INTRODUCTION
Pseudomonas aeruginosa is a pathogen that can survive in different environments (Logan et al., 2017). It is also described as an important human pathogen, responsible for nosocomial infections (Ruiz-roldán et al., 2018). It is reported that multidrug-resistant (MDR) P. aeruginosa infections were associated with cost increase per patient in comparison with those nonresistant ones (Morales et al., 2012). Children are much more sensitive to disease-causing bacteria than adults, whereas no descriptions exist about characterization of P. aeruginosa isolates from children (Ruiz-roldán et al., 2018).
However, the treatment of those infections has become of importance as a result of its antibiotic resistance behavior (Pachori et al., 2019). Nowadays, P. aeruginosa displays resistance to a variety of antibiotics, including different classes (Alnour and Ahmed-Abakur, 2017), besides a large number of virulence factors (toxin A, alkaline protease, elastase, exoenzymes, and pyocyanin production) that play important roles in the pathogenicity process (Jazayeri et al., 2016). Bacterial biofilm formation in both medical and industrial fields causes big problems, as these structures resist the antibiotics and biocides treatments, besides reducing the host immune-responses action (Lima et al., 2018). Pseudomonas aeruginosa intrinsic resistance includes efflux pump expression that expels antibacterial agents outside the cell and produces antibiotic inactivating enzymes (Sachdeva et al., 2017). Because of all this, this study was aimed at studying antibiotic resistance profile, resistance mechanisms, and virulence factors in P. aeruginosa collected from children.
MATERIALS AND METHODS
Collection and identification P. aeruginosa strains
The study was conducted with 12 P. aeruginosa selected out of 136 Pseudomonas species that were collected from clinical samples routinely sent to the microbiology laboratory in a tertiary care pediatric Cairo University hospital in 3 months from October 2019 to December 2019. All the collected isolates were identified according to the standard procedures and confirmed by growing on Cetrimide agar and examined under ultraviolet to see the fluorescence of their colonies (Singh et al., 2015). The standard strain P. aeruginosa (American Type Culture Collection 15442) was used as control.
Antibiotics susceptibility tests
The bacterial antibiotic susceptibility profile was tested using Kirby–Bauer disk diffusion technique (Clinical and Laboratory Standards Institute (CLSI), 2019), using the commercial antibacterial agents (Oxoid, UK): ciprofloxacin (CIP, 5 μg), meropenem (MEM, 10 μg), gentamycin (CN, 10 μg), amikacin (AK, 30 μg), cefotaxime/cloxacillin (CTC, 40 μg), cefotaxime (CTX, 30 μg), cefoxitin (FOX, 30 μg), ceftriaxone (CRO, 30 μg), and cefepime (FEP, 30 μg).
Detection of virulence factors
Biofilm ability using microtiter plate assay
Biofilm formation
Pseudomonas aeruginosa isolates were screened for their biofilms formation ability by microtiter plate according to the method described (Mohamed et al., 2018c). 100 μl P. aeruginosa suspension of 0.5 McFarland was added to sterile brain heart infusion broth (100 μl) in wells of 96-well microtiter plate (Cellstar®, Greiner Bio-One) and then incubated at 37°C. The assay was done in triplicate.
Biofilm quantification using crystal violet (CV) assay
After 18 hours incubation period, wells were evacuated and gently washed. Pseudomonas aeruginosa biofilms formed on the walls were dried and stained with 1% CV for 20 minutes. After properly washing and drying, the stained P. aeruginosa biofilm was reconstituted in absolute ethanol, and its absorbance was measured at 630 nm using microplate reader (Stat Fax-2100; GMI, Inc.,). Optical density cut-off value (O.Dc) was calculated as mentioned by (Mohamed et al., 2020b).
Pyocyanin production
Phenotypic visual detection
Phenotypic detection was done as described with minor modifications. Briefly, the bacterial suspension (0.5 McFarland) was swabbed on Pseudomonas-Agar P media plates supplemented with 1.5% glycerol. After incubation for 24 hours, agar was cut into small pieces, and pyocyanin was extracted with chloroform after shaking. The chloroform color changed to dark green or blue and then was re-extracted into 1 ml of HCl (0.2N) to give a pink to a deep red solution, which was considered positive results (Feghali and Nawas, 2018).
Quantitative of pyocyanin
Pigment quantification was done depending on the absorbance of pyocyanin in acidic solution, at which solution was measured, and the concentrations expressed as μg/mL of pigment produced by the culture supernatant were calculated by using the optical density reading at 520 nm multiplying by 17.072 (Khadim and Marjani, 2019).
Cluster analysis
Based on the phenotypic traits like antibiotics susceptibility results and virulence factors (biofilm formation ability and pyocyanin production) of different P. aeruginosa strains, hierarchical cluster analysis was performed using Statistical Package for the Social Sciences software (SPSS Inc. v. 12). The antibiotic susceptibility results for each were coded as “1” resistant and “0” nonresistant. The positive results of biofilm formation and pyocyanin production for each strain were coded as “1” where the negative ones were coded “0.” The analysis was presented graphically to find the similarity among strains, based on average linkage, and the branch length represents the distance between the strains (Mohamed et al., 2018a).
Phenotypic detection of efflux pump activity
Minimum inhibitory concentration (MIC)
Resistance to CIP was confirmed by MIC evaluation using an antibiotic agar dilution test (Al Rashed et al., 2020). CIP stock solution was prepared according to manufacturer recommendation. Antibiotic final concentration range of 2 mg/l up to 512 mg/l and 0.5 McFarland standard bacterial turbidity were used. A volume of 10 μl of each inoculum was pipetted into serial agar plates. After incubation for 18 hours at 37°C, MIC was read. The interpretation of MIC values was done according to CLSI recommendations (Clinical and Laboratory Standards Institute (CLSI), 2019).
Ethidium bromide-(EtBr-) agar cartwheel method
Tryptic soy agar plates containing EtBr concentrations (Sigma-Aldrich) of 0–3 mg/l were swabbed with bacterial suspension (0.5 MacFarland), making cartwheel pattern (Osman et al., 2018). All plates were examined under the UV light, after the incubation period of 18 hours at 37ºC. EtBr concentration at which the swabbed culture produces fluorescence was recorded. The higher the concentration of EtBr required for the appearance of fluorescence considered, the greater the EP activity. P. aeruginosa standard strain is considered a negative control.
Detection of MβLs
This test was done using the combined imipenem/ethylenediaminetetraacetic acid (EDTA) disk method (Alkhudhairy and Al-Shammari, 2020). Single imipenem disk and imipenem disk (10 μg) supplemented with 10 μl EDTA (750 μg) were placed on the plate and incubated for 18 hours period at 37°C. A 7 mm inhibition increase of the imipenem/EDTA disk than the single imipenem disk was considered a positive result.
Genotypic detection of resistance mechanisms
Total bacterial DNA extraction was done as mentioned by Mohamed and Khalil (2020). Detection of resistance genes was done using primers mentioned in Table 1. Cycling conditions for the efflux pump regulatory genes and both blaIMP and blaVIM genes were performed as mentioned. The amplified DNA was electrophoresed using 1.6% agarose gel stained with EtBr, and the bands were visualized under ultraviolet trans-illuminator (Alkhudhairy and Al-Shammari, 2020; Osman et al., 2018).
RESULTS
In this study, 12 P. aeruginosa strains were included, at which endotracheal aspirate (50%) was the most predominant source among our isolates, followed by sputum (33.33%) and urine (16.67%). High resistance rates (100%) toward cephems antibiotics (including CTX, FOX, and CRO) and carbapenems antibiotic (MEM) were observed (Table 2). On the other hand, imipenem and AK were the most effective antibiotics by sensitivity percentages of 41.7 and 33.3, respectively. Out of 12 strains, 8 (66.7%) were able to form a biofilm at which they were categorized as moderate (25%) and weak (41.7%); besides, 4 isolates (33.3%) were found non-biofilm formers. Three isolates showed positive results through the visual pyocyanin test (Table 3), and by spectrophotometer, pyocyanin concentrations of 1.23, 1.1, and 0.836 μg/ml were detected.
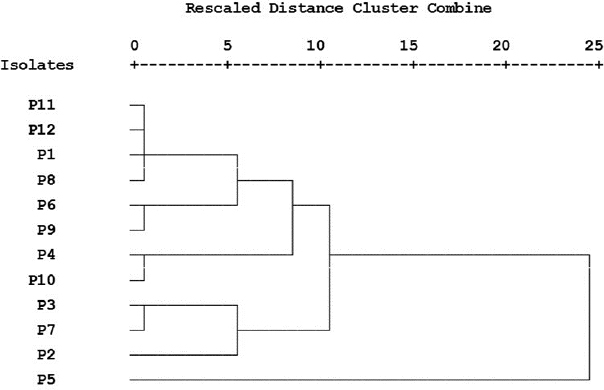
Cluster analysis of converted antimicrobial susceptibility and virulence factors production data was performed and presented in a dendrogram (Fig. 1) showing the difference between the used P. aeruginosa strains.
CIP susceptibility was confirmed among all isolates using agar dilution method, at which isolates were categorized at breakpoint recommended by CLSI, one isolate was categorized as not resistant with MIC value <2 μg/ml, one isolate was categorized as resistant with MIC value 4 μg/ml, three isolates were categorized as resistant with MIC value 16 μg/ ml, two isolates were categorized as resistant with MIC value 32 μg/ml, two isolates were categorized as resistant with MIC value 128 μg/ml, and three isolates were categorized as resistant with MIC value 256 μg/ml. By using the EtBr-agar Cartwheel method, results showed that isolates express efflux pump activity by different levels. The ATCC strain was found to fluoresce at EtBr concentration of 0.5 mg/l (negative control). The minimum concentration at which the adapted isolates showed fluorescence was 1 mg/l for isolates P1, P2, P3, and P4, while it was 1.5 mg/l for isolate P10, but isolate P8 was 2.5 mg/l, and isolates P11 and P12 were 3 mg/l; however, the isolates P6, P7 and P9 were more than 3 mg/l. The previous results revealed that isolates P6, P7, and P9 have a high efflux pump activity. Seven isolates (58.3%) were defined as imipenem resistance using the cut-off value of 1.5 cm according to CLSI recommendation using disk diffusion method. By using imipenem/EDTA combined disk test, the increase in inhibition zone with the imipenem/EDTA disk 7 mm or more than the imipenem disk alone was observed in five isolates [positive metallo-β-lactamase (MBL)] (Table 4).
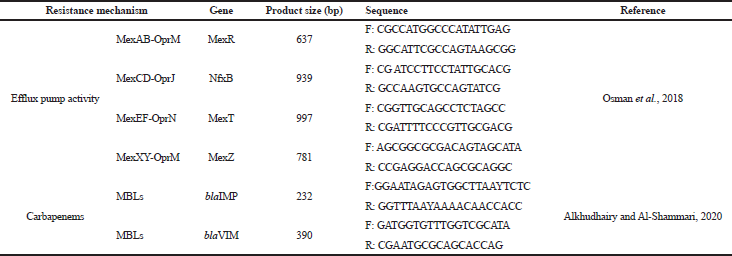 | Table 1. Primer sequences used in this study. [Click here to view] |
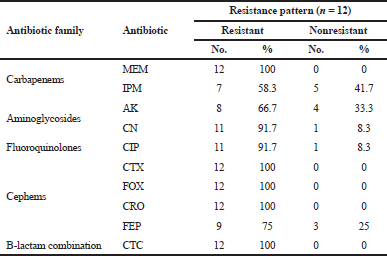 | Table 2. Resistance percentages of P. aeruginosa toward different antibiotics. [Click here to view] |
All CIP-resistant isolates (n = 11) were subjected to genotypic detection for efflux pump genes presented in Table 1. Both MexR and MexZ genes were detected in all isolates (n = 11). NfxB gene and MexT gene were found in 3 isolates and 5 isolates, respectively. All genes were found collectively in three isolates (P3, P4, and P11). Concerning carbapenem genes, seven isolates (Imipenem-resistant) including five isolates (positive MBL) that previously showed positive results on phenotypic screening and two isolates (negative MBL) were subjected to blaVIM and blaIMP genes detection. Results showed that blaVIM gene was found only in one isolate (Table 4).
DISCUSSION
Pseudomonas aeruginosa is commonly known to cause healthcare-acquired infections showing high rates of mortality. Recently, many studies had been conducted to study bacterial infections among children, especially P. aeruginosa (Bitsori et al., 2012; Hassuna et al., 2015; Logan et al., 2017). We found that the frequency of P. aeruginosa in children infections reported differs according to the region. In Omani’s study published in 2015, among children uropathogens, the low frequency of P. aeruginosa was reported (2.86%) (Sharef et al., 2015). Similar low frequencies of 2.3% and 5.9% were also reported in Saudi Arabia among children with urinary tract infection (Alshamsam et al., 2009; Hameed et al., 2019) and 11.54% among children diagnosed with respiratory tract infections (Walid et al., 2016). In the United Arab Emirates, P. aeruginosa (36%) was the most common bacteria in CF children (Redha and Panickar, 2016). In 2018, a study conducted on children patients revealed that P. aeruginosa was the most prevalent by frequently 16.67%; those bacteria were found to be MDR. The authors suggest that this may be due to the high prevalence of the use of invasive devices during the period of hospitalization (El-Nawawy et al., 2018).
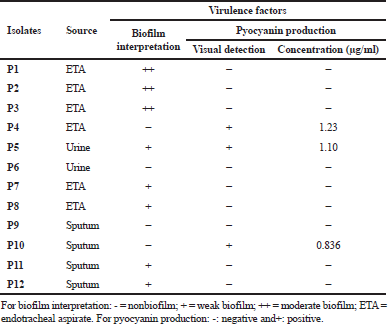 | Table 3. Source and virulence factors distribution among P. aeruginosa strains. [Click here to view] |
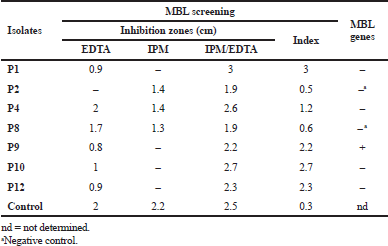 | Table 4. MBL screening using imipenem/EDTA combined disk test (CDT) and the occurrence of blaVIM and blaIMP genes. [Click here to view] |
 | Figure 1. Hierarchical cluster analysis of different strains of Pseudomonas aeruginosa. [Click here to view] |
Besides the fact that P. aeruginosa is naturally resistant to many antimicrobial agents, they easily acquire resistance to new ones (Alnour and Ahmed-Abakur, 2017). This antimicrobial behavior significantly limits therapeutic options (Hirsch and Tam, 2010). Herein, high percentages of resistance to several antibiotics were detected (Table 2), at which a 100% resistance toward cephems antibiotics was observed; our results are in line with those reported about children (Hassuna et al., 2015; Pourakbari et al., 2016), but opposite to those reported among Spanish children (Ruiz-roldán et al., 2018) and Saudi children at which P. aeruginosa was found to be more sensitive to a range of antibiotics (Hameed et al., 2019).
Genetically, P. aeruginosa virulence is known to be multifactorial and combinatorial (Wendt et al., 2017). Such bacterial pathogens always share common mechanisms for their abilities to adhere, invade, survive host defenses, and cause infection (Wilson et al., 2002). The pathogenesis of P. aeruginosa infection is somehow related to its ability to synthesize some virulence factors (pyocyanin; proteases) and to form biofilms (Maisetta et al., 2020). In this study, biofilm formation ability was studied as one factor of the important virulence factors presented in P. aeruginosa, and as a result, 66.7% were able to form a biofilm at which their ability is ranging between moderate and weak. During our research journey, only a few types of research studied the biofilm formation among P. aeruginosa isolates from children’s infections. Ralte et al. (2019) reported that, among different bacterial species isolated from children, the P. aeruginosa strain showed a strong ability to produce a biofilm. Biofilm formation P. Aeruginosa ability is an important process for bacterial colonization for persistence (Vallet et al., 2004), making it a problematic issue especially for patients requiring mechanical ventilation and catheterization (Alnour and Ahmed-Abakur, 2017). Inside those biofilms, bacteria became more resistant and difficult to eradicate than the planktonic ones (Mohamed et al., 2018b, 2020a); therefore, this mode of bacterial growth is strongly linked to various diseases at which many bacterial infections are biofilm-related (Mohamed et al., 2019, 2020c).
Three of our isolates are shown to produce pyocyanin pigment after 24 hours with different concentrations (Table 3). P. aeruginosa synthesizes a characteristic chloroform-soluble blue redox-active secondary metabolite called pyocyanin (Khadim and Marjani, 2019), which is considered an important virulence factor since it has a role to induce oxidative stress (Jazayeri et al., 2016). Cluster analysis was applied to detect the similarity of our P. aeruginosa strains phenotypic traits (Fig. 1). This analysis was performed to survey the prevalence of bacterial antibiotic susceptibility and complete phenotypic classification (Berrazeg et al., 2013).
It is reported that rates of antibiotic resistance among children P. aeruginosa infection are rising nationally (Logan et al., 2017). This bacteria can acquire new resistance mechanisms (Ruiz-roldán et al., 2018); the main ones for P. aeruginosa were mutation in target sites, changes in membrane permeability, the transmission of plasmid resistance genes, and efflux mechanism (Bejestani et al., 2015). In our study, two resistance mechanisms (efflux pump and MBL) were studied phenotypically and some genes have been detected genotypically. All CIP-resistant isolates showed higher MIC for EtBr than the standard strain using the phenotypic method. Reports of efflux pump percentages among children’s P. aeruginosa infections were also found. Among children’s P. Aeruginosa studied by Pourakbari et al. (2016), 62% showed an increased expression level of efflux pump genes. Helmy and Kashef (2017) reported that 23.5% of P. aeruginosa isolates showed multidrug efflux-mediated resistance. Many phenotypic methods have been used nowadays to detect MBL producing bacteria, which are majorly based on the ability to inhibit MBLs usingmetal chelators (Sachdeva et al., 2017). Our data identified that 41.67% of the P. aeruginosa isolates are MBL-positive using the combined imipenem/EDTA disk test method. In the United States, 11.3% of Pseudomonas was found to be carbapenem-resistant (Logan et al., 2017). Bejestani et al. (2015) reported that the frequency of imp gene was detected in 3.3% P. aeruginosa isolates; on the other hand, the vim gene was not detected in any of MBL-positive isolates (0%), at which this low frequency comes in line with our results.
CONCLUSION
This study highlights the high frequency of antibiotic resistance, besides the production of important virulence factors (biofilm formation and pyocyanin production). Also, the gene occurrence of antibiotic resistance mechanisms was reported among our P. aeruginosa from children. This virulent bacterial behavior is alarming which needs attention to the way we use antibiotics. However, studies with a larger number of P. aeruginosa children isolates are recommended.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Amr Mohamed Basha, Department of Microbiology, National Organization for Drug Control and Research, Egypt, for sharing carbapenem primers to be used in this work.
ETHICAL APPROVAL
This study involved microbes isolated in microbiology laboratory as a part of continuous routine laboratory work and was determined not to be human subject’s research. The study did not involve clinical data for patients and did not involve laboratory animals or invasive procedures.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
FUNDING
None.
REFERENCES
Al Rashed N, Joji RM, Saeed NK, Bindayna KM. Detection of overexpression of efflux pump expression in fluoroquinolone-resistant Pseudomonasaeruginosa isolates. Int J Appl Basic Med Res, 2020; 10:37–42. CrossRef
Alkhudhairy MK, Al-Shammari MMM. Prevalence of metallo-β-lactamase-producing Pseudomonas aeruginosa isolated from diabetic foot infections in Iraq. New Microbes New Infect, 2020; 35:100661. CrossRef
Alnour TM, Ahmed-Abakur EH. Multidrug resistant Pseudomonas (P) aeruginosa: medical impact, pathogenicity, resistance mechanisms and epidemiology. JSM Microbiol, 2017; 5:1046.
Alshamsam L, Al Harbi A, Fakeeh K, Al Banyan E. The value of renal ultrasound in children with a first episode of urinary tract infection. Ann Saudi Med, 2009; 29:46–9. CrossRef
Bejestani FB, Hakemi-vala M, Momtaheni R, Bagheri Bejestani O, Gholami M. The frequency of imp and vim Genes among Pseudomonas aeruginosa isolates from children ’ s medical center of Tehran. Arch Clin Infect Dis, 2015; 10:e20991. CrossRef
Berrazeg M, Drissi M, Medjahed L, Rolain JM. Hierarchical clustering as a rapid tool for surveillance of emerging antibiotic- resistance phenotypes in Klebsiella pneumoniae strains. J Med Microbiol, 2013; 62:864–74. CrossRef
Bitsori M, Maraki S, Koukouraki S, Galanakis E. Pseudomonas Aeruginosa urinary tract infection in children: risk factors and outcomes. J Urol, 2012; 187:260–4. CrossRef
Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 29th edition. Clinical and Laboratory Standards Institute, Wayne, PA, 2019.
El-Nawawy A, Ashraf GA, Antonios MAM, Meheissen MA, El-Alfy MMR. Incidence of multidrug-resistant organism among children admitted to pediatric intensive care unit in a developing country. Microb DRUG Resist, 2018; 24:1198–206. CrossRef
Feghali PAR El, Nawas T. Extraction and purification of pyocyanin: a simpler and more reliable method. MOJ Toxicol, 2018; 4:417–22. CrossRef
Hameed T, Al Nafeesah A, Chishti S, Al Shaalan M, Al Fakeeh K. Community-acquired urinary tract infections in children: resistance patterns of uropathogens in a tertiary care center in Saudi Arabia. Int J Pediatr Adolesc Med, 2019; 6:51–4. CrossRef
Hassuna NA, Mohamed AHI, Eleuoon SMA, Rizk HAA. High prevalence of multi- drug resistant Pseudomonas aeruginosa recovered from infected burn wounds in children. Arch Clin Microbiol, 2015; 6:1–7.
Helmy OM, Kashef MT. Different phenotypic and molecular mechanisms associated with multidrug resistance in gram-negative clinical isolates from Egypt. Infect Drug Resist, 2017; 10:479–98. CrossRef
Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res, 2010; 10:441–51. CrossRef
Jazayeri JA, Nguyen K, Kotsanas D,Schneiders F, Tan C, Jazayeri M, Armstrong D. Comparison of virulence factors in Pseudomonas aeruginosa strains isolated from cystic fibrosis patients. J Med Microbiol Diagnosis, 2016; 5:1000242. CrossRef
Khadim MM, Marjani MFAL. Pyocyanin and biofilm formation in Pseudomonas aeruginosa isolated from burn infections in Baghdad, Iraq. Jordan J Biol Sci, 2019; 12:31–5.
Lima JLDC, Alves LR, Jacomé PRLA, Bezerra Neto JP, Maciel MAV, Morais MMC. Biofilm production by clinical isolates of Pseudomonasaeruginosa and structural changes in LasR protein of isolates non biofilm-producing. Braz J Infect Dis, 2018; 22:129–36. CrossRef
Logan LK, Gandra S, Mandal S, Klein EY, Levinson J, Weinstein RA, Laxminarayan R. Multidrug- and carbapenem-resistant Pseudomonasaeruginosa in children, United States, 1999–2012. J Pediatric Infect Dis Soc, 2017; 6:352–9. CrossRef
Maisetta G, Grassi L, Esin S, Kaya E, Morelli A, Puppi D, Piras M, Chiellini F, Pifferi M, Batoni G. Targeting Pseudomonasaeruginosa in the sputum of primary ciliary dyskinesia patients with a combinatorial strategy having antibacterial and anti-virulence potential. Int J Mol Sci, 2020; 21:69. CrossRef
Mohamed MSM, Mostafa HM, Mohamed SH, Abd El-Moez SI, Kamel Z. Combination of silver nanoparticles and vancomycin to overcome antibiotic resistance in planktonic/biofilm cell from clinical and animal source. Microb DRUG Resist, 2020a; http://doi.org/10.1089/mdr.2020.0089.
Mohamed SH, Elshahed MMS, Saied YM. Evaluation of Honey as an antibacterial agent against drug-resistant uropathogenic E.coli strains. Res J Pharm Technol, 2020c; 13(8):3720–4. CrossRef
Mohamed SH, Khalil MS. DNA sequencing of bla SHV genes among uropathogenic Klebsiellapneumoniae harboring bla CTX-M-15. Int J Pharm Res, 2020; 12:443–9.
Mohamed SH, Khalil MS, Azmy M. In vitro efficiency of ampicillin, thymol and their combinations against virulence strains of Klebsiellapneumoniae. Int J Pharm Res, 2019; 11:315–21.
Mohamed SH, Khalil MS, Mohamed MSM, Mabrouk MI. Prevalence of antibiotic resistance and biofilm formation in Klebsiellapneumoniae carrying fimbrial genes in Egypt. Res J Pharm Technol, 2020b; 13(7):3051–8. CrossRef
Mohamed SH, Mohamed MSM, Khalil MS, Azmy M, Mabrouk MI. Combination of essential oil and ciprofloxacin to inhibit/eradicate biofilms in multidrug-resistant Klebsiellapneumoniae. J Appl Microbiol, 2018b; 125:84–95. CrossRef
Mohamed SH, Mohamed MSM, Khalil MS,Mohamed WS, Mabrouk MI. Antibiofilm activity of papain enzyme against pathogenic Klebsiellapneumoniae. J Appl Pharm Sci, 2018a; 8:163–8. CrossRef
Mohamed SH, Salem D, Azmy M, Fam NS. Antibacterial and antibiofilm activity of cinnamaldehyde against carbapenem-resistant Acinetobacterbaumannii in Egypt: in vitro study. J Appl Pharm Sci, 2018c; 8:151–6. CrossRef
Morales E, Cots F, Sala M, Comas M, Belvis F, Riu M, Salvadó M, Grau S, Horcajada JP, Montero MM, Castells X. Hospital costs of nosocomial multi-drug resistant Pseudomonasaeruginosa acquisition. BMC Health Serv Res, 2012; 12:122. CrossRef
Osman ME, El-hendawy HH, El-all SMA, Hassan AM, Mahmoud DE. Correlation between resistance of Pseudomonasaeruginosa to benzalkonium chloride and expression of efflux pump genes. J Appl Pharm Sci, 2018; 8:44–52. CrossRef
Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonasaeruginosa in intensive care unit; a critical review. Genes Dis, 2019; 6:109–19. CrossRef
Pourakbari B, Yaslianifard S, Yaslianifard S, Mahmoudi S, Keshavarz-Valian S, Mamishi S. Evaluation of efflux pumps gene expression in resistant Pseudomonasaeruginosa isolates in an Iranian referral hospital. Iran J Microbiol, 2016; 8:249–56.
Ralte Z, Naina P, Amladi A, John M, Anndan S, Varghese AM. Determination of biofilm-forming capacity of otopathogens isolated from discharging ears in children with chronic otitis media. Indian J Med Microbiol, 2019; 37:442–5. CrossRef
Redha KY Al, Panickar J. Bacteriology among CF children in United Arab Emirates. J Cyst Fibros, 2016; 15:S70. CrossRef
Ruiz-roldán L, Bellés A, Bueno J, Azcona-Gutiérrez JM, Rojo-Bezares B, Torres C, Castillo F, Sáenz Y, Seral C. Pseudomonasaeruginosa isolates from Spanish children: occurrence in faecal samples, antimicrobial resistance, virulence, and molecular typing. Biomed Res Int, 2018; 2018:8060178. CrossRef
Sachdeva R, Sharma B, Sharma R. Evaluation of different phenotypic tests for detection of metallo -β- lactamases in imipenem-resistant Pseudomonasaeruginosa. J Lab Physicians, 2017; 9:249–53. CrossRef
Sharef SW, El-Naggari M, Al-Nabhani D, Al Sawai A, Al Muharrmi Z, Elnour I. Incidence of antibiotics resistance among uropathogens in Omani children presenting with a single episode of urinary tract infection. J Infect Public Health, 2015; 8:458–65. CrossRef
Singh I, Jaryal SC, Thakur K, Sood A, Grover PS, Bareja R. Isolation and characterization of various Pseudomonasspecies from distinct clinical specimens. IOSR J Dent Med Sci, 2015; 14:80–4.
Vallet I, Diggle SP, Stacey RE, Cámara M, Ventre I, Lory S, Lazdunski A, Williams P, Filloux A. Biofilm formation in Pseudomonasaeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J Bacteriol, 2004; 186:2880–90. CrossRef
Walid J, Moshref L, Alzanbagi M, Moshref R, Baabdullah W, Aldigs E, AlAhmadi T, Kari J, Alharbi S, Safdar O, Felemban O. Antibiotic resistance pattern in healthy children diagnosed with community acquired respiratory tract infections in King Abdulaziz University Hospital. J Med Microbiol Diagn, 2016; 5:1000232. CrossRef
Wendt M, Silva BCJ De, Heo G. Virulence factors and antimicrobial resistance of Pseudomonasaeruginosa isolated from Pet Turtles. Asian J Anim Vet Adv, 2017; 12:205–11. CrossRef
Wilson JW, Schurr MJ, LeBlanc CL, Ramamurthy R, Buchanan KL, Nickerson CA. Mechanisms of bacterial pathogenicity. Postgrad Med J, 2002; 78:216–24. CrossRef