INTRODUCTION
Molecules with either temporary or permanent positive net charges are known as organic cations. Due to the fact that most organic cations are non-membrane permeating, they depend mainly on membrane transporters, like organic cation transporters (OCTs), for their intestinal absorption, tissue distribution, and biliary, renal, and intestinal excretions (Han et al., 2013).
Human organic cation transporter 1 (hOCT1) protein is an influx transporter and is encoded by the SLC22A1 gene. It belongs to the solute carrier family which is a superfamily of transporters (He et al., 2009; Makhtar et al., 2018). OCT1, OCT2, and OCT3 are the three isoforms of OCT (Ahmadimoghaddam et al., 2013, p. 1; Sala-Rabanal et al., 2013, p. 3). OCT1 has a wide tissue distribution and is expressed in epithelial cells and some neurons. It is expressed predominantly in the hepatocytes in humans (Lozano et al., 2013). OCT2 has a more restricted expression pattern than OCT1 or OCT3. OCT2 is expressed in epithelial cells and neurons (Zhao, 2013). OCT2 is strongly expressed in the basolateral membrane of epithelial cells in the kidney and also in various other organs, including small intestine, lung, skin, brain, and choroid plexus (Brosseau and Ramotar, 2019). OCT3 is not only expressed in epithelial cells and neurons but also in muscle cells and glial cells. In humans, the strongest expression was found in skeletal muscle, liver, placenta, and heart; however, OCT3 is also expressed in many other organs, including brain, and in some cancer cell lines (Kucheryavykh et al., 2014). It plays a significant role in the hepatic uptake of several xenobiotics, including therapeutic agents like metformin (Glucophage), imatinib (Gleevec), levodopa, amantadine (Symmetrel), pramipexole (Mirapex), and pindolol (Visken) (Lozano et al., 2013; Umamaheswaran et al., 2014).
Significant interindividual variability is noted in terms of drug response [i.e., pharmacokinetics (PKs), pharmacodynamics (PDs), safety, and efficacy] when an identical drug is administered. This variation can be a result of polymorphism in the genes coding for a drug transporter that leads to the formation of different phenotypes. These phenotypes vary greatly in their expression level, protein folding, membrane location, and transporter efficiency (Gharavi and Hassan, 2018).
OCT1 is encoded by the SLC22A1 gene, which consists of 11 exons and is mapped to 6q26 chromosome, spanning approximately 37 kb. Human gene OCT1 is highly polymorphic, and in various populations, multiple polymorphisms have been described (Chen et al., 2010; Umamaheswaran et al., 2011). Up until now, there have been 19 non-synonymous SLC22A1 single nucleotide polymorphisms (SNPs) that have been identified. In OCT1, 41C>T, 181C>T, 292T>C, 566C>T, 659G>T, 848C>T, 859C>G, 1022C>T, 1201G>A, 1256delATG, and 1393G>A have all been associated with functional changes resulting in alterations in PK substrates. Influx transporters move drugs into the epithelium from the blood. In this way, ADME as well as the disposition of endogenous substrates are affected by influx transporters (Sissung et al., 2012).
In the coding region of the transporter gene, SNPs may results in the same (synonymous; SNP) or altered (non-synonymous; nsSNP) protein being expressed (Gharavi and Hassan, 2018), and it may also affect both drug therapy and organ-specific toxicity (Sissung et al., 2012). SNPs in the OCT1 gene are recognized as a possible mechanism which explains interindividual drug response variation (Du Plessis et al., 2015; Fattah et al., 2017).
Genetic factors and other environmental factors, such as chemicals and radiation exposure, lifestyle factors, such as drinking, smoking, and exercise, and physiological factors, such as age, sex, liver and kidney functions, pregnancy, and starvation, are the causative factors for drug response variations (Ahmed et al., 2016).
The presence of nsSNPs in human OCT1 will impair the drug transport process and may alter the bioavailability of substrates. While hOCT1 is expressed in renal tubules, genetic variations may impair the elimination of drugs and precipitate the risk of toxicity. Hence, high-risk patients (e.g., patients with Chronic Kidney Disease (CKD), on multiple OCT substrates, etc.) should be screened for transporter polymorphisms so as to ensure patient safety and better therapeutic care.
MATERIALS AND METHODS
Target Modeling
The high-resolution crystal structure of hOCT1 was modeled using iTASSER server. iTASSER server predicts the protein structure by a hierarchical iterative threading assembly refinement approach (available at: https://zhanglab.ccmb.med.umich.edu/I-TASSER/). The primary sequence of the retrieved structure showed the presence of methionine and proline at positions 408 and 341, respectively (wild type variants). The mutant variants bearing valine at position 408 and leucine at position 341 were modeled using Swiss-PDB viewer. The retrieved and modeled three-dimensional (3D) structures were pre-processed further as per the standard methods (Roy et al., 2010; Yang and Zhang, 2015; Yang et al., 2015).
Energy Minimization
The 3D structures of the target proteins were minimized using the YASARA server that uses YASARA force fields to compute energy functions called knowledge-based potentials (available at: http://www.yasara.org/minimizationserver.htm) (Krieger et al., 2009).
Protein Conformation and Stability
Energy minimized 3D structures were subjected to conformational stability analysis. Dihedral angles and atomic contacts were analyzed through Ramachandran plot using Swiss-PDB viewer (Guex and Peitsch, 1997).
Active Site Prediction
The drug-binding domain of the hOCT1 was determined using the Prankweb Server, which uses P2 rank, a template-free machine-learning method based on local chemical neighborhood ligand ability (available at: http://prankweb.cz/) (Jendele et al., 2019).
Molecular Docking Analysis
Molecular docking analysis was carried out using AutoDock 4.2.6. The drug transporting domain of hOCT1 was predefined by a 3D grid using the AutoGrid program. The genetic algorithm was adopted for conformer search with 25,000 evaluations and 27,000 generations. The interaction of the docked ligands with the amino acid residues in the active site of the target were visualized using Pymol 2.3 (Schrodinger, LLC, New York, NY). Two-dimensional ligand interactions maps were constructed using LeView software to gain deeper insights on the differences in substrate binding to wild type and modeled variants of hOCT1 (Samuel Gideon George and Dhivya, 2016).
RESULTS AND DISCUSSION
The hOCT1 is a transmembrane protein that helps in influx of various endogenous substances and xenobiotics into the cells (Boxberger et al., 2014). Non-synonymous polymorphisms in the human SLC22A1 gene are associated with decreased influx transport of a diverse spectrum of therapeutic agents, including metformin (Glucophage), acyclovir (Zovirax), ranitidine (Zantac), imatinib (Gleevec), etc., leading to therapeutic failure (Koepsell, 2013). In this study, using in silico methods, we analyzed the impact on the transport of various hOCT1 substrates due to non-synonymous polymorphism (1022C>T).
The mutant variants of hOCT1, Met408Val and Pro341Leu, were modeled via the iTASSER database, which uses a mathematical scoring function known as C-score to estimate the predicted quality of the sample. C-score, expanded as confidence score, is computed based on the significance of the threading template alignments and the convergence parameters of the structure assembly simulations. Two models were constructed for each mutant variant and those with the highest C-scores were selected to be the target (+0.97 and +1.2 for Met408Val and Pro341Leu variants, respectively).
In order to mimic the biological target protein, the iTASSER predicted models were subjected to energy minimization. The pre- and post-minimization potential energies of the wild type and mutant variants are shown in Table 1.
Protein conformation and stability of the energy minimized targets were analyzed through dihedral angles and atomic contacts by Ramachandran plot. Ninety-eight percent of the residues of were within the allowed region, exception being glycine and proline for wild type, and Met408Val and Pro341Leu variants were suggestive of minimal steric collisions and reliable spatial geometry for the modeled structures (Aanandhi and George, 2015; Ramakrishnan and Ramachandran, 1965). Ramachandran plot of energy minimized structures is shown in Figure 1.
In order to determine the effect of SNPs on the drug-binding and transmembrane domains of hOCT1, Prankweb-predicted active sites of wild type and variants were analyzed visually to determine the difference in residues forming the active site. Computational studies have shown that the transmembrane helices 1 and 2 of hOCT comprise cysteine residues and potential glycosylation sites, including asparagine, serine, and threonine (Dakal et al., 2017). We observed that Met408Val and Pro341Leu variations in hOCT altered the conformation of transmembrane helices, disrupting the glycosylation sites and cysteine residues which are crucial for drug binding and uptake. In addition, putative phosphorylation sites comprising serine, threonine, and tyrosine in transmembrane helices 6 and 7 are either absent or disrupted in the variants with Met408Val and Pro341Leu mutations. Thus, it is evident that non-synonymous polymorphisms in the hOCT decrease the drug uptake and transport process by altering the conformation of transmembrane helices (Mani, 2017). Differences in transmembrane helices of wild type and mutant proteins are shown in Figure 2.
The effect of conformational changes due to non-synonymous polymorphism in the transmembrane domain on substrate binding was investigated through molecular docking analysis. Ligand interaction and binding conformations were analyzed in terms of the following parameters: binding energy (ΔG Kcal/mol), inhibitory constant (kI), conformational orientation of the ligand in the active site, hydrogen bonding, π–π interactions, and root mean square deviation (Morris et al., 1998). The molecular docking analysis of hOCT1 substrate with wild type, Val408, and Leu341 is shown in Table 2.
The presence of Met408Val and Pro341Leu polymorphisms will decrease the ability of OCT1 to establish a week hydrogen bond with ligands which are crucial for substrate uptake, and thereby alter the transport of the drugs across the cell membrane. Hydrogen-bonding interactions with the active site residues of wild type, Val408, and Leu341 are shown in Table 3.
The 3D docked conformation of hOCT1 substrates with the wild type, Val408, and Leu341 is shown in Figure 3.
Using one-way analysis of variance (ANOVA), statistically significant differences were observed between the binding energies of wild type, Val408, and Leu341. At 95% confidence interval, a p-value of 0.0069 was obtained. The summary of the binding energies of hOCT1 with the wild type, Val408, and Leu341 is shown in Figure 4.
 | Table 1. Energy Minimization of the wild type, Val408, and Leu341 variants. [Click here to view] |
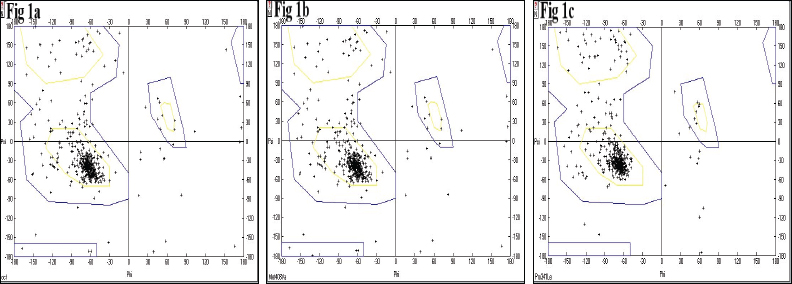 | Figure 1. Ramachandran plot of modeled energy-minimized proteins. Represent (1a) Wild Type, (1b) Val408 and (1c) Leu341 variants, respectively. [Click here to view] |
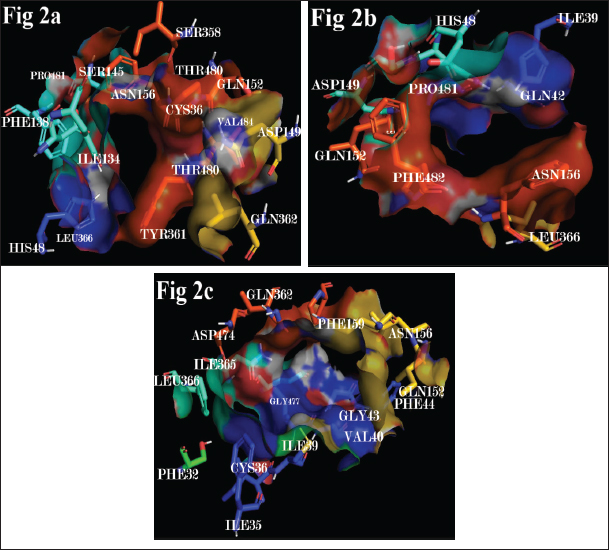 | Figure 2. Human OCT transmembrane domains involved in substrate uptake and transport of (2a) Wild type, (2b) Val408 and (2c) Leu341. [Click here to view] |
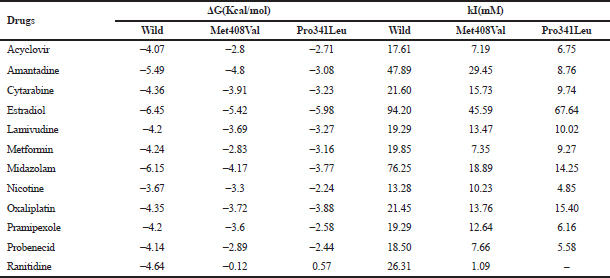 | Table 2. Molecular docking analysis of organic cation transporter substrates with the Wild Type, Val408, and Leu341. [Click here to view] |
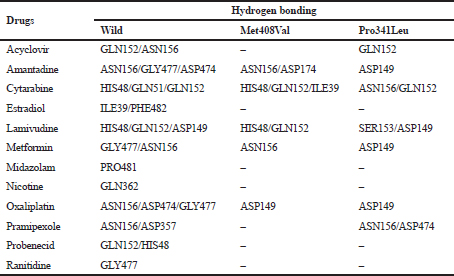 | Table 3. Hydrogen-bonding analysis of OCT substrates with the wild type, Val408, and Leu341. [Click here to view] |
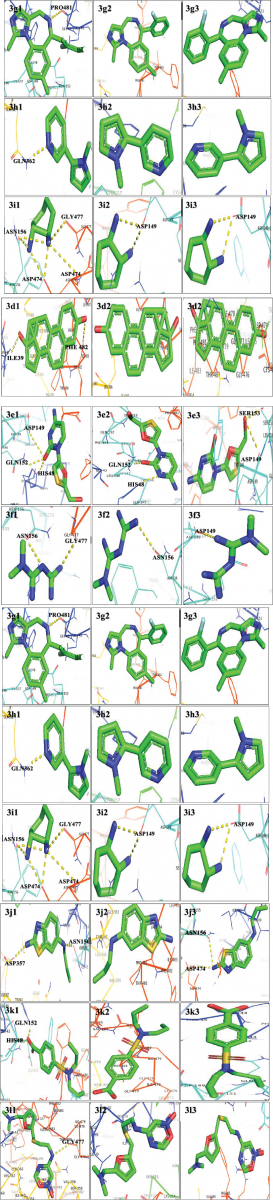 | Figure 3. Three-dimensional complexes of wild and modeled mutant hOCT structures with docked substrates. 3a1, 3a2 and 3a3 represent Acyclovir bound to wild, Met408Val, and Pro341Leu variants, respectively, 3b1, 3b2 and 3b3 represent Amantadine bound to wild, Met408Val, and Pro341Leu variants, respectively, 3c1, 3c2 and 3c3 represent Cytarabine bound to wild, Met408Val, and Pro341Leu variants, respectively. 3d1, 3d2 and 3d3 represent Estradiol bound to wild, Met408Val, and Pro341Leu variants, respectively, 3e1, 3e2 and 3e3 represent Lamivudine bound to wild, Met408Val, and Pro341Leu variants, respectively, 3f1, 3f2 and 3f3 represent Metformin bound to wild, Met408Val, and Pro341Leu variants, respectively. 3g1, 3g2 and 3g3 represent Midazolam bound to wild, Met408Val, and Pro341Leu variants, respectively, 3h1, 3h2 and 3h3 represent Nicotine bound to wild, Met408Val, and Pro341Leu variants, respectively, 3i1, 3i2 and 3i3 represent Oxaliplatin bound to wild, Met408Val, and Pro341Leu variants, respectively, 3j1, 3j2 and 3j3 represent Pramipexole bound to wild, Met408Val, and Pro341Leu variants, respectively, 3k1, 3k2 and 3k3 represent Probenecid bound to wild, Met408Val, and Pro341Leu variants, respectively, 3l1, 3l2 and 3l3 represent Ranitidine bound to wild, Met408Val, and Pro341Leu variants, respectively. [Click here to view] |
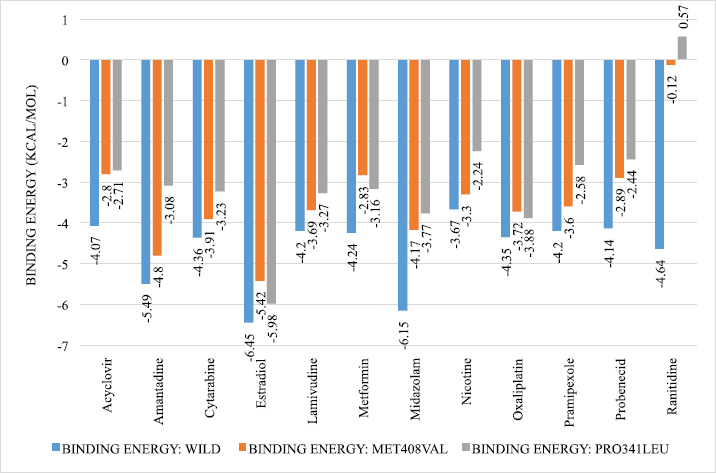 | Figure 4. Summary of binding energies of OCT substrates with the wild type, Val408, and Leu341. p value = 0.0069, 95% CI, p value retrieved through one-way ANOVA. [Click here to view] |
CONCLUSION
The binding energy of hOCT1 substrates to the mutants (M408V and P341L) was found to be less than that of the wild type. This indicates that the presence of functional non-synonymous polymorphism in the hOCT1 significantly alters the binding of the ligand. It also helps to obtain information about the PKs and PDs of a drug. Hence, patients who have this SNP (1022C>T and 1222A>G) would have decreased hOCT1 function which would result in therapeutic failure or subtherapeutic drug response. It also provides a personalized treatment regimen in patients who have such SNPs.
ACKNOWLEDGMENT
The authors extend their gratitude to the management of Krupanidhi College of Pharmacy for providing them with all the facilities that were required. They are also thankful to the principal of Krupanidhi College of Pharmacy for his excellent research guidance and support. They are also very thankful to the access to Pymol 2.3 (Schrodinger, LLC, New York, NY) for visualizing the docked complexes.
CONFLICTS OF INTEREST
Authors declared that they do not have any conflicts of interest.
FUNDING
None.
REFERENCES
Aanandhi MV, George PSG, Structure affinity relationship and characterization of benzoporphyrins as potent inhibitors of yap oncoprotein through in silico experiments. Int J Pharm Pharm Sci, 2015; 7(3):278–84.
Ahmadimoghaddam D, Zemankova L, Nachtigal P, Dolezelova E, Neumanova Z, Cerveny L, Ceckova M, Kacerovský M, Micuda S, Staud F. Organic Cation Transporter 3 (OCT3/SLC22A3) and multidrug and toxin extrusion 1 (MATE1/SLC47A1) transporter in the placenta and fetal tissues: expression profile and fetus protective role at different stages of gestation1. Biol Reprod, 2013; 88(3):55; doi:10.1095/biolreprod.112.105064
Ahmed S, Zhou Z, Zhou J, Chen SQ. Pharmacogenomics of drug metabolizing enzymes and transporters: relevance to precision medicine. Genomics Proteomics Bioinformatics, 2016; 14:298–313; doi:10.1016/j.gpb.2016.03.008
Boxberger KH, Hagenbuch B, Lampe JN. Common drugs inhibit human organic cation transporter 1 (OCT1)-mediated neurotransmitter uptake. Drug Metab Dispos, 2014; 42:990–95; doi:10.1124/dmd.113.055095
Brosseau N, Ramotar D. The human organic cation transporter OCT1 and its role as a target for drug responses. Drug Metab Rev, 2019; 51:389–407. doi:10.1080/03602532.2019.1670204
Chen L, Takizawa M, Chen E, Schlessinger A, Segenthelar J, Choi JH, Sali A, Kubo M, Nakamura S, Iwamoto Y, Iwasaki N, Giacomini KM. Genetic polymorphisms in organic cation transporter 1 (OCT1) in Chinese and Japanese populations exhibit altered function. J Pharmacol Exp Ther, 2010; 335:42–50; doi:10.1124/jpet.110.170159
Dakal TC, Kumar R, Ramotar D. Structural modeling of human organic cation transporters. Comput Biol Chem, 2017; 68:153–63. https://doi.org/10.1016/j.compbiolchem.2017.03.007
Du Plessis M, Pearce B, Jacobs C, Hoosain N, Benjeddou M. Genetic polymorphisms of the organic cation transporter 1 gene (SLC22A1) within the cape admixed population of South Africa. Mol Biol Rep, 2015; 42:665–72; doi:10.1007/s11033-014-3813-2
Fattah S, Shinde AB, Matic M, Baes M, van Schaik RHN, Allegaert K, Parmentier C, Richert L, Augustijns P, Annaert P. Inter-subject variability in OCT1 activity in 27 batches of cryopreserved human hepatocytes and association with OCT1 mRNA expression and genotype. Pharm Res, 2017; 34:1309–19; doi:10.1007/s11095-017-2148-9
Gharavi R, Hassan HE. Genomics and drug transporters and application in drug discovery, delivery, and development. In: Pathak Y (ed.). Genomics-driven healthcare. Springer Singapore, Singapore, pp 133–75, 2018. Available via https://doi.org/10.1007/978-981-10-7506-3_8
Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 1997; 18: 2714–23; doi:10.1002/elps.1150181505
Han T. (Kevin), Everett RS, Proctor WR, Ng CM, Costales CL, Brouwer KLR, Thakker DR. Organic cation transporter 1 (OCT1/mOct1) is localized in the apical membrane of Caco-2 cell monolayers and enterocytes. Mol Pharmacol, 2013; 84:182–9; doi:10.1124/mol.112.084517
He L, Vasiliou K, Nebert DW. Analysis and update of the human solute carrier (SLC) gene superfamily. Hum Genomics, 2009; 3:195–206; doi:10.1186/1479-7364-3-2-195
Jendele L, Krivak R, Skoda P, Novotny M, Hoksza D. PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res, 2019; 47:W345–49; doi:10.1093/nar/gkz424
Koepsell H, Schmitt BM, Gorboulev V. Organic cation transporters. Rev Physiol Biochem Pharmacol, 2003; 150:36–90; doi:10.1007/s10254-003-0017-x
Koepsell H. The SLC22 family with transporters of organic cations, anions and zwitterions. Mol Aspects Med, 2013; 34:413–35; doi:10.1016/j.mam.2012.10.010
Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins Struct Funct Bioinform, 2009; 77:114–22; doi:10.1002/prot.22570
Kucheryavykh LY, Rolón-Reyes K, Kucheryavykh YV, Skatchkov S, Eaton MJ, Sanabria P, Wessinger WD, Inyushin M, Glioblastoma development in mouse brain: general reduction of OCTs and mislocalization of OCT3 transporter and subsequent uptake of ASP+ substrate to the nuclei. J Neurosci Neuroeng, 2014; 3:3–9; doi:10.1166/jnsne.2014.1091
Lozano E, Herraez E, Briz O, Robledo VS, Hernandez-Iglesias J, Gonzalez-Hernandez A, Marin JJG. 2013. Role of the plasma membrane transporter of organic cations OCT1 and its genetic variants in modern liver harmacology. Biomed Res Int, 2013; 2013:1–13; doi:10.1155/2013/692071
Makhtar SM, Husin A, Baba AA, Ankathil R. Genetic variations in influx transporter gene SLC22A1 are associated with clinical responses to imatinib mesylate among Malaysian chronic myeloid leukaemia patients. J Genet, 2018; 97:835–842; doi:10.1007/s12041-018-0978-9
Mani RK. Structural insights into warfarin resistance due to single nucleotide polymorphisms in vitamin K epoxide reductase complex I. J Pharm Res, 2017; 11(3), pp. 261–266.
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem, 1998; 19:1639–62; doi:10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B.
Ramakrishnan C, Ramachandran GN. Stereochemical criteria for polypeptide and protein chain conformations. Biophys J, 1965; 5:909–33. CrossRef
Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc, 2010; 5:725–38; doi:10.1038/nprot.2010.5
Sala-Rabanal M, Li DC, Dake GR, Kurata HT, Inyushin M, Skatchkov SN, Nichols CG. Polyamine transport by the polyspecific organic cation transporters OCT1, OCT2, and OCT3. Mol Pharm, 2013; 10:1450–58; doi:10.1021/mp400024d
Samuel Gideon George P, Dhivya K. Effect of GLY 16 ARG single nucleotide polymorphism on agonist binding to beta 2 adrenergic receptor – a structural pharmacogenomic approach. Int J Pharm Sci Res, 2016. 7(10), pp. 4064–4073. doi: 10.13040/IJPSR.0975-8232.7(10).4064-73.
Sissung TM, Troutman SM, Campbell TJ, Pressler HM, Sung H, Bates SE, Figg WD. Transporter polymorphisms affect normal physiology, diseases, and pharmacotherapy. Discov Med, 2012; 13:19–34.
Umamaheswaran G, Arunkumar A, Shewade D, Praveen R, Das A, Adithan C. Genetic analysis of OCT1 gene polymorphisms in an Indian population. Indian J Hum Genet, 2011; 17:164; doi:10.4103/0971-6866.92094
Umamaheswaran G, Kumar DK, Adithan C. Distribution of genetic polymorphisms of genes encoding drug metabolizing enzymes & drug transporters - a review with Indian perspective. Indian J Med Res, 2014; 139:27–65.
Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods, 2015; 12:7–8; doi:10.1038/nmeth.3213
Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res, 2015; 43:W174–81; doi:10.1093/nar/gkv342
Zhao FQ. Octamer-binding transcription factors: genomics and functions. Front Biosci, 2013; 18:1051–71. CrossRef