample buffer and transferred to the vertical slab space electrophoresis system (height 10 cm, width 10 cm, and thickness 0.6 mm). The electrophoresis process was running at 60 mA for 3–4 hours at room temperature (Celebioglu et al., 2017) and a protein marker (Fermentas, Lithuania) was used as a molecular weight standard. Finally, the pattern of protein bands was visualized with Coomassie Brilliant Blue R250 described by Fluka USA.
Bioactivity test
Toxicity and antidengue test
Toxicity and the antidengue tests were conducted using Brine Shrimp Lethality (BSLT) and 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) method, respectively. BSLT was used to test the cytotoxic according to Sugrani et al. (2019b), and Ahmad et al. (2019). Observation data were tabulated, and probit analysis was carried out to get the value of the Lethal Concentration 50 (LC50). Antidengue test of protein fraction and its hydrolysates was carried out against Vero cells infected with the DENV-2 virus. Cytotoxic tests were carried out using Viral ToxGlo™ Assay kit (Promega) based on Karimi et al. (2016). The measurements of cells on the viability percentage (%) and the 50% cytotoxic concentration (CC50) value (μg/ml) were calculated from the percentage of inhibition at 20 μg/ml samples administration, based on the probit analysis (Ahmad et al., 2019).
Cytotoxicity test
Cytotoxicity of peptide fraction was carried out quantitatively using the MTT method (Boomi et al., 2019). HeLa were preserved in DMEM with 10% fetal bovine serum (FBS) and antibiotic-antimycotic solution. The cell was allowed to grow at a temperature of 37°C with a continuous supply of 5% CO2. HeLa cells (2 × 103 cells/100 μl) inserted into 96-well plates contained 200 μl DMEM media and 10% FBS. The plates were incubated under conditions similar to 5% CO2. After 24 hours of culture, the used media were removed. Fresh DMEM media with determined concentrations (200; 100; 50; 25; 12.5; 6.25; 3.13; and 1.56 μg/ml) and hydrolysate and peptide samples were added and incubated again under conditions similar to 5% CO2. The used media were replaced with 10 μl MTT solutions for each well (5 μg/ml) and incubated for 4 hours at 37°C in a humid incubator with 5% CO2. After 4 hours, the MTT was removed, and a 100 μl DMSO was added to all the wells. Spectral absorbance was observed at 570 nm. Cells themselves except the addition of peptides/hydrolysates were considered as controls.
Identification of bioactive protein hydrolysates fraction
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
The pepsin peptide fraction <3 kDa was dissolved in acetonitrile and then filtered by a Millipore filter unit with a size of 0.2 μm. A total of 10 μl of sample filtrate was injected into the LC-ESI-QTOF instrument system. LC-MS analysis was carried out by the UPLC-MS [ACQUITY UPLC®H-Class System (waters, Milford, MA)] equipped with a binary pump. LC was connected to a QTOF mass spectrometer equipped with an ESI ionization source. MS used Xevo G2-X2 QT with positive ionization mode. The ESI parameters used in full scan mode from m/z 50–3,000 were carried out with a source temperature of 50°C (column) and 25°C (room). The UPLC column used CORTECS C8 Column (2.7 μm, 2.1 mm × 100 mm). The effluents used were H2O plus 5 mM ammonium formate and acetonitrile plus 0.05% formic acid, flow velocity: 0.2 ml/minute (gradient) (Wang and Zhang, 2016).
Database searching and in silico activity prediction
The analysis resource and immune epitope database (IEDBe) were used for antidengue prediction (Guevarra et al., 2020). The AntiCP was used for anticancer activity prediction (Tyagi et al., 2013). Prot Param was used to calculate various chemical and physical parameters of the isolated peptide fraction (Gasteiger et al., 2005).
RESULT AND DISCUSSION
Protein isolation and protein hydrolysate production
In this study, protein isolation from the exophytic bacteria E. agglomerans SB 5 (1) derived from Sargassum sp. (Ahmad et al., 2019) was conducted by using a combined method (mechanical and nonmechanical) with Tris–HCl buffer media pH 8.3. The results showed that the extraction yield of 20% protein (PEA) was 7.50%, with a protein content of 27.78 mg/ml (Table 1). The fraction of protein content (22.78 mg/ml) was higher than the level of crude extract (4.45 mg/ml) of the exophytic bacteria E. agglomerans SB 5(1) derived from Sargassum sp. Compared with our previous study, the combined method obtained higher yield protein (7.50%) than those previously reported in the same resources, which only obtained 2.30%–3.50% of the protein fraction (Ahmad et al., 2019; Karim et al., 2020).
Furthermore, the isolated protein was hydrolyzed using the commercial enzyme by pepsin (EC 3.4.23.1) to evaluate its biological waste. Enzymatic hydrolysis was carried out to release bioactive bacterial peptides. Protein compounds from algae symbiont bacterial were valuable sources of bioactive peptides, trapped in sequences of primary amino acids, which are released through enzymatic hydrolysis during food digestion in the digestive tract (Beaulieu et al., 2015). The DH of PEA1 was more than 10%. According to Morris et al. (2008), such a DH has the potential to be used to produce pharmaceutical products.
Bioactivity test
Toxicity and antidengue potential activity
The toxicity test results of the protein fraction (PEA) and pepsin hydrolysate (PEA1) can be seen in Table 2. The smaller the toxicity value, the more the significant toxicity activity. Based on this result, PEA1 was continued in the next stage. Overall, the toxicity value, as shown in Table 2, showed that protein and its hydrolysate fractions have different values. Toxicity value of PEA (protein fraction), PEA1 fraction (pepsin hydrolysate), PEA1a fraction (pepsin peptide fraction >10 kDa), and PEA1b fraction (pepsin peptide fraction 5–10 kDa) showed high toxic levels. Meanwhile, PEA1c fraction (pepsin peptide fraction 3–5 kDa) and PEA1d fraction (pepsin peptide fraction <3 kDa) showed remarkably high toxic levels. The various toxicity values of the protein fraction and its hydrolysate are due to the biological activity. Peptides with small molecular weights often exhibit substantial biological activities (Vandanjon et al., 2007). Pepsin peptide fraction <3 kDa (PA1d) was the potential peptide with the lowest LC50 (1.77 μg/ml) compared to other fractions.
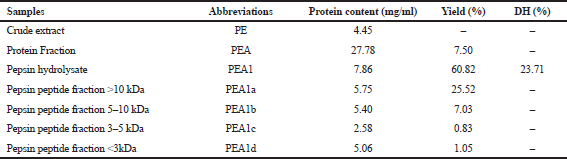 | Table 1. The DH, the protein content fraction, hydrolysate, and peptides of exophytic bacteria E. agglomerans SB 5 (1) from Sargassum sp. [Click here to view] |
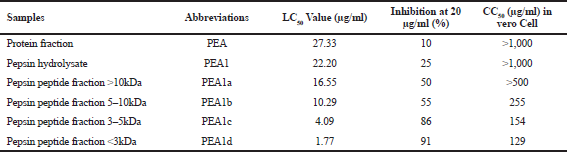 | Table 2. Summary of protein and its hydrolysate for toxicity and antidengue potential after the administration of protein and its hydrolysates fraction. [Click here to view] |
The antidengue properties of the protein and its hydrolysate fractions were confirmed by cytotoxicity test using Vero cells infected with the DENV-2 virus. The antidengue activity of protein fraction and its hydrolysate against Vero cells indicated the potential antidengue. The highest inhibition percentage and CC50 were PEA1d (91 %, 129 μg/ml, respectively), followed by PEA1c (86%, 154 μg/ml, respectively). Some peptide sequences exhibited an antiviral effect on dengue in cell culture. For instance, peptide sequence FWFTLIKTQAKQPARYRRFC has been reported as antidengue and displayed an IC50 value of 7 μM in a focus-forming unit assay (Costin et al., 2010).
Anticancer potential activity
Based on toxicity and antidengue results, it was found that PEA1d is a potential isolated protein. The PEA1d was then tested further in its cytotoxic activity by using the MTT method against HeLa cells as a model of cervical cancer cells. The cytotoxic activity can be observed from the absorbance value (Table 3), which was used to calculate the viability percentage of HeLa cells (Fig. 1). The results showed that PA1d could inhibit HeLa cell growth with an IC50 value of 0.75 μg/ml, considered to be a very toxic level. According to E-Kobon et al. (2016), anticancer peptide activity is influenced by several things, including amino acid composition and physicochemical properties such as charge, aliphatic index, and amphipathic and structural or confirmation form.
SDS-PAGE electrophoresis
The profile of the extracted protein, protein hydrolysate, and peptide from the exophytic bacteria E. agglomerans SB 5 (1) derived from Sargassum sp. was determined by running SDS-PAGE (Fig. 2). Estimated MW for each protein was carried out by comparing the distance between sample mobility with protein markers in the SDS-PAGE chromatogram.
Electrophoresis results showed that crude extracts (PE) had scattered and thick bands at various molecular weights. The thickest crude extract tapes were at MW 173.33 kDa, and after fractionation with 20% ammonium sulfate, 20% protein fraction (PEA) was seen having an MW of 27.67 kDa. These results indicated that ammonium sulfate successfully fractionated crude protein samples. This 20% protein fraction was then hydrolyzed using the pepsin enzyme to become protein hydrolysate (PEA1), which appears to have a band at MW of 20.02 kDa, but with a thinner or faded band color, indicating that the hydrolysis successfully occurred. Ultrafiltration of protein hydrolysate showed PEA1c, and PEA1d had low molecular weight at MW of 4.25 kDa and 2.98 kDa, respectively.
Identification of bioactive peptides
LC-MS/MS was chosen for further characterization of peptides. The results of peptide patterns contained in the PEA1d fraction are shown in Figures 3 and 4. Peptide identification was carried out using MASCOT mass fingerprint peptide software (de novo sequencing). De novo sequence peptide was determined from MS/MS spectra data (Fig. 4), fragments PEA1d-1 (344.2310; 345.2341; 346.2357), PEA1d-2 (388.2568; 389.2601; 390.2616), PA1d-3 (432.2827; 436.2856; 434.23; 476.3097; 477.3118; 478.3138), PEAd1-4 (476.3097; 477.3118; 478.3138; 497.2363; 504.3391; 519.3161), PEAd1-5 (520.3341; 521.3376; 522.3398), PEAd1-6 (564.3606; 565.3635; 566.3657; 567.3682), and PEA1d-7 (608.3867; 609.3898; 6109.389). A total of seven sequences of pepsin peptide fraction <3kDa were identified by the MASCOT program (score >60%) with NCBIprot as a protein sequence database (Table 4). All identified peptide fragments had come from bacteria-derived proteins. All peptide fragments had low molecular masses of less than 3 kDa. Of seven peptides, three peptide fragments were negatively charged, three peptide fragments were positively charged, and one was a neutral fragment.
In our study, seven peptide fragment sequences were obtained from the enzymatic hydrolysis process of a protein fraction, namely, MGFIVLGRLEAFVDLCHL (PEA1d-1), MQLDYLECFKGVLGEALRQLGRR (PEA1d-2), NSLKATLCLSLTLALAPSL (PEA1d-3), LADGLDSYLELPLFTAVAIL (PEA1d-4), MEELSRNFYIVFIIIGLYSMF (PEA1d-5), SGACTFGMVFLTGCGLF (PEA1d-6), and VRSIFVRVQLEALYLCRAIFHDVVF (PEA1d-7). All of the discovered peptide sequences seemed to be rich in leucine amino acid residue, especially in PEA1d3 and PEA1d4 fractions, which contained the leucine-rich repeat motifs, and these motifs were not observed in the other five peptide sequences. Thus, these leucine-rich repeat motifs in PEA1d3 and PEA1d4 may contribute to antidengue and anticancer properties (Tables 2, 3, and 5). According to Brogden (2005), leucine-rich repeat motifs had an effect on the unique bioactivity of proteins and peptides. Search results on the NCBIprot and UniProt/SWISS-PROT protein and peptide databases of all peptide sequences (PEA1d-1–PEA1d-7), especially four leucine-rich repeat motifs in PEA1d3 and PEA1d4, did not show any report of these peptides; therefore, these were the first reported peptide sequences.
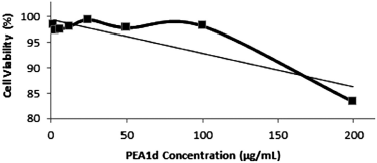 | Figure 1. The cytotoxic effect of PEA1d peptides on the viability of HeLa cells. [Click here to view] |
 | Table 3. The data of cytotoxic activity against HeLa cell cancer from PEA1d peptides. [Click here to view] |
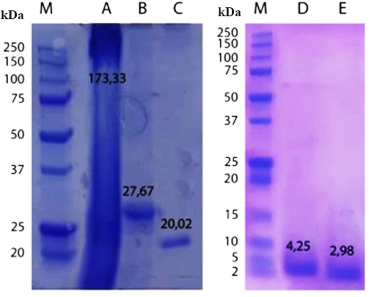 | Figure 2. SDS-PAGE chromatogram of protein and its hydrolysate fractions. Marker (M), crude extract (A), protein fraction 20% (B), protein hydrolysate (C), PEA1c peptides 3–5kDa (D), and PEA1d peptides <3kDa (E). [Click here to view] |
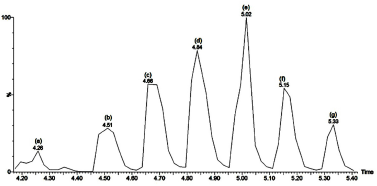 | Figure 3. Liquid chromatography (LC) chromatogram of PEA1d-1 to PEA1d-7 peptide fraction: (a) PEA1d-1, (b) PEA1d-2, (c) PEA1d-3, (d) PEA1d-4, (e) PEA1d-5, (f) PEA1d-6, and (g) PEA1d-7. [Click here to view] |
On the other hand, out of the seven peptide fragments, PEA1d-3 had the highest cationic charge among the others (+2). Cationic charge (positive) determined the antidengue (Guevarra et al., 2020; Schmidt et al., 2010) and anticancer activity (E-Kobon et al., 2016; Hajisharifi and Mohabathar, 2013) of a peptide because the positive charge of the peptide becomes the initial step of electrostatic interaction with cell membranes (cancer, bacteria, or virus) which are negatively charged (E-Kobon et al., 2016; Hajisharifi and Mohabathar, 2013). However, the positive charge of active peptides is known to have limits. The seven peptides were further analyzed using in silico antidengue prediction of the analysis resource and IEDB (http://www.iedb.org/assay/15063), also anticancer prediction using AntiCP (https://webs.iiitd.edu.in/raghava/anticp) (Table 5). Although in vitro and in vivo assays have been used, these are both financially and time costly and require more sampling. Conversely, in silico analysis can reduce costs, speed up quantification time, and minimize the use of test samples (Sucher, 2014).
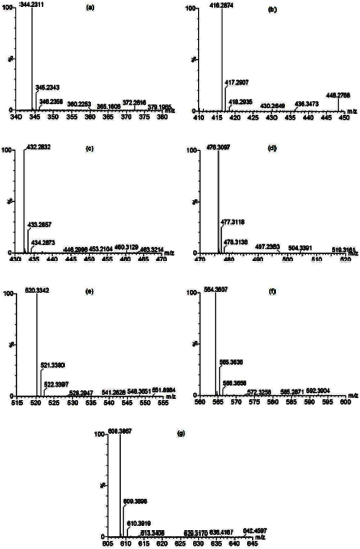 | Figure 4. MS/MS spectra of PEA1d-1 to PEA1d-7 peptide fraction: (a) PEA1d-1, (b) PEA1d-2, (c) PEA1d-3, (d) PEA1d-4, (e) PEA1d-5, (f) PEA1d-6, and (g) PEA1d-7. [Click here to view] |
There were two peptides based on the in silico analysis, which is potentially responsible for the antidengue and anticancer activities from peptides of PEA1d-3 (NSLKATLCLSLTLALAPSL) and PEA1d-4 (LADGLDSYLELPLFTAVAIL). Interestingly, both of these peptide sequences consist of four leucine-rich repeat motifs (L in a bold and underlined letter), and this specific motif may contribute to the physiological function of peptides, including antidengue and anticancer properties. The exact mechanism of antidengue and anticancer peptides was not identified in this study and would require further work. According to Wang (2020), frequently active peptides were cationic peptides with +3 to +8 charges. Research by Hajisharifi and Mohabatkar (2013) on P38 and P22 peptides from E. coli protein, P38 peptides which have a cationic charge +7 active as anticancer and P22 which has a charge +10, showed no activity.
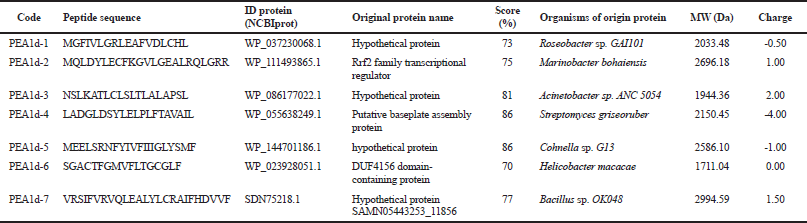 | Table 4. Results of PEA1d peptide identification with MASCOT program. [Click here to view] |
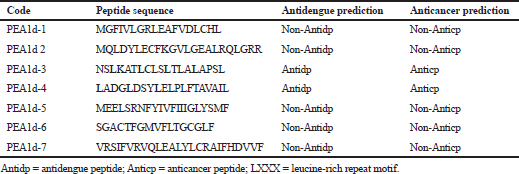 | Table 5. The amino acid sequences and antidengue and anticancer activity prediction of PEA1d-1 to PEA1d-7 peptides fragment. [Click here to view] |
Based on the aforementioned facts, it would be very early to state the sequence of PA1d-3 peptides that most contribute to the activity of the pepsin peptide fraction <3kDa (PA1d). Further research is required whether peptides from PA1d play an individual role or synergize with other peptides as Beaulieu et al. (2015) stated that the existence of bioactivity of peptides probably can be caused by synergism between several peptides.
CONCLUSION
The PEA1d fraction (pepsin peptide fraction <3kDa) which showed potential antidengue activity and cytotoxic activity with the CC50 value of 129 μg/ml against Vero cells and IC50 value of 0.749 μg/ml against HeLa cells is considered to be highly toxic. Seven peptide sequences were obtained (PEA1d-1–PEA1d-7) in the PEA1d fraction, but just two novel peptide sequences were identified and predicted to be the source of antidengue and cytotoxic activity, namely, PEA1d-3 (NSLKATLCLSLTLALAPSL) and PEA1d-4 (LADGLDSYLELPLFTAVAIL). It can be concluded that these peptides can be potent candidates for new drug production, especially for antidengue and anticancer from natural products, and developed as synthetic peptides in the future.
ACKNOWLEDGMENTS
The authors would like to express special thanks and gratitude to KEMENRISTEK (Directorate of Research and Development)/BRIN Indonesia for the fund, research contract number: “1516/UN4.22/PT.01.03/2020”, Applied Research (PT) scheme the for the 2nd year. They also thank Dr. Muh. Ade Artasasta and Syadza Firdausiah, MSc., for their writing assistance and proofreading of this manuscript.
AUTHOR’S CONTRIBUTION
Ahmad A and Massi MN gathered data and were conceptor of this study. Ahmad A, Sugrani A, and Karim H contributed to the study design. Asmi N, Wahid I, Ahmad A, and Karim H contributed to data collection. Ahmad A, Sugrani A contributed to manuscript writing. Ahmad A, Sugrani A, Asmi N, and Wahid I edited the final version of the manuscript.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
FUNDING
None.
REFERENCES
Ahmad A, Massi MN, Wahid I, Arfah R, Karim H. Anti-dengue potential of bioactive protein from exophytic bacteria that are symbiotic with Sargassum sp. J Phys Conf Ser, 2019; 1341:032012; doi:10.1088/1742-6596/1341/3/032012 CrossRef
Asmi N, Ahmad A, Karim H, Massi MN, Natsir H. Antibacterial effect of protein and protein hydrolysates isolated from bacteria Enterobacter hormaechei associated with marine algae Sargassum sp. Rasayan J Chem, 2020; 13(3):1606–11. CrossRef
Beaulieu L, Bondu S, Doiron K, Rioux LE, Turgeon SL. Characterization of antibacterial activity from protein hydrolysates of the macroalga Saccharina longicruris and identification of peptides implied in bioactivity. J Funct Foods, 2015; 17:685–97. CrossRef
Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, William Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature, 2013; 496(7446):504–7. CrossRef
Boomi P, Ganesan RM, Poorani G, Prabu HG, Ravikumar S, Jeyakanthan J. Biological synergy of greener gold nanoparticles by using Coleus aromaticus leaf extract. Mater Sci Eng C, 2019; 99:202–10. CrossRef
Brogden KA. Antimicrobial peptides: pore former or metabolic inhibitor in bacteria. Nat Rev Microbiol, 2005; 3:238–50. CrossRef
Celebioglu HU, Olesen SV, Prehn K, Lahtinen SJ, Brix S, Hachem MA, Svensson B. Data regarding the growth of Lactobacillus acidophilus NCFM on different carbohydrates and recombinant production of elongation factor G and pyruvate kinase. Data Brief, 2017; 14:118–22. CrossRef
Chew MF, Poh KS, Poh CL. Peptides as therapeutic agents for dengue virus. Int J Med Sci, 2017; 14(13):1342–59. CrossRef
Cian RE, Martinez-Augustin O, Drago SR. Bioactive properties of peptides obtained by enzymatic hydrolysis from protein byproducts of Porphyracolumbina. Food Res Int, 2012; 49:364–72. CrossRef
Costin JM, Jenwitheesuk E, Lok SM, Hunsperger E, Conrads KA, Fontaine KA, Rees CR, Rossmann MG, Isern S, Samudrala R, Michael SF. Structural optimization and de novo design of dengue virus entry inhibitory peptides. PLoS Neglected Trop Dis, 2010; 4:e721; doi:10.1371/journal.pntd.0000721 CrossRef
E-kobon T, Thongararm P, Roytrakul S, Meesuk L, Chumnanpuen P. Prediction of anticancer peptides against MCF-7 breast cancer cells from the peptidomes of Achatina fulica mucus fractions. Comput Struct Biotechnol J, 2016; 14:49–57. CrossRef
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook, Humana Press, Totowa, NJ, pp 571–607, 2005.
Guevarra LA Jr, Boado KJO, Ceñidoza FBB, Imbao MRLM, Sia MJG, Dalmacio LMM. A synthetic peptide analog of in silico-predicted immunogenic epitope unique to dengue virus serotype 2 NS1 antigen specifically binds immunoglobulin G antibodies raised in rabbits. Microbiol Immunol, 2019; 1–9. 2020; 64(2):153–61.
Guo Q, Cui SW, Wang Q, Young JC. Fractionation and physicochemical characterization of psyllium gum. Carbohydr Polym, 2008; 73(1):35–43. CrossRef
Hajisharifi Z, Mohabatkar H. In silico prediction of anticancer peptides by TRAINER tool. Mol Biol Res Commun, 2013; 2:39–45.
Hrobowski YM, Garry RF, Michael SF. Peptide inhibitors of dengue virus and west nile virus infectivity. Virol J, 2005; 2:49; doi:10.1186/1743-422X-2-49 CrossRef
Karim H, Ahmad A, Asmi N, Natzir R, Massi MN, Hatta M, Kabo P, Djide MN, Minhajat R, Hasyim Z, Ibrahim M. Optimization of enzyme activity of L-Asparaginase derived from Enterobacter agglomerans SB 221 bacterial symbiont of Sargassum sp. Rasayan J Chem, 2020; 13:1571–9. CrossRef
Karimi A, Kamalabadi MM, Rafieian-Kopaei M, Amjad L, Salimzadeh L. Determination of antioxidant activity, phenolic contents and antiviral potential of methanol extract of Euphorbia spinidens bornm (Euphorbiaceae). Trop J Pharm Res, 2016; 15(4):759–64. CrossRef
Koishi AC, Zanello PR, Bianco VM, Bordignon J, Santos CND. Screening of dengue virus antiviral activity of marine seaweeds by an In Situ enzyme-linked immunosorbent assay PLoS One, 2012; 7(12):e51089; doi:10.1371/journal.pone.0051089 CrossRef
Manuel J, Cornelia M, Wijaya W. Utilization of Eucheuma cottonii and Eucheuma spinosum flour in narrow-barred Spanish mackerel meatballs. KnE Life Sci, 2015; 12–8; doi:10.18502/kls.v1i0.78 CrossRef
Morris HJ, Almarales A, Carrillo O, Bermúdez RC. Utilisation of chlorella vulgaris cell biomass for the production of enzymatic protein hydrolysates. Bioresour Technol, 2008; 99:7723–9; doi:10.1016/j.biort ech.2008.01.080 CrossRef
Pan X, Zhao Y Q, Hu F Y, Chi C F, Wang B. Anticancer activity of a hexapeptide from skate (Raja porosa) cartilage protein hydrolysate in HeLa Cells. Mar Drugs, 2016; 14(8):153; doi:10.3390/md14080153 CrossRef
Rodrigues EG, Dobroff AS, Taborda CP, Travassos LR. Antifungal and antitumor models of bioactive protective peptides. An Acad Bras Ciênc, 2019; 81:503–20. CrossRef
Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Phatogens, 2010; 6(4):e1000851; doi:10.1371/journal.ppat.1000851 CrossRef
Sucher NJ. Searching for synergy in silico, in vitro, and in vivo. Synergy, 2014; 1(1):30–43. CrossRef
Sugrani A, Ahmad A, Djide MN, Natsir H. Biofunctional protein fraction from red algae (Rhodophyta) Eucheuma spinosum as an antibacterial and anticancer drug agent. Int Res J Pharm, 2019a; 10:64–9. CrossRef
Sugrani A, Ahmad A, Djide MN, Natsir H. Toxicological evaluation and antibacterial activity of crude protein extract from endophytic bacteria associated with Algae Eucheuma spinosum. J Phys Conf Ser, 2019b; 1341:032006; doi:10.1088/1742-6596/1341/3/032006 CrossRef
Sugrani A, Ahmad A, Djide MN, Natsir H. Two novel antimicrobial and anticancer peptides prediction from vibrio sp. strain ES25. J Appl Pharm Sci, 2020; 10(08), 058–66.
Tyagi A, Kapoor P, Kumar R, Chaudhary K, Gautam A, Raghava GPS. In silico models for designing and discovering novel anticancer peptides. Sci Rep, 2013; 2984(3):1–7. CrossRef
Uluko H, Zhang S, Liu L, Tsakama M, Lu J, Lv J. Effects of thermal, microwave, and ultrasound pretreatments on antioxidative capacity of enzymatic milk protein concentrate hydrolysates. J Funct Foods, 2015; 18:1138–46. CrossRef
Vandanjon L, Johannsson R, Derouiniot M, Bourseau P, Jaouen P. Concentration and purification of blue whiting peptide hydrolysates by membrane processes. J Food Eng, 2007; 83:581–9. CrossRef
Wang G. The antimicrobial peptide database provides a platform for decoding the design principles of naturally occurring antimicrobial peptides. Protein Sci, 2020; 29(1):8–18; doi: 10.1002/pro.3702 CrossRef
Wang X, Zhang X. Isolation and identification of anti-proliferative peptides from Spirulina platensis using three-step hydrolysis. J Sci Food Agric, 2016; 97:918–22. CrossRef
World Health Organization. Dengue and severe dengue, 2020. Available via https://www.who.int/news-room/fact-sheets/ detail/dengue-and-severe-dengue (Accessed 28 July 2020).
Wouthuyzen S, Herandarudewi SM, Komatsu T. Stock assessment of brown seaweeds (Phaeophyceae) along the Bitung-Bentena Coast, North Sulawesi Province, Indonesia for alginate product using satellite remote sensing. Procedia Environ Sci, 2016; 33:553–61. CrossRef
Yasuhara-Bell J, Lu Y. Marine compounds and their antiviral activities. Antiviral Res, 2010; 86:231–40. CrossRef