INTRODUCTION
In the recent scenario worldwide, bacteria is reported as a major and intensive causative factor of infections in marine living beings, like finfish and shellfish (Austin, 2012). One of the negative impacts of bacterial pathogens includes severe mass mortalities, which lead to considerable economic losses and also cause potential consequences on the health and survival of aquatic creators. Finfish bacterial pathogens include Aeromonas sp., Pseudomonas sp., and Vibrio sp., (Austin and Austin, 2007). Vibrio sp. is reported as a harmful shrimp pathogen, especially Vibrio harveyi causes massive mortalities in shrimp (Aguirre-Guzman et al., 2004; Austin and Zhang, 2006; Vandenberghe et al., 2003). Many researchers have reported that the mass mortality of shrimp is by bacterial aquatic pathogens (Beaz-Hidalgo et al., 2010; Kesarcodi-Watson et al., 2009; Lambert, 1998; Prado et al., 2005; Travers et al., 2015). The most vital remedy to overcome this mass mortality is the application of antibiotics and it has now been extensively used in aquaculture indiscriminately, including finfish, shrimp, and shellfish cultures (Armstrong et al., 2005; Bermdez-Almada and Espinosa-Plascenci, 2012; Cabello, 2006; Holbach et al., 2015; Prado et al., 2010; Santhosh et al., 2016). As many antibiotics play an effective roll in controlling bacteria, some bacteria are developing a resistance that is making many antibiotics less effective (Albuquerque Costa et al., 2015; Fernandez-Alarcon et al., 2010; Miranda and Zemelman, 2002; Sandrawati et al., 2020). With regard to the least effective antibiotics, search and utilization of novel and effective antibiotics have extensively increased public health concern (Sapkota et al., 2008). Even though there are many alternate biological compounds available against bacterial pathogens, microalgae-derived compounds have identified as a positive and effective impact over other biological compounds. The advantage of microalgae against aquatic pathogens includes readily available, eco-friendly, cost-effective, and least side effect (Citarasu, 2012). So, the demand for identification of potential microalgae and large-scale production of effective microalgae have increased drastically.
Algae are excellent sources for bioactive compounds and numerous pharmaceutical compounds have been isolated and identified with effective clinical applications (Tandeau de Marsac and Houmard, 1993). However, several new compounds have been continuously isolated from marine sources, the majority are yet to be identified and characterized (Blunt et al., 2008). Therefore, microalgae represent a unique opportunity to discover novel metabolites. The rate of finding metabolites already obtained from other biological sources is less in microalgae as related to other bioresources (Mc Gee et al., 2020; Olaizola, 2003). Triggering of secondary metabolites can induce the medical and pharmacological efficacy of microalgae (Bone and Mills, 2013; Guedes et al., 2011) and there are many challenges in the synthesis of clinically significant compounds from algae (Kaushik and Chauhan, 2008). Therefore, there is great enthusiasm for the development and manufacture of various compounds that can potentially be used as functional ingredients, such as carotenoids, phycocyanins, polyphenols, fatty acids, and polyunsaturated compounds (Plaza et al., 2010).
Bioactive compounds synthesized from microalgae can be directly obtained from primary metabolism or secondary metabolism. Synthesized compounds have antimicrobial, antioxidant, and anti-inflammatory properties, with potential pharmaceutical applications (Harun et al., 2010; Ibanez and Cifuentes, 2013; Markou and Nerantzis, 2013; Smee et al., 2008). So, most of the researchers are interested in the isolation, extraction, purification, and characterization of the bioactive compounds from microalgae. Among which, the characterization of the bioactive compounds is the most promising aspect. Generally, thin-layer chromatography, high-performance liquid chromatography (HPLC), Fourier transform infrared (FTIR), and gas chromatography-mass spectrometry (GC-MS) methods are used. Among these, FTIR has proved to be a well-accepted and effective method used for the identification of bioactive compounds since it offers a simple reparation sample and high analysis speed (Noh et al., 2016). The present study aims to characterize marine microalgal bioactive compounds using FTIR and to screen their potential in antimicrobial activity against aquatic pathogens.
MATERIALS AND METHODS
Collection of marine microalgae
Three marine microalgal species, such as Chlorella sp., Pavlova sp., and Cheatocerous sp., were collected from the Planktology Division of Centre for Marine Science and Technology, Manonmaniam Sundaranar University, Rajakkamangalam, Tamil Nadu, India. The collected algal samples were then brought to the laboratory for further studies. Then, the stock was maintained in Walne’s media. The cultured flasks were placed in front of the tube lights of 1,000 lux light intensity. The temperature range was between 28°C and 30°C.
The composition of Walne’s media included macronutrients, such as potassium nitrate – 100 gm; sodium di-hydrogen orthophosphate − 20.0 gm; Ethylene diamine tetraacetic acid (EDTA), sodium salt −45.0 gm; boric acid – 33.48 gm; ferric chloride – 1.30 gm; and manganese chloride −0.36 gm. They were then dissolved in 1 l of distilled water and named as stock solution ‘A’. The micronutrients consisted of zinc chloride – 4.2 gm; copper sulphate – 4.0 gm; cobalt chloride – 4.0 gm; and ammonium molybdate – 1.8 gm. All were dissolved in 1 l distilled water. Vitamins such as thiamine (vitamin B1) – 200 mg and cyanocobalamine (vitamin B12) −10 mg were dissolved in 100 ml of water to prepare the stock solution ‘C’ and stored in a refrigerator. Solutions A, B, and C were prepared in separate reagent bottles. For the preparation of the working solution, 1 ml of solution ‘A’, 0.5 ml of solution ‘B’, and 0.1 ml of solution ‘C’ were individually taken and added in 1 l of filtered and sterilized seawater.
Mass culture and harvest of algal biomass
The seawater was enriched with the required quantity of Walne’s media. Then, 10–20% of the inoculums of log-phase culture were transferred into the mass culture tank with 15 l media. Finally, the culture tanks were placed under diffused sunlight with continuous aeration. Algal cells in the stationary phase were harvested from the culture by batch centrifugation at 8,000 rpm for 10 minutes. The resultant algal pastes were sundried and the algal powder was packed in an airtight bag and stored in the refrigerator until the extraction process.
Extraction of antimicrobial compounds
Five grams of algal powder were taken in each glass bottle and then 12.5 ml of methanol, ethanol, acetone, and chloroform solvents was added to each tube. The algal powder was soaked with solvents and kept for 1 week in a rotary shaker at 150 rpm. After that, the extract was collected by filtering the extract through Whatman No.1 filter paper and the remaining compounds of cellular materials were again extracted by grinding with the respective solvents in a sterile mortar and pestle. The extract was collected through Whatman No.1 filter paper and evaporated under room temperature for the complete removal of the solvent and stored at 4°C for further studies.
Antimicrobial activity
Collection of bacterial pathogens
In the present study, the extracted antimicrobial compounds were assayed against aquatic pathogens like V. harveyi, Vibrio parahaemolyticus, Pseudomonas aeroginosa, Aeromonas hydrophila, and Staphylococcus aureus. The aquatic pathogens were obtained from Microbiology Laboratory of Centre for Marine Science and Technology, M.S. University, Rajakkamangalam, Tamil Nadu, India.
Disk diffusion method
The three different algal extracts of different solvent systems (extracts) were screened against selected aquatic pathogens like V. harveyi, V. parahaemolyticus, P. aeroginosa, A. hydrophila, and S. aureus. The above-mentioned bacterial species were inoculated into the nutrient broth and incubated at 37°C overnight. After the incubation period, the Muller Hinton agar medium (beef extract – 2.00 gm, acid hydrolysate of casein – 17.50 gm, starch – 1.50 gm, agar – 17.00 gm, and distilled water – 1,000 ml) was prepared and poured in to sterilized petri plates and allowed to solidify. The swabs were prepared from various stock cultures of pathogens and spread over the agar plate. Then, the petriplate was allowed for 20 minutes and with the help of a well cutter (0.85 cm) the wells were made on the petri plate. Then, the crude extracts were loaded into the wells and the plates were incubated at 37°C for 24 hours. The lack of microbial growth was determined by measuring the diameter of the zone of inhibition. After determining the antimicrobial activity of the extract obtained from a particular solvent against the selected pathogens, the algal extracts which showed the maximum antimicrobial activity in terms of zone of inhibition were selected for further studies.
Phytochemical analysis
The algal extracts were screened for the presence of biologically active compounds. Chemical tests including carbohydrate, flavonoids, terpenoids, sterols, phenolic compounds, and alkaloids were carried out using the aqueous extracts of the powdered algae, using standard procedures described by Harborne (1973), Trease and Evans (1978), and Kokate (1999).
Purification and characterization of compounds
Column chromatography
The column was packed with hexane using silica gel 60–120 mesh size as a matrix at a two-third volume. Approximately 5 ml of the algal extract was loaded as dried slurry on the top of the silica gel column. The column with non-polar solvent was eluted first and the polarity was gradually increased with acetone: an 80:20% hexane was added first and then the percentage of hexane was gradually increased up to 100%. Each elution had two volumes of the silica column. The entire fractions were collected in different tubes. Once the elution reached mid-polarity, then the percentage of hexane decreased and increased the polarity by increasing the concentration of methanol. After collecting all the fractions, the collected ellutants were concentrated and store at 4°C to reduce the evaporation.
Thin-layer chromatography
TLC is one type of chromatographic technique. The TLC chamber is a specially designed chamber, with a jar and a lid. Pour The solvent (2:2:1) ratio of methanol, chloroform and acetone into a developing container to a depth of just less than 0.5 cm. Cut The TLC sheet horizontally to a convenient size of 5 × 10 cm. This was the line for a spot on the plate. The sample to be analyzed was added to the plate in a process called spotting. After developing the chromatogram, the TLC plate was housed into the developing chamber and then the mobile phase was allowed to run through the adsorbent phase up to three-quarters of the plate. Remove The plate from the TLC chamber and allow it to dry and mark the position of the solvent front. Observe The plate under a UV lamp and circle each spot with a pencil.
Calculate the Rf value for each spot as follows:
Fourier transform infra red spectroscopic analysis
The samples were analyzed qualitatively for the active compounds by the FTIR method described by Kemp (1991). The frequency of spectra set to analysis was between 400 and 5,000 cm−1 wave number and the vibration spectrum was recorded as a graphical chart. The instrument used for FTIR analysis was Shimadzu, Japan.
Antimicrobial activity of bioactive compounds
Determination of minimum inhibitory concentration (MIC)
Ten milliliters of Mueller Hinton Broth was taken in 10 test tubes and sterilized at 121°C for 10 minutes. MIC was used to study the lowest inhibitory concentration of eluted antimicrobial fractions against the selected pathogens. The quantities of algal extract used were 10, 20, 40, 60, 80, 100, and 120 μg. The pathogens were inoculated into the broth and incubated at 37°C for 24 hours. After 24 hours, the turbidity was read at 600 nm using UV spectrophotometer.
Determination of minimum bactericidal concentration (MBC)
The MBC was determined by plating the culture from the antimicrobial treated tube. The MBC is the lowest concentration which inhibited bacterial growth when subcultured in the Mueller Hinton Agar. The plates were incubated at 37°C overnight and evaluated by comparing them with the control plates containing bacteria without test compounds.
Toxicity studies
Cytotoxic effects on Artemia (LD-50)
One gram of decapsulated Artemia cysts was allowed in 1 l of seawater of 28 ppt taken in to a cylindrical hatching device illuminated with fluorescent light. The cysts were kept in suspension using strong aeration; and after 20–24 hours, the cysts hatched out into instar I nauplii stage.
Hundred numbers of Artemia nauplii of the instar II stage were taken in each tube in a group of six numbers of hatching cones containing 10 ml of seawater. The algal extract of 10–30 μg was added to the hatching cones in a way that one tube will get one particular concentration. Every 30 minutes, the percentage of mortality was noted. The particular concentration at which the 50% of A. nauplii died at a particular time was considered as LD-50 of the particular concentration.
Algal toxicity test
Two hundred microliters of different algal cultures (Nannochloropsis sp., Chlorella sp., Tetraselmis sp., and Isochyrysis sp.,) were taken into wells present in a microtitre plate. The active compound of 10–120 μg was added in the well and allowed for incubation. To count the cytotoxic effect of the compound on algal cells, the pipetted sample was introduced into the counting grid of the hemocytometer gently. The samples spread uniformly inside the coverslip and a thin film of the culture was added and the cell distributed uniformly. The number of cells counted under the microscope and the total cell was calculated using the following formula:
The counting continued up to 7 days. The number of cells that existed in each well was counted and recorded. From the results, the decrease in the cells counted was determined and referred to as the cytotoxic effect of the compound.
Statistical analysis
Each experiment was carried out in triplicates. The mean and standard deviation (SD) were calculated and data are expressed as ± SD of the triplicates. The statistical analysis for each experiment was carried out using one way analysis of variance, followed by Tukey’s multiple comparison tests with statistical significance set at p < 0.05. All the analyses were carried out with Statistical Package for the Social Sciences version 11.5.
RESULTS AND DISCUSSION
Screening of potential microalgae using agar well diffusion method
The antibacterial efficacy of three microalgae, such as Pavlova sp., Chlorella sp., and Chaetoceros sp., was tested against five aquatic bacterial pathogens, namely V. harveyi, V. parahaemolyticus, P. aeroginosa, A. hydrophila, and S. aureus. All three microalgal samples were extracted using four different solvents: methanol, acetone, ethanol, and methanol:chloroform. Among the three microalgal extracts, acetone extraction of Pavlova sp. showed 17 mm zone of inhibition, followed by Chaetoceros sp. and Chlorella sp.
In Pavlova sp., acetone extract controlled all the five pathogens tested, whereas the methanolic extract controlled S. aureus and V. harveyi, the ethanolic extract controlled V. parahaemolyticus, P. aeroginosa, A. hydrophila, and S. aureus, and the methanol:chloroform extract controlled V. parahaemolyticus, P. aeroginosa, and A. hydrophila. The results are given in Table 1. Similar to the present study, Moorthi and Balasubramanian (2015) also conducted comparative studies of different solvents using microalgae Sargassum muticum. The present result is also correlated with the studies of the antibacterial potential of Sargassum wightii and Caulerpa scalpelliformis (Rosaline et al., 2012; Jebasingh et al., 2014). In their study, they demonstrated the attribution of the ability of polar acetone which binds the hydrophilic phlorotannins in marine microalgae, and they also stated the efficacy of antibacterial activity. On the contrary, the results in Hetta et al., (2012) study proved that methanol and water extracts of several species of algae were effective against pathogens such as Escherichia coli, Salmonella typhimurium, and Streptococcus faecalis. Thus, our results proved that acetone was the best solvent for extracting the antibacterial agents from Pavlova sp. The possible reason for the present result is that the interaction of the functional groups on the microbial surface is greatly influenced by the polarity of algal extraction which is extracted using acetone.
Phytochemical analysis
In this study, phytochemicals analysis of acetone extraction of Pavlova sp. revealed the presence of various types of phytochemicals, which include carbohydrate, flavonoids, terpenoids, sterols, phenolic compounds, and alkaloids (Table 2). Fernandes et al., (2020) explored the phytochemical diversity in Pavlova pinguis J.C and reported 72 compound distributions, which include 29 fatty acids, 14 sterols, 13 fatty alcohols, and 16 other lipophilic compounds. Bhagavathy et al., (2011) reported microbial growth inhibition and phytochemical analysis of Chlorococcum humicola and they stated the presence of phytochemicals, including alkaloids, flavonoids, fatty acids, and saponins, using ethyl acetate extraction of microalgae. In Sun chlorella extracts, phytochemicals, such as flavonoids, reducing sugar, carotenoids, tannins, alkaloids, anthraquinones, and quinines, were reported (Geetha et al., 2010). The phytochemical analysis of the present study revealed that the presence of diverse chemical compounds could be the reason for its efficiency against aquatic pathogens.
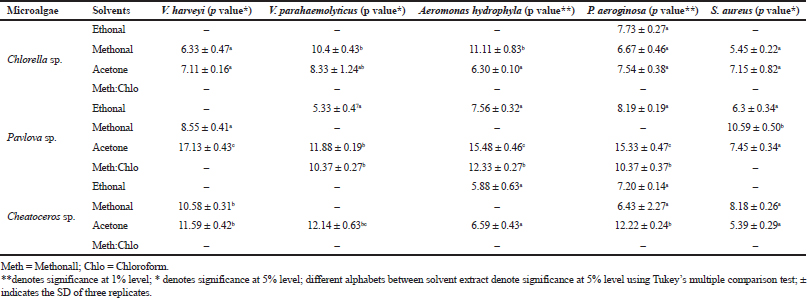 | Table 1. Antibacterial activity of microalgal extract against selected pathogens. [Click here to view] |
Effect of purified compounds for antimicrobial activity
Column and thin-layer chromatography
The potent algal extract of Pavlova sp. was purified through column chromatography. During the purification process, nine fractions were collected and they were screened against the same bacterial pathogens. Among the tested fractions, fraction 1 (acetone) had minimum inhibitory activity and it controlled only two pathogens. The activities ranged from 10.74 to 12.68 mm against V. parahaemolyticus and A. hydrophila. Fraction 2 (hexane:acetone) had maximum inhibitory activity against V. harveyi, V. parahaemolyticus, P. aeruginosa, and S. aureus with the activity of 11.37, 13.04, 11.26, and 12.35 mm, respectively. The minimum zone formation was found in 50:50 of hexane:acetone against P. aeroginosa (12.63 mm), A. hydrophila (14.70 mm), and S. aureus (13.23 mm). The maximum zone formation occurred in a 70:30 ratio of hexane:acetone extract; the inhibition zones recorded were 11.22, 0.66, 10.38, and 10.38 mm against the bacterial pathogens V. parahaemolyticus, P. aeruginosa, S. aureus, and A. hydrophila, respectively. The 100% of hexane fraction showed 10.85 and 10.68 mm antibacterial zone against V. parahaemolyticus and V. harveyi, respectively. The 100% of hexane, methonal:hexane (30:70), methonal:hexane (50:50), methonal:hexane (70:30) fractions showed no activity (Table 3).
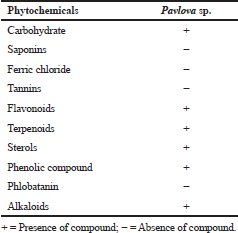 | Table 2. Phytochemical screening of crude extract of Pavlova sp. [Click here to view] |
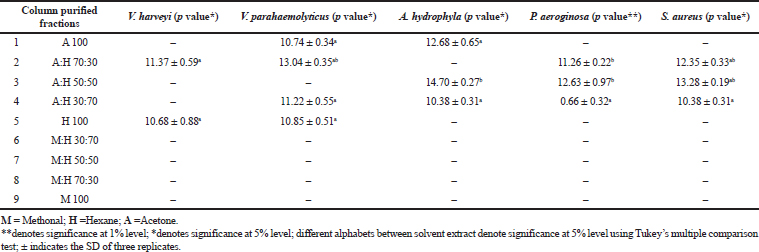 | Table 3. Antibacterial activity of partially purified column fractions of Pavlova sp. using different solvents. [Click here to view] |
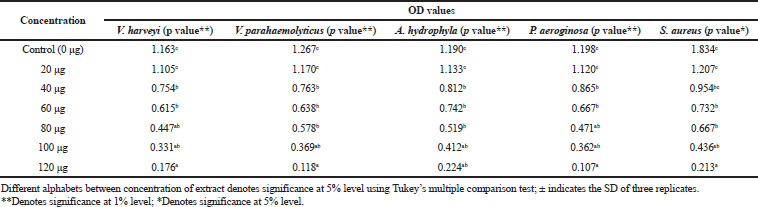 | Table 4. Determination of MIC. [Click here to view] |
The fraction which had the maximum inhibitory activity was selected for compound characterization by TLC, and TLC plated results showed two fractions of separation. Rf values of the separated spots were calculated as 0.923 and 0.635. Based on the Rf values, they were predicted as carotenoid compounds and other photosynthetic pigments. The role of the carotenoid compound in controlling diseases and their potential in antimicrobial, anti-inflammatory, and antioxidant properties have already been elaborately reported by de Morais et al., (2015).
Determination of MIC and MBC for purified compound
Among the test sample of column purified active fraction concentrations ranging from 10 to 140 μg/ml, the concentration of 10–80 μg did not exhibit any inhibitory activity, whereas 120 μg/ml concentration of acetone extract of Pavlova sp. showed minimum inhibitory activity (Table 4). As for MBC, concentrations ranging from 100 to 140 μg/ml were studied and the results showed that there was no bacterial colony observed in 140 μg/ml (Table 5). Saleh and Mariri (2017) reported that the highest zone of inhibition of bacteria was observed with the lowest MIC of 26.7 μg/ml and the lowest MBC of 53.3 μg/ml with Dilophus spiralis, which was extracted from chloroform. In another study, Chandrasekaran et al., (2014) showed that the inhibitory effect of Ulva lactuca against bacteria using various solvents and their result showed a higher range of MIC and MBC values: MIC values ranged between 250 and 500 μg/ml and MBC values ranged between 500 and 1,000 μg/ml. Similar to the present study, a positive influence of the acetone extraction of microalgae was reported by Kolanjinathan and Stella (2009). They stated that acetone was the effective solvent for the extraction of microalgae, such as Sargassum myricystum, Turbinaria conoides, Hypnea musiformis, Gracilaria edulis, and Halimedia gracilis for antimicrobial activity. Thus, in the present study, the result of MIC and MBC proved that acetone extraction of Pavlova sp. can be explored as promising bacteriostatic agents.
Structural analysis of column fractionated extract of Pavlova sp. by FTIR analysis
FTIR spectrum was used to identify the functional group of the active components based on the peak value in the region of infrared radiation. FTIR results are tabulated in Table 6. The first column fraction showed a broadband peak at 2,960.83cm−1, this may be due to the hydrogen group (C-H group) having the bond structure Csp3-H or the amino group (N-H group) having the bond structure R2NH2; R3NH+; and RCO2H. It also showed the cumulated system of R2C=N=N with the intensity at 51.146 (Fig. 1).
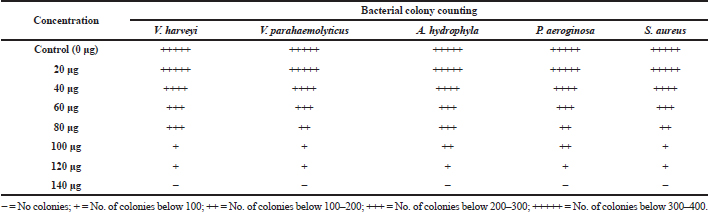 | Table 5. Determination of minimum bacterial concentration. [Click here to view] |
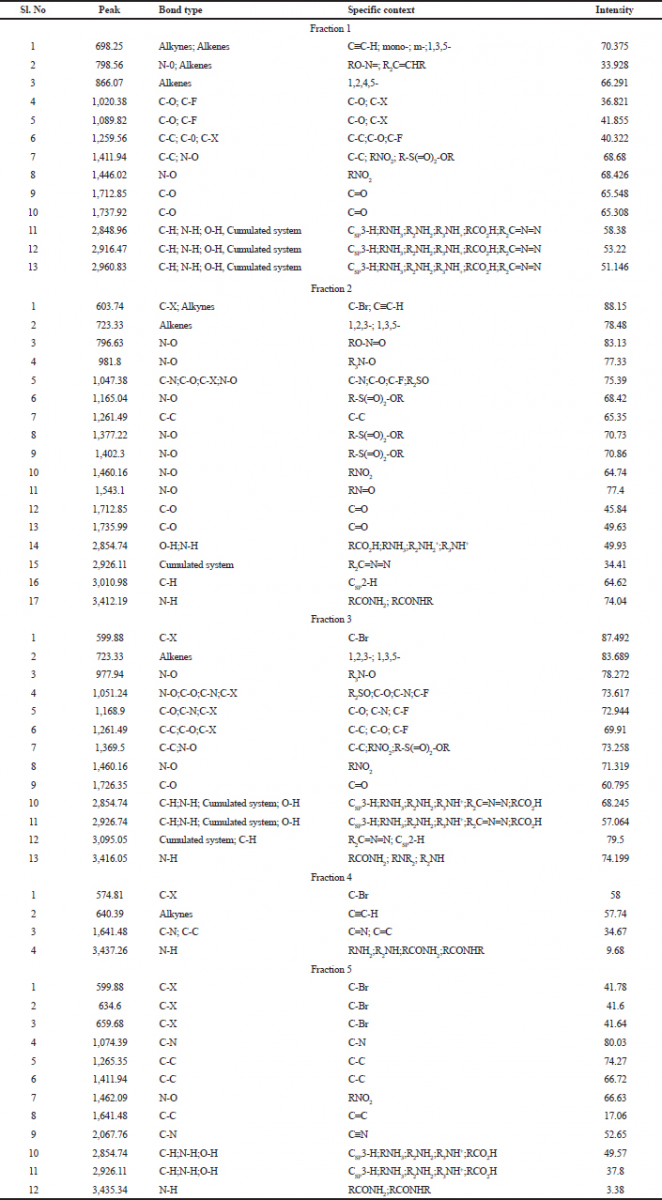 | Table 6. FTIR analysis acetone extraction of column purified fraction 1–5 of Pavlova sp. [Click here to view] |
The second column fraction was the active fraction and it showed 17 peaks. The broadband peak is seen at 3,412.19 cm−1. This may be due to the amino group (N-H group) having the bond structure: RCONH2 and RCONHR. These show the possibility of primary (or) secondary amine (or) an amide (or) substituted amine with an intensity of 74.04. The second broad peak is seen at 3,030.98 cm−1 is due to the hydrogen group (C-H) having the bond structure Csp2-H, with the intensity of 64.62. The broad peak at 1,377.22 cm−1 is due to the nitro group (N-O) having the bond structure R-S(O)2-OR, where the intensity was 70.73. It also showed the cumulated system of R2C=N=N with an intensity of 34.41. The broad peak 1,047.38cm−1 was due to the keto group (C-O group) having the bond structure C-O, cyanide group (C-N group) having the bond structure C-N, halogen group (C-X group) having the bond structure C-F, and the nitro group (N-O group) having the bond structure R2SO with the intensities of 7.39. The third column fraction showed the broadband peak at 3,416.05 cm−1. This may be due to the amino group (N-H group) having the bond structure: R2NH2; R3NH+; and RCO2H. These show the possibility of primary (or) secondary amine (or) an amide (or) substituted amine with an intensity of 74.1999. The peaks at a range from 22,854.74 to 3,009.05 cm−1 were due to the hydrogen group (C-H group) having the bond structure Csp3-H (or) amino group (N-H group) having the bond structure R2NH2; R3NH+; and RCO2H. It also showed the cumulated system of R2C=N=N with an intensity of 68.245–79.5. Other peaks between 1,051.24 and 1,261.49 cm−1 were due to the keto group (C-O group) having the bond structure C-C, cyanide group (C-N group) having the bond structure C-N, halogen group (C-X group) having the bond structure C-F with the intensities of 72.944–73.258 (Figure 1).
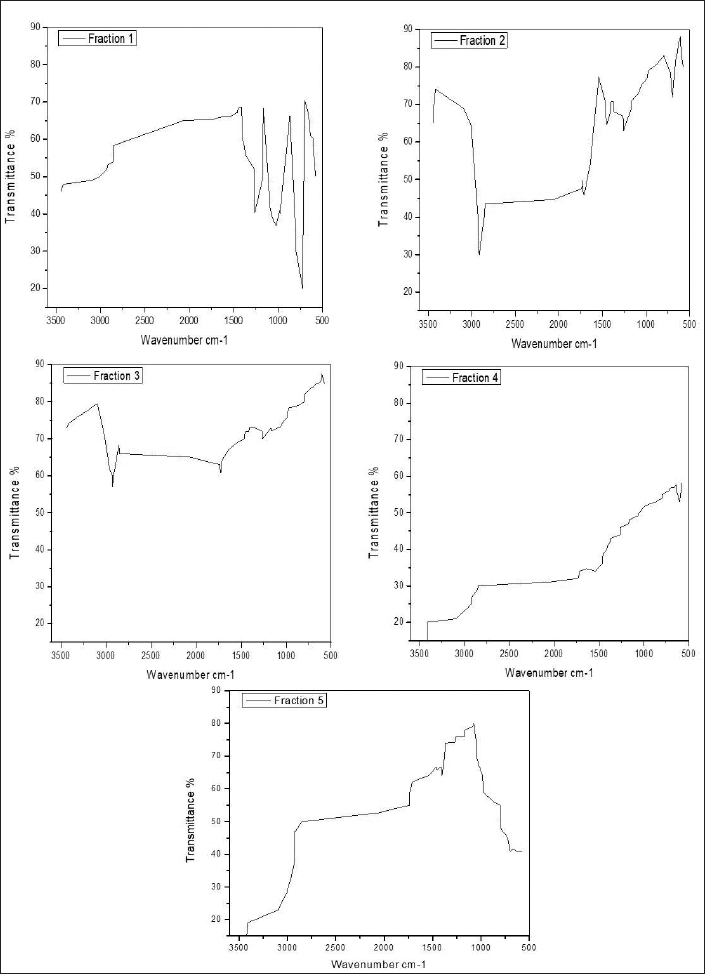 | Figure 1. FTIR analysis of bioactive compound purified fraction (1–5) of Pavlov sp. [Click here to view] |
The fourth column fraction showed a broadband peak of 3,437.26 cm−1. This may be due to the amino group (N-H group) having the bond structure R2NH2; R3NH+; and RCO2H with the intensity of 9.68. Some other peaks ranged from 574.81 to 1,641.48 cm−1 due to the halogen group (C-X group) having the bond structure C-Br, alkynes group having the bond structure CspC-H, carboxyl group (C-C group) having the bond structure C=C, and cyanide group (C-N group) having the bond structure C=N with the intensities of 34.67–58. The fifth column fraction showed a broadband peak at 3,435.34 cm−1. This may be due to the amino group (N-H group) having the bond structure RCONH2 and RCONHR with an intensity of 3.38. Another two broadband peaks between 2,854.74 and 2,926.11cm−1 were due to the amino group (N-H group) having the bond structure R2NH2; R3NH+; and RCO2H or hydrogen group (C-H group) having the bond structure Csp3-H or O-H group having the bond structure RCO2H with the intensities of 49.57 and 37.8. The peak at 1,462.09cm−1 was due to the nitro group (N-O group) having the bond structure RNO2 (Figure 1).
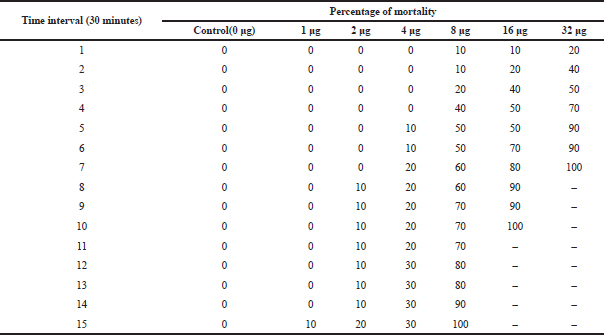 | Table 7. LD-50 of active column fraction of compound isolated from Pavlova sp. on A. nauplii based on time interval. [Click here to view] |
The result of the FTIR analysis shows a vast number of chemical groups, such as O-H, N-H, C-H, N-O, C-Br, and C=O, and many other compounds. This result confirms the presence of bioactive compounds, such as amines, alkane, carboxylic acid, sulfoxide, alcohol, boron, etc. These compounds could be the reason for the antibacterial efficiency of Pavlova sp., and the present statement is in agreement with the findings of de Morais et al., (2015). In the present study, peak position changes were observed between five different fractions. This positional flotation of peaks may due to the influence of temperature, pH, and concentration of the isolated compounds.
Toxicity analysis
Cytotoxic effect on A. nauplii (LD-50)
The cytotoxic effect of compounds on A. nauplii was studied. During the literature survey, it was demonstrated that the early developmental stages of Artemia are highly vulnerable to toxins (Meyer et al., 1982; Solis et al., 1993). In the present study, the percentage of lethality of Artemia is directly proportional to the concentration of algal extract. During this study, nearly 50% of mortality was observed in 32 μg/ml at 3–4 hours (Table 7). Kumar et al., (2012) also revealed that lethality was found to be directly proportional to the concentration of extract and the maximum mortality rate was observed at 100 nM concentrations, while 50% mortality was observed at 10 nM concentrations. Kladi et al., (2006) reported 100% of inhibition of hatching after 24 hours at a lower concentration (100 μg/ml) in Laurencia brandenii algal fraction. Brine shrimp lethality test was effective and simple in detecting antibacterial compounds and in this study, and Pavlova sp. extraction showed promising cytotoxic activity against A. nauplii.
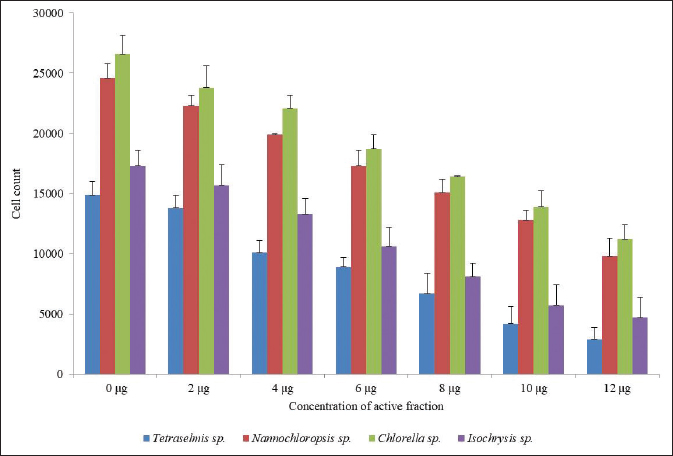 | Figure 2. Algal toxicity assay of active fraction of compounds isolated from Pavlova sp., against different algae. [Click here to view] |
Algal toxicity assay
The algal toxicity assay of the active fraction of four different marine microalgae (Tetraselmis sp., Nannochloropsis sp., Chlorella sp., and Isochrysis sp.,) was studied. 200 μl of freshly cultured microalgae was treated with an active fraction of different concentrations for 24 hours. The number of live algal cells reduced when the concentration of the active compounds was increased. Twelve micrograms per milliliter concentration of active fraction killed the algal cells when compared with control. This result proved that the higher the concentration of algicidal microalgae, the higher the alginolytic rate and the more significant the alginolytic effect. The cell count was 2,900 in Tetraselmis sp., 9,800 in Nannochloropsis sp., 11,200 in Chlorella sp., and 4,700 in Isochrysis sp., But in the control it was 14,900 in Tetraselmis sp., 24,600 in Nannochloropsis sp., 26,600 in Chlorella sp., and 17,300 in Isochrysis sp. (Figure 2). A inhibitory phenomena between microalgal cells have been reported in the past; Bagchi et al., (1999) originally proposed that natural algaecides could effectively be applied in the control of toxic algal blooms like Isochrysis galbana, Dunaliella salina, Platymonas elliptica, Chlorella vulgaris, Chaetoceros muelleri, Chlorella gracilis, Nitzschia closterium, and Phaeodactylum tricornutum. However, Pratt et al., (1942) were the first to report that growth of C. vulgaris was depressed by a compound (chlorellin) which is secreted into the medium, and which inhibits their growth and the growth of other species. The present result suggested that Pavlova sp. is an effective source of the active compound with antialgal activity and this effect may be due to the interference of chlorophyll and protein synthesis. In addition, our result is evident with the findings of Fernandes et al., (2020), which state that Pavlova sp. has a high content of unsaturated fatty acids, long-chain aliphatic alcohols, and stigmasterol. These compounds might also be the reason for its efficiency as algicidal.
CONCLUSION
The present study demonstrates the in vitro antimicrobial properties of Pavlova sp., and partial characterization of compounds was carried out using FTIR analysis. It also concludes that marine microalgae are a potential source of novel bioactive compounds. The extracts in different solvents exhibited different antimicrobial activities. The present investigation reveals the existence of biologically active compounds in the extractions of Pavlova sp. and they can be exploited for the production of novel and important bioactive compounds which are of use in the pharmaceutical industry. Furthermore, complete characterization of the pharmaceutically potential bioactive compound using HPLC and GC-MS and in-vivo studies to explore the new drugs against multi-resistant aquatic pathogens are very essential.
ACKNOWLEDGMENTS
The authors thank the Planktology and Aquaculture Division, Centre for Marine Science and Technology, Manonmaniam Sundaranar University, Rajakkamangalam for providing laboratory and technical assistance with the microalgal culture.
AUTHORS’ CONTRIBUTIONS
Subilal George conceptualized the idea, designed the experiments, drafted the manuscript, and conducted part of the experiment. Ajan Chellappan and Praba Thangamani conducted part of the experiment; Selvaraj Thangaswamy and Citarasu Thavisimuthu assisted in Statistics; Mariavincent Michaelbabu supervised the work and assisted in the drafting of the manuscript. All authors approved the manuscript, agreed to the authorship and submission of the manuscript for peer review.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
FUNDING
None.
REFERENCES
Aguirre-Guzman G, Mejia Ruiz H, Ascencio F. A review of extracellular virulence product of Vibrio species important in diseases of cultivated shrimp. Aquac Res, 2004; 35:1395–404. CrossRef
Albuquerque Costa R, Araujo RL, Souza OV, dos Fernandes Vieira RHS. Antibiotic-resistant Vibrios in farmed shrimp. BioMed Res Int, 2015; 2015:1–5. CrossRef
Armstrong SM, Hargrave BT, Haya K. Antibiotic use in finfish aquaculture: modes of action, environmental fate, and microbial resistance. Environmental Effects of Marine Finfish Aquaculture, Canada, pp 341–57, 2005. CrossRef
Austin B. Infectious disease in aquaculture: prevention and control. Elsevier, Amsterdam, Netherlands, 2012. CrossRef
Austin B, Austin DA. Bacterial fish pathogens: disease of farmed and wild fish. Springer Science & Business Media, Berlin, Germany, 2007.
Austin B, Zhang XH. Vibrio Harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol, 2006; 43:119–24. CrossRef
Bagchi S, Raychaudhuri P, Nevins JR. Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: a novel mechanism for E1A trans-activation. Cell, 1999; 24:659–69. CrossRef
Beaz-Hidalgo R, Dieguez AL, Cleenwerck I, Balboa S, Doce A, de Vos P, Romalde JL. Vibrio Celticus Sp. Nov., a new Vibrio species belonging to the splendidus clade with pathogenic potential for clams. Syst Appl Microbiol, 2010; 33:311–5. CrossRef
Bermdez-Almada MC, Espinosa-Plascenci A. The use of antibiotics in shrimp farming. In Health and Environment in Aquaculture, IntechOpen, London, UK, 2012. CrossRef
Bhagavathy S, Sumathi P, Jancy Sherene Bell I. Green algae Chlorococcum humicola-a new source of bioactive compounds with antimicrobial activity. Asian Pac J Trop Biomed, 2011; 1:S1–7. CrossRef
Blunt JW, Copp BR, Hu WP, Munro HGM, Northcote PT, Michelp R. Marine natural products. Nat Prod Rep, 2008; 25:35. CrossRef
Bone K, Mills SY. Principles and practice of phytotherapy. Elsevier Health Sciences, Amsterdam, Netherlands, pp 11–12, 2013.
Cabello FC. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol, 2006; 8:1137–44. CrossRef
Chandrasekaran M, Venkatesalu V, Adaikala Raj G, Krishnamoorthy S. Antibacterial activity of Ulva Fasciata against multidrug resistant bacterial strains. Int Lett Nat Sci, 2014; 19:40–51. CrossRef
Citarasu T. Natural antimicrobial compounds for use in aquaculture. Infect Dis Aquac, 2012; 419–56. CrossRef
de Morais MG, Vaz Bda S, de Morais SEG, Costa JA. Biologically active metabolites synthesized by microalgae. Biomed Res Int, 2015; 2015:1–15. CrossRef
Fernandes T, Martel A, Cordeiro N. Exploring Pavlova pinguis chemical diversity: a potentially novel source of high value compounds. Sci Rep, 2020; 10:339. CrossRef
Fernandez-Alarcon C, Miranda CD, Singer RS, Lopez Y, Rojas R, Bello H, Dominguez M, Gonzalez-Rocha G, Detection of the floRGene in a diversity of florfenicol resistant gram-negative Bacilli from freshwater salmon farms in Chile. Zoonoses Public Health, 2010; 57:181–8. CrossRef
Geetha BV, Navasakthi R, Padmini E. Investigation of antioxidant capacity and phytochemical composition of Sun Chlorella-an In vitro study. J Aquac Res Dev, 2010; 1(104):2. CrossRef
Guedes AC, Amaro HM, Xavier Malcata F. Microalgae as sources of high added-value compounds-a brief review of recent work. Biotechnol Prog, 2011; 27:597–613. CrossRef
Harborne JB. Phytochrome. Phytochem, 1973; 12:1836–7. CrossRef
Harun R, Manjinder S, Gareth MF, Michael KD. Bioprocess engineering of microalgae to produce a variety of consumer products. Renew Sust Energy Rev, 2010; 14:1037–47. CrossRef
Hetta MH, Abdo SM, Samhan FA, El Din RAS, Ali GH. Phytochemical and antibacterial study of five freshwater algal species. Asian J Plant Sci, 2012; 11:109–16. CrossRef
Holbach M, Robert R, Boudry P, Petton B, Archambault P, Tremblay R. Scallop larval survival from erythromycin treated broodstock after conditioning without sediment. Aquaculture, 2015; 437:312–7. CrossRef
Ibanez E, Cifuentes A. Benefits of using algae as natural sources of functional ingredients. J Sci Food Agric, 2013; 93:703–9. CrossRef
Jebasingh SEJ, Rajesh RP, Raja P, Lakshmikandan M. Presence of antibacterial activity in feather star, Capillaster multiradiatus extract against human and fish bacterial pathogens. Proc Natl Acad Sci India Sect B Biol Sci, 2014; 85:271–6. CrossRef
Kaushik P, Chauhan A. In vitro antibacterial activity of laboratory grown culture of Spirulina platensis. Indian J Microbiol, 2008; 48:348–52. CrossRef
Kemp W. Infrared spectroscopy. Org Spectrosc, Third Edition, 1991; 19–99. CrossRef
Kesarcodi-Watson A, Kaspar H, Lategan MJ, Gibson L. Two pathogens of greenshell™ Mussel larvae, Perna canaliculus: Vibrio Splendidus and V. Coralliilyticus/neptunius-like isolate. J Fish Dis, 2009; 32:499–507. CrossRef
Kladi M, Xenaki H, Vagias C, Papazafiri P, Roussis V. New cytotoxic sesquiterpenes from the red algae Laurencia obtusa and Laurencia microcladia. Tetrahedron, 2006; 62:182–189. CrossRef
Kokate CK. Phytochemical methods. Phytother, 1999; 78:126–9.
Kolanjinathan K, Ganesh P, Govindarajan M. Antibacterial activity of ethanol extracts of seaweeds against fish bacterial pathogens. Eur Rev Med Pharmacol Sci, 2009; 13:173–7.
Kumar P, Senthamil Selvi S, Lakshmi Prabha A, Selvaraj M, Macklin Rani L, Suganthi P, Sarojini Devi B, Govindaraju M. Synthesis of silver nanoparticles from Sargassum tenerrimum and screening phytochemicals for its antibacterial activity. Nano Biomed Eng, 2012; 4(1):12–6. CrossRef
Lambert C, Nicolas JL, Cilia V, Corre S. Vibrio Pectenicida sp. Nov., a pathogen of scallop (Pecten Maximus) larvae. Int J Syst Bacteriol, 1998; 48:481–7. CrossRef
Markou G, Nerantzis E. Microalgae for high-value compounds and biofuels production: a review with focus on cultivation under stress conditions. Biotechnol Adv, 2013; 31:1532–42. CrossRef
Mc Gee D, Archer L, Smyth TJ, Gerard TA, Fleming N. Touzet. Bioprospecting and LED-based spectral enhancement of antimicrobial activity of microalgae isolated from the West of Ireland. Algal Res, 2020; 45:101704. CrossRef
Meyer B, Ferrigni N, Putnam J, Jacobsen L, Nichols D, McLaughlin J. Brine shrimp: a convenient general bioassay for active plant constituents. Planta Med, 1982; 45:31–4. CrossRef
Miranda CD, Zemelman R. Bacterial resistance to oxytetracycline in chilean salmon farming. Aquaculture, 2002; 212:31–47. CrossRef
Moorthi PV, Balasubramanian C. Antimicrobial properties of marine seaweed, Sargassum muticum against human pathogens. J Coast Life Med, 2015; 3:122–5.
Noh C, Hafizah C, Azmin M, Fadhillah N, Asnawi AL. A review: fourier transform infrared application on bioactive compound. In 4th International Conference on Biotechnology Engineering (ICBioE), Kuala Lumpur, Malaysia, 2016, p 1–5.
Olaizola M, Commercial development of microalgal biotechnology: from the test tube to the marketplace. Biomol Eng, 2003; 20:459–66. CrossRef
Plaza M, Santoyo S, Jaime L, Garcia-Blairsy Reina G, Herrero M, Senorans FJ, Ibanez E. Screening for bioactive compounds from algae. J Pharm Biomed Anal, 2010; 51:450–5. CrossRef
Prado S, Romalde JL, Barja JL. Review of probiotics for use in bivalve hatcheries. Vet Microbiol, 2010; 145:187–97. CrossRef
Prado S, Romalde JL, Montes J, Barja JL. Pathogenic bacteria isolated from disease outbreaks in shellfish hatcheries. First description of Vibrio neptunius as an oyster pathogen. Dis Aquat Organ, 2005; 67:209–15. CrossRef
Pratt R. Studies on Chlorella vulgaris. V. Some properties of the growth-inhibitor formed by Chlorella cells. American J Bot, 1942; 1:142–8. CrossRef
Rosaline XD, Shanmugavel S, Rajendran K, Janarthanan S. Screening of selected marine algae from the coastal Tamil Nadu, South India for antibacterial activity. Asian Pac J Trop Biomed, 2012; 2:S140–6. CrossRef
Saleh B, Al Mariri A. Antimicrobial activity of the marine algal extracts against selected pathogens. J Agric Sci Tech, 2017; 19(5):1067–77.
Sandrawati N, Hati SP, Yunita F, Putra AE, Ismed F, Tallei TE, Hertiani T, Handayani D. Antimocrobial and cytotoxic activities of marine sponge-derived fungal extracts isolated from Dactylospongia sp. J App Pharm Sci, 2020; 10(4):028–33. CrossRef
Santhosh S, Dhandapani R, Hemalatha N. A review on potential biotechnological applications of microalgae. J App Pharm Sci, 2016; 6(8):179–84. CrossRef
Sapkota A, Amy R. Sapkota, Margaret K, Janelle B, McKenzie S, Walker, P, Lawrence R. Aquaculture practices and potential human health risks: current knowledge and future priorities. Environ Int, 2008; 34:1215–26. CrossRef
Smee D, Bailey K, Wong M, Okeefe B, Gustafson K, Mishin V, Gubareva L. Treatment of influenza a (H1N1) virus infections in mice and ferrets with cyanovirin-N. Antivir Res, 2008; 80:266–71. CrossRef
Solis P, Wright C, Anderson M, Gupta M, Phillipson J. A microwell cytotoxicity assay using Artemia salina (Brine Shrimp). Planta Med, 1993; 59:250–2. CrossRef
Tandeau de Marsac N, Houmard J. Adaptation of cyanobacteria to environmental stimuli: new steps towards molecular mechanisms. FEMS Microbiol Lett, 1993; 104:119–89. CrossRef
Travers MA, Boettcher Miller K, Roque A, Friedman CS. Bacterial diseases in marine bivalves. J Invertebr Pathol, 2015; 131:11–31. CrossRef
Trease GE, Evans WC. Pharmacology. 11th edition, Bailliere Tindall Ltd, London, UK, pp 60–75, 1978.
Vandenberghe F, Thompson L, Gomez-Gil B, Swings J. Phenotypic diversity amongst Vibrio isolates from marine aquaculture systems. Aquaculture, 2003; 219:9–20. CrossRef