INTRODUCTION
The sterility of parenteral products is a key attribute for product safety (Hussong, 2010). An appropriate cleaning standard must be maintained to ensure the sterile production in the controlled environments, where this type of product is manufactured (Xavier et al., 2013). Hertroys (1997) addressed the use of cleanrooms as an essential requirement for pharmaceutical production. The cleanliness controls of cleanrooms associated with microbiological monitoring and control are a part of the industry’s quality assurance routine (Xavier et al., 2017). These control measures aim to maintain the quality and purity required for the parenteral products (Pacheco and Pinto, 2010).
The American (USP, 2012) and Brazilian (Brasil, 2010) Pharmacopoeias establish the requirements for the control and monitoring of controlled environments. The United States Pharmacopoeia also determines that if microorganisms are detected in drug substances, excipients, water for pharmaceutical use, the manufacturing environment, intermediates, and finished drug products in exceeded levels, they must undergo species-level characterization. This may include identification and strain typing, as appropriate. This information provides data for investigations on contaminations and the understanding of the area’s microbiota.
Sandle (2011) showed two general categories of microbial identification: those that examine phenotypic characteristics and those that examine the genotypic composition, including the nature of the microorganisms’ constituent nucleic acids and genes.
One of the methodologies of microbial identification by proteomic methods cited in the United States Pharmacopoeia and described by Arnold and Reilly (1998), Barbosa et al. (2019), and Patel (2015) is the MALDI-TOF mass spectrometry (matrix-assisted laser desorption ionization-time of flight). This methodology is based on the protein profile obtained by MALDI-TOF MS, which combines the acquired sequence with a database of known profiles, such as a proteomic fingerprint (Ojima-Kato et al., 2014; Porte et al., 2017). This technique has already been used for fungal and yeast identification providing a rapid, accurate, and competitive analysis (Patel, 2015; Xavier et al., 2019; Zhao et al., 2018).
As genotypic methods, the techniques that use the sequences of representative elements based on PCR are widely used for microbial identification. In filamentous fungi and yeasts, 18S rDNA, 5.8S rDNA, and 28S rDNA genic regions are the target regions for these methods. According to Bellemain et al. (2010), among these genic regions, there are also conserved DNA intergenic regions, known as internal transcribed spacer (ITS), which can be used for genetic differentiation. As these regions accumulate more mutations when compared to coding regions, they can be used for the infrageneric studies (Baldwin et al., 1995; Spier et al., 2008).
In the genotypic method of restriction fragment length polymorphism analysis using polymerase chain reaction (PCR-RFLP), the PCR-amplified fragments are digested with one or more specific restriction endonucleases, followed by agarose gel electrophoresis (Bai, 2014; Farber et al., 2001; Gandra et al., 2008), whereas evaluation of the obtained fragments may be used to detect intra- and interspecific polymorphisms.
The PCR-RFLP method has been described in the literature as a fast and efficient method capable of discriminating fungal and yeast species in routine laboratory identification (Fatima et al., 2017; Gharaghani et al., 2018; Mirhendi et al., 2006).
The objective of this study was to identify the filamentous fungi and yeasts isolated from an industrial environment by the MALDI-TOF MS proteomic approach and to analyze the PCR-RFLP capacity in discriminating inter- and intraspecific differences in these isolates.
MATERIALS AND METHODS
Sampling and identification of filamentous fungi and yeasts
The fungal and yeast samples used in this study came from the environmental monitoring and surfaces of drug production areas and microbiological quality control laboratories of a pharmaceutical industry located in southeastern Minas Gerais, Brazil. The isolates in question were selected from the available samples stored from January 2018 to January 2019. The sampling method consisted of active and passive air sampling and surface sampling. All the isolates had been previously collected to irradiated tryptic soy agar culture medium plates (BioMérieux) incubated at 30°C–35°C for 3 days. After this period, the microorganisms were kept in a cold (temp) chamber and were periodically transferred according to the internal procedures of the industry where they were isolated. Besides the isolates, a standard strain of Aspergillus brasiliensis ATCC16404 and two strains of Candida albicans ATCC10231 were used in this study.
Identification of isolates by the MALDI-TOF MS method
The standard strains and selected microorganisms presumably identified by microscopic methods such as filamentous fungi and yeasts were reactivated in irradiated tryptic soy agar culture medium (BioMérieux) incubated at 30°C–35°C for 3 days. After the incubation period, they were subjected to species-level identification procedures by the MALDI-TOF proteomic approach.
Since they relate to fungal/yeast samples, which present a more resistant cell wall as compared to bacteria, the samples were prepared through the Bruker Daltonics 70% formic acid ribosomal protein extraction method, for later identification by the MALDI-TOF MS methodology. The reactivated colonies were individually added to tubes containing tryptic soy broth, which were kept under constant rotation at the room temperature until the demonstration of enough biological material to perform extraction. After growth in the tubes, they were rested on a bench for approximately 10 minutes for the fungi to settle to the bottom of the tubes. About 1.5 ml of the supernatant from each tube was transferred to microtubes, which were centrifuged for 2 minutes at 13,000 rpm. After centrifugation, all supernatants from the microtubes were removed, and 1 ml of sterile deionized water was added to each pellet formed on centrifugation, which was then subjected to vortex agitation and further centrifugation, whereas such steps were repeated twice. To each microtube, 300 μl of sterile deionized water and 900 μl of EtOH were then added, followed by vortex agitation and centrifugation for 2 minutes at 13,000 rpm. All supernatants were removed, and the pellets were oven-dried in an inverted position at 37°C for 10 minutes. After drying, the amount of 70% formic acid needed to cover and suspend the pellets was added. The same amount of acetonitrile was added to the microtubes which were subjected to agitation and centrifugation. Finally, 1 μl of the supernatant of each tube was added to the spots of a Microflex LT equipment stainless steel plate. After the material was dried on the plate, 1 μl of HCCA matrix (a saturated solution of α-cyano-4-hydroxycinnamic acid from supplier Bruker Daltonics) was applied to the spots. It was necessary to wait for the complete drying of the spots on the plate before placing it into the analysis equipment.
The mass spectra of the samples were obtained and classified using FlexControl 3.4 and MBT-RTC 4.0 software programs, respectively, on Bruker Daltonics GmbH’s Microflex LT mass spectrometer, equipped with an N2 laser operating at a wavelength of 337 nm and frequency of 60 Hz. The positively charged intact proteins and peptides were electrostatically accelerated (at about 20 kV) on a 96-cm time-of-flight analyzer. The ions were measured in a positive linear model with instrument parameters optimized in the range of 1,900–20,000 m/z. Before sample acquisition, a calibrator named BTS— Bacterial Test Standard (E. coli DH5 alpha, Bruker Daltonics)—was acquired and compared to a standard spectrum, with a standard deviation result <300 ppm. All spectra acquired in the analysis were compared to the system database, and a score representing the degree of similarity to the standard was given to each result. The scores ≥2,000 indicate a reliable identification to the species level, scores ≥1,700 and <2,000 indicate reliable identification to the genus level, and scores <1,700 do not represent the reliable results for release.
DNA extraction from isolates for genetic analysis
The isolates previously identified by MALDI-TOF were added to microtubes, and their DNA was extracted using PrepMan™ Ultra Sample Preparation Reagent Kit (Thermo Fisher Scientific, USA) according to the manufacturer's instructions. The quality of the DNA extracted for subsequent PCR analysis was verified in 1.0% agarose gel stained with ethidium bromide.
Characterization of genetic profile by PCR-RFLP
To obtain DNA fragments from the samples of approximately 1,000 base pairs, 5'-TTGGTCATTTAGAGGAAGTAA-3' and 5'-CCGTGTTTCAAGACGG-3' oligonucleotide sequences, named OligoITS1F and OligoLR3, described by Raja et al. (2017), were selected. Primers ITS1 5'-TCCGTAGGTGAACCTGCGG-3' and 5'- TACTACCACCAAGATCT-3', named OligoITS1 and OligoLR7, respectively, described by Fujita et al. (2001) and from Duke University (Vilgalys, 2017), were also used to obtain 2,000 base pair amplicons. Two pairs of primers were defined to obtain two different sizes for more robust results.
Fragments obtained with the oligonucleotides selected for the study comprise fungal universal gene regions 18S rDNA, 5.8S rDNA, 28S rDNA, ITS1, and ITS2, as shown in Figure 1.
Reactions were performed in a mix containing 2× PlatinumTM HotStart Green Master Mix (Thermo Fisher Scientific, USA), MgCl2 (2.5 mM), 10 μM of each primer, and 50 ng of yeast DNA in a final reaction volume of 50 μl. The PCR was performed on an Applied Biosystems Veriti Thermal Cycler. The amplification conditions followed the following parameters: an initial denaturation cycle at 98°C for 30 seconds, followed by 30 denaturation cycles at 98°C for 10 seconds, annealing at 54°C for 30 seconds, extension at 72°C for 2 minutes, and final extension at 72°C for 10 minutes. The amplicons were visualized in 1.5% agarose gel stained with ethidium bromide and photodocumented.
For restriction analysis of approximately 2,000-bp PCR products, 15 μl of PCR product (containing approximately 500 ng of DNA), 10U of BsuRI enzyme (Thermo Fisher Scientific), 2.5 μl of digestion buffer (10×), 1 μl of bovine serum albumin (0.2 μg/μl), and 5.5 μl of sterile water were used in a digestion system. The digestion system was incubated in a dry bath in an Applied Biosystems Veriti Thermal Cycler at 37ºC for 2 hours. After the incubation period, 15 μl of the restriction enzyme-digested PCR product was electrophoresed in 3% agarose gel stained with ethidium bromide, visualized, and photodocumented.
 | Figure 1. Schematic representation of the fungal and yeast ribosomal genes containing the target sites of oligonucleotide annealing and expected PCR amplification sizes used in this study. [Click here to view] |
For restriction analysis of approximately 1,000-bp PCR products, 15 μl of PCR product (containing approximately 500 ng of DNA), 10U of TaqαI enzyme (Promega Corporation), 2.5 μl of digestion buffer (10×), 1 μl of bovine serum albumin (0.2 μg/μl), and 5.5 μl of sterile water were used in a digestion system. The digestion system was incubated in a dry bath in an Applied Biosystems Veriti Thermal Cycler at 65ºC for 2 hours. After the incubation period, 15 μl of the restriction enzyme-digested PCR product was electrophoresed in 3% agarose gel stained with ethidium bromide, visualized, and photodocumented.
RESULTS AND DISCUSSION
In this study, the MALDI-TOF method could identify only 68.42% (13/19) of the samples to the species level (MALDI-TOF real-time identification score ≥2,000). However, this methodology enabled the genus-level identification of all isolates (19/19) and the three standard strains included in this study (Figure 2). The most prevalent microorganisms among the isolated yeast samples were Candida guilliermondii (04/07), followed by Rhodosporidium toruloides (02/07) and Rhodotorula mucilaginosa (01/07). Among the filamentous fungi samples, the genus Aspergillus was the most common (03/09), followed by Penicillium (02/09). The remaining fungi and yeasts identified and their frequencies are shown in Figure 2.
The MALDI-TOF MS methodology is a technique that has recently been used worldwide to perform the inexpensive and reliable real-time identification of different types of microorganisms (Lau et al., 2019; Patel, 2015; Schubert and Kostrzewa, 2017; Xavier et al., 2019; Zhao et al., 2018). The use of solvents, such as formic acid, ethanol, and acetonitrile, in the sample preparation facilitates cell disruption and, consequently, the release of ribosomal proteins, leading to more accurate measurements by this technique (Intra et al., 2018). As reviewed by Angeletti (2017), the MALDI-TOF MS is a technological innovation that allows rapid and accurate microbial identification. The provision of a comprehensive reference spectra database for MALDI-TOF MS is essential to obtain results through this analysis (Freiwald and Sauer, 2009; Normand et al., 2017; Zhao et al., 2018).
 | Figure 2. Identification and frequencies of filamentous fungi and yeasts by the MALDI-TOF method. [Click here to view] |
 | Table 1. Isolation site and sampling method of filamentous fungi and yeasts isolated from industrial environment selected for the study. [Click here to view] |
Among the microorganisms identified in our study, isolated from the environmental monitoring programs of a pharmaceutical industry, collected according to Table 1, we observed the prevalence of C. guilliermondii, Aspergillus, Penicillium, and R. toruloides (Fig. 2). Despite the difficulties in finding published data to compare with our results, mainly because pharmaceutical companies usually do not disclose the microbiota of their production areas, but Xavier et al. (2017), by sampling an industrial environment, identified the genera Penicillium spp., Scopulariopsis spp., and Chaetomium spp., as we have also did here. The identification results show the occurrence of fungi and yeasts that are commonly found in the environment or the human microbiota. The yeast C. guilliermondii is found in the microbiota of human skin and mucous membranes, whereas its isolation as a pathogenic microorganism is not common, and it lives in symbiosis (Belkaid and Segre, 2014; Marcos-Zambrano et al., 2017; Pasqualotto et al., 2006). The filamentous fungi genera Aspergillus and Penicillium are commonly found on earth (in the soil), with more than 300 accepted species (Bennett, 2010; Park et al., 2017; Perrone and Susca, 2017). They are responsible for decomposing various organic substances in nature and are also largely used in industrial production (Cairns et al., 2018; Houbraken et al., 2014; Park et al., 2017; Perrone and Susca, 2017). The yeast R. toruloides is known for its lipid and chemical production (Singh et al., 2018), presenting a great bioengineering potential (Otoupal et al., 2019).
Besides the characterization by MALDI-TOF MS, the ITS regions of the samples were analyzed by the PCR-RFPL method, to assess genetic diversity among the isolates, which is performed by PCR amplification followed by restriction analysis of the amplified fragments (Hierro et al., 2004). For the PCR-RFLP analysis, a pilot experiment was performed, and two samples of C. guilliermondii (Iso18 and Iso19) isolated from different plates and sampling points were selected (Table 1). The PCR method allowed amplification of an expected fragment of approximately 2,000 bp corresponding to a fungal universal genic region (18S rDNA, 5.8S rDNA, 28S rDNA, ITS1, and ITS2) (Fig. 3, panel A). The restriction profile analysis of the 2,000-bp region digested with BsuRI revealed a homogeneous profile among the C. guilliermondii isolates tested (120, 140, 400, 500, and 610 bp) (Fig. 3, panel B; Table 2). Subsequently, we sought to analyze all the isolates and strains of this study (Table 1) by PCR-RFLP. Figure 4 shows the amplification profile of the isolates and strains by the PCRs to detect the 2,000- and 1,000-bp genic universal regions (Fig. 1). Some samples revealed weak or no amplification of the expected 2,000-bp fragment (Iso2, Iso7, Iso10, Iso12, Iso13, and Iso14) (Fig. 4, panel A). In the PCR performed to obtain the 1,000-bp fragment, only two samples did not show any amplification pattern (Iso5 and Iso10) (Fig. 4, panel B). This result can be attributed to problems that occurred in the DNA extraction step, for instance, low DNA yield. However, it was decided to proceed with the hydrolysis of the 2,000 and 1,000 bp fragments of the amplified samples.
The 2,000-bp PCR products were digested with BsuRI and Taqα1 restriction enzymes to verify the ability of PCR-RFLP methods to characterize inter- and intraspecific genetic variability among the ATCC strains and isolates. The restriction analysis of the 2,000-bp PCR digested with both enzymes revealed a heterogeneous profile among the samples (Fig. 5, panels A and B). However, some isolates showed profiles, whose digestion products could not be clearly defined in 3% agarose gel. Bands presenting higher intensity in the gel were considered for reading, whereas those with poor definition were disregarded. The restriction profile analysis of the 2,000-bp fragment digested with BsuRI enzyme generated eight different profiles for the 13 microorganisms tested. The isolates such as Iso8, Iso9, Iso18, and Iso19—corresponding to C. guilliermondii species—presented the same restriction profile (120, 140, 400, 500, and 610 bp). Two isolates identified as Penicillium spp. also presented the same restriction profile, sharing the 250, 390, and 500 bp bands (Fig. 5, panel A; Table 2).
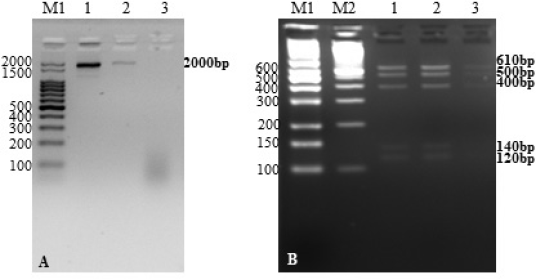 | Figure 3. Results of PCR-RFLP optimization of the fungal Universal region. Panel A: PCR of the fungal universal region. M1: 100 base pair molecular mass marker (Ludwig biotechnologies). 1) Iso18 (C. guilliermondii). 2) Iso19 (C. guilliermondii). 3) Iso18 (C. guilliermondii). The 2,000-bp amplified fragment is indicated to the right of the 1.8% agarose gel. Panel B: Restriction profile of PCR products digested with BsuRI enzyme. M1: mid-range molecular mass marker (Cellco); M2: 100 base pair molecular mass marker (Invitrogen). 1) Iso18 (C. guilliermondii). 2) Iso19 (C. guilliermondii). 3) Iso18 (C. guilliermondii). Restriction profiles of 120, 140, 400, 500, and 610 base pairs are indicated to the right of the 3% agarose gel. [Click here to view] |
Although the PCR-RFLP technique was used here only for genetic diversity analysis, the profile of Iso9, identified as Aspergillus versicolor, was the same as that presented by Aspergillus spp./Iso17 (identified only to the genus level by MALDI-TOF MS). However, the poor definition of the bands of Iso17 digested with Taqα1 enzyme does not allow us to infer that the Aspergillus spp. could correspond to A versicolor species. The restriction profile analysis of the 2,000-bp fragment digested with Taqα1 enzyme generated nine different profiles for the 11 microorganisms tested. The isolates such as Iso8, Iso9, Iso18, and Iso19, corresponding to C. guilliermondii species, presented the same restriction profile (140, 200, 300, 400, and 500 bp). Two isolates identified as Penicillium spp. also presented the same restriction profile (90, 140, 150, 350, 400, and 500 bp) sharing 250, 390, and 500 bp bands (Fig. 5, panel B; Table 2).
The 1,000-bp PCR products were also digested with BsuRI and Taqα1 restriction enzymes to verify the ability of PCR-RFLP methods to analyze inter- and intraspecific genetic variability among the ATCC strains and isolates. The restriction analysis of the 1,000-bp PCR digested with BsuRI and Taqα1 enzymes revealed a heterogeneous profile among the samples (Fig. 6, panels A and B; Table 3). A digestion of the 1,000-bp fragment with BsuRI resulted in nine different profiles for 15 microorganisms tested. The isolates of the same species of C. guilliermondii (Iso8 and Iso11) presented the same band profile (150, 350, and 500 bp). The homogeneous profiles were also observed among Penicillium spp., Rhodosporidium spp., and C. albicans species (Fig. 6, panel A; Table 3). Digestion of the 1,000-bp fragment with Taqα1 resulted in 11 profiles for 15 microorganisms tested. The homogeneous profiles were also observed among C. guilliermondii, Penicillium spp., R. toruloides, and C. albicans species (Fig. 6, panel B; Table 3).
 | Table 2. Size in base pairs obtained by PCR-RFLP (2,000-bp fragment digested with BsuRI and Taqα1). [Click here to view] |
 | Figure 4. PCR of the fungal universal region. Panel A: Amplification of the 2,000-bp fragment of different species of filamentous fungi and yeasts. M: Mid-range molecular mass marker (Cellco); 1) Iso1 (R. toruloides); 2) Iso3 (Penicillium spp.); 3) Iso4 (Penicillium spp.); 4) Iso5 (R. mucilaginosa); 5) CP1 (C. albicans); 6) Iso8 (C. guilliermondii); 7) Iso9 (A. versicolor); 8) Iso11 (C. guilliermondii); 9) Iso15 (Scopulariopsis spp.); 10) Iso16 (Chaetomium spp.); 11) Iso17 (Aspergillus spp.). M: Mid-Range Molecular Mass Marker (Cellco). Panel B: Amplification of the 2000-bp fragment of different species of filamentous fungi and yeasts. M: Mid-range molecular mass marker (Cellco); 1) Iso1(R. toruloides); 2) Iso2 (R. toruloides); 3) Iso3 (Penicillium spp.); 4) Iso4 (Penicillium spp.); 5) CP1 (C. albicans); 6) CP2 (C. albicans); 7) Iso8 (C. guilliermondii); 8) Iso9 (A. versicolor); 9) Iso11 (C. guilliermondii); 10) Iso12 (Moniliella spp.); 11) Iso13 (Aspergillus unguis); 12) Iso14 (Paecilomyces lilacinus); 13) Iso15 (Scopulariopsis spp.); 14) Iso16 (Chaetomium spp.); and 15) Iso17 (Aspergillus spp.). [Click here to view] |
According to Raja et al. (2017) and Schoch et al. (2012), the ITS region is one of the markers that present a higher probability of correct identification for a broad group of fungi. They consider the ITS region as an appropriate fungal barcode. The sequences of the ITS1 and ITS2 regions and 5.8S and 28S rDNA genes were used by Benedetti et al. (2016) to correctly identify the yeast species.
 | Figure 5. Restriction profile of 2,000-bp PCR products digested with BsuRI and Taqα1 enzymes. Panel A: Restriction profile of PCR products digested with BsuRI enzyme. M: Mid-range molecular mass marker (Cellco); 1) Iso1 (R. toruloides); 2) Iso3 (Penicillium spp.); 3) Iso4 (Penicillium spp.); 4) Iso5 (R. mucilaginosa); 5) CP1 (C. albicans); 6) Iso8 (C. guilliermondii); 7) Iso9 (A. versicolor); 8) Iso11 (C. guilliermondii); 9) Iso15(Scopulariopsis spp.);10) Iso16 (Chaetomium spp.); 11) Iso17 (Aspergillus spp.). Panel B: Restriction profile of digestion with Taqα1 enzyme. M: Mid-range molecular mass marker (Cellco); 1) Iso1 (R. toruloides); 2) Iso3 (Penicillium spp.); 3) Iso4 (Penicillium spp.); 4) Iso5 (R. mucilaginosa); 5) CP1 (C. albicans); 6) Iso8 (C. guilliermondii); 7) Iso9 (A. versicolor); 8) Iso11 (C. guilliermondii); 9) Iso15 (Scopulariopsis spp.);10) Iso16 (Chaetomium spp.); 11) Iso17 (Aspergillus spp.). [Click here to view] |
The restriction profile analysis of the 2,000-bp fragment digested with BsuRI enzyme generated eight different profiles for 13 microorganisms tested. The isolates such as Iso8, Iso9, Iso18, and Iso19, corresponding to C. guilliermondii species, presented the same restriction profile (120, 140, 400, 500, and 610 bp). No difference was found in the restriction profiles of the two Penicillium spp. samples related to 250-, 390-, and 500-bp bands (Fig. 5, panel A; Table 2). It was also possible to verify that the band profiles of the A. versicolor isolate were identical to the profile presented by Aspergillus spp., suggesting that they are the same microorganism. The restriction profile analysis of the 2,000-bp fragment digested with Taqα1 enzyme generated nine different profiles for 11 microorganisms tested. The isolates such as Iso8, Iso9, Iso18, and Iso19, corresponding to C. guilliermondii species, presented the same restriction profile (140, 200, 300, 400, and 500 bp). The two isolates identified as Penicillium spp. also presented the same restriction profile (90, 140, 150, 350, 400, and 500 bp) sharing the 250, 390, and 500 bp bands (Fig. 5, panel B; Table 2).
Digestion of the 1,000-bp fragments with BsuRI enzyme generated nine different profiles for 15 microorganisms tested. C. guilliermondii species (Iso8 and Iso11) presented the same band profile (150, 350, and 500 bp). The homogeneous profiles were also observed among Penicillium spp., Rhodosporidium spp., and C. albicans species (Fig. 6, panel A; Table 3). Digestion of the 1,000 bp fragment with Taqα1 resulted in eleven profiles for 15 microorganisms tested. Homogeneous profiles were also observed among C. guilliermondii, Penicillium spp., R. toruloides, and C. albicans species (Fig. 6, panel B; Table 3). Thus, it can be concluded that there was no significant difference in the tests whether performed with 1,000- or 2,000-bp fragments.
 | Figure 6. Restriction profile of 1000 bp PCR products digested with BsuRI and Taqα1 enzymes. Panel A: Restriction profile of PCR products digested with BsuRI enzyme. M: Mid-Range Molecular Mass Marker (Cellco); 1) Iso1 (R. toruloides); 2) Iso2 (R. toruloides); 3) Iso3 (Penicillium spp.); 4) Iso4 (Penicillium spp.); 5) CP1 (C. albicans); 6) CP2 (C. albicans); 7) Iso8 (C. guilliermondii); 8) Iso9 (A. versicolor); 9) Iso11 (C. guilliermondii); 10) Iso12 (Moniliella spp.); 11) Iso13 (A. unguis); 12) Iso14 (P. lilacinus); 13) Iso15 (Scopulariopsis spp.); 14) Iso16 (Chaetomium spp.); and 15) Iso17 (Aspergillus spp.). Panel B: Restriction profile of digestion with Taqα1 enzyme. M: Mid-Range Molecular Mass Marker (Cellco); 1) Iso1 (R. toruloides); 2) Iso2 (R. toruloides); 3) Iso3 (Penicillium spp.); 4) Iso4 (Penicillium spp.); 5) CP1 (C. albicans); 6) CP2 (C. albicans); 7) Iso8 (C. guilliermondii); 8) Iso9 (A. versicolor); 9) Iso11 (C. guilliermondii); 10) Iso12 (Moniliella spp.); 11) Iso13 (A. unguis); 12) Iso14 (P. lilacinus); 13) Iso15 (Scopulariopsis spp.); 14) Iso16 (Chaetomium spp.); and 15) Iso17 (Aspergillus spp.). [Click here to view] |
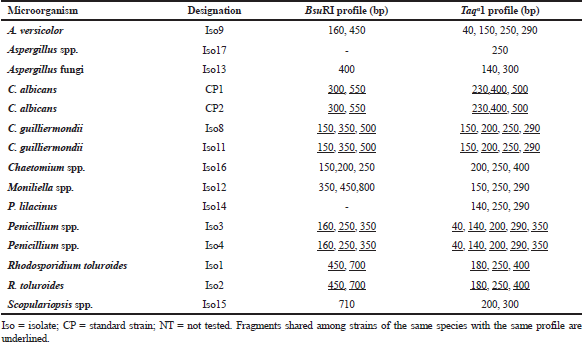 | Table 3. Size in base pairs obtained by PCR-RFLP (1000-bp fragment digested with BsuRI and Taqα1). [Click here to view] |
Kordalewska et al. (2018) proved the efficiency of the PCR-RFLP method for advanced species-level identification of the filamentous fungus Scopulariopsis. This same method was used by Robledo-Leal et al. (2018) to identify approximately 60 isolates of Candida yeast.
CONCLUSION
The MALDI-TOF MS method identified all samples of filamentous fungi and yeasts to the species or genus level. The PCR-RFLP method of the ITS regions, performed with fragments of two sizes and two different restriction enzymes, was demonstrated to be capable of the interspecific analysis of the yeast and fungi samples analyzed in this study.
ACKNOWLEDGMENTS
The authors are grateful to the Biotechnology Graduate Program of the State University of Montes Claros, Minas Gerais, Brazil, and Novo Nordisk Produção Farmacêutica do Brasil for the incentive and support in performing sample identification by MALDI-TOF MS.
AUTHORS’ CONTRIBUTIONS
Luciana Nobre Leite, Mauro aparecido de Sousa Xavier, and Alessandra Rejane Ericsson de Oliveira Xavier designed the study. Luciana Nobre Leite, Josiane dos Santos, Léia Cardoso, Frederico Santos Barbosa, Rosimar Fonseca dos Santos, and Soraya Aparecida Maia Dias performed the experiments. Luciana Nobre Leite, Felipe José Nobre Lelis, Mauro aparecido de Sousa Xavier, and Alessandra Rejane Ericsson de Oliveira Xavier performed data analysis. Alessandra Rejane Ericsson de Oliveira Xavier and Luciana Nobre Leite wrote the manuscript. All authors read, edited, and approved the final manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest related to the publication of this paper.
FINANCIAL SUPPORT
None.
REFERENCES
Angeletti S. Matrix assisted laser desorption time of flight mass spectrometry (MALDI-TOF MS) in clinical microbiology. J Microbiol Meth, 2017; 138:20–9. CrossRef
Arnold RJ, Reilly JP. Fingerprint matching of E. coli strains with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of whole cells using a modified correlation approach. Rapid Commun Mass SP, 1998; 12:630–6. CrossRef
Bai FY. Association of genotypes with infection types and antifungal susceptibilities in Candida albicans as revealed by recent molecular typing strategies. Mycology, 2014; 5:1–9. CrossRef
Baldwin BG, Sanderson MJ, Porter JM, Martin F, Wojciechowski CSC, Donoghue MJ. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Missouri Bot Gard, 1995; 82:247–77. CrossRef
Barbosa FS, Leite LN, Xavier MAS, Xavier AREO. Genomic and proteomic approach as a tool to discrimination of Escherichia coli strains of biotechnological interest. J Appl Biol Biotechnol, 2019; 7:48–54. CrossRef
Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science, 2014; 346:954–9. CrossRef
Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol, 2010; 10: 189. CrossRef
Benedetti VP, Savi DC, Aluizio R, Adamoski D, Kava-Cordeiro V, Galli-Terasawa LV, Glienke C. Analysis of the genetic diversity of Candida isolates obtained from diabetic patients and kidney transplant recipients. Mem Inst Oswaldo Cruz, 2016; 111: 417-422. CrossRef
Bennett, J. 2010. An overview of the genus Aspergillus. In: Aspergillus: Molecular Biology and Genomics, Portland, Caiser Academic Press.
Brasil. 2010. Brazilian Pharmacopeia. 5.ed., v. 1. [Online] Available: http://portal.anvisa.gov.br/documents/33832/2845829/FBrasileira+Volume+1+INGLES+com+alerta.pdf/f9a3480f-b71e-4a30-b459-d07368eaf6ee. [Accessed 2 March 2018].
Cairns TC, Nai C, Meyer V. 2018. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol Biotechnol, 2018; 5: 13. CrossRef
Farber JM, Gendel SM, Tyler KD, Boerlin P, Landry WL, Fritschel SC, Barret TJ. 2001. Molecular typing and differentiation. In Compendium of Methods for the Microbiological Examination of Food, Washington, DC, American Public Health Association. CrossRef
Fatima A, Bashir G, Wani T, Jan A, Kohli A, Khan MS. Molecular identification of Candida species isolated from cases of neonatal candidemia using polymerase chain reaction-restriction fragment length polymorphism in a tertiary care hospital. Indian J Pathol Micr, 2017; 60: 61-65.
Freiwald A, Sauer S. 2009. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat Protoc, 2009; 4: 732-742. CrossRef
Fujita SI, Senda Y, Nakaguchi S, Hashimoto T. Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J Clin Microbiol, 2001; 39: 3617-3622. CrossRef
Gandra EA, Gandra TKV, Mello WSD, Godoi HDS. 2008. Molecular techniques applied to food microbiology/ Técnicas moleculares aplicadas a microbiologia de alimentos. Acta Sci-Technol, 2008; 30: 109-118. CrossRef
Gharaghani M, Taghipour S, Halvaeezadeh M, Mahmoudabadi AZ. Candiduria; a review article with specific data from Iran. Turk J Urol, 2018; 44:445–52. CrossRef
Hertroys R, Van Vught PA, Van De Donk HJ. Moving towards a (microbiological) environmental monitoring programme that can be used to release aseptically produced pharmaceuticals: a hypothesis, a practical programme, and some results. PDA J Pharm Sci Technol, 1997; 51:52–9.
Hierro N, Gonzalez A, Mas A, Guillamon JM. New PCR-based methods for yeast identification. J Appl Microbiol, 2004; 97:792–801. CrossRef
Houbraken J, De Vries RP, Samson RA. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol, 2014; 86:199–249.
Hussong D. Sterile products: advances and challenges in formulation, manufacturing and regulatory aspects—a regulatory review perspective. AAPS PharmSciTech, 2010; 11:1482–4. CrossRef
Intra J, Sarto C, Tiberti N, Besana S, Savarino C, Brambilla P. Genus-level identification of dermatophytes by MALDI-TOF MS after 2 days of colony growth. Lett Appl Microbiol, 2018; 67:136–43. CrossRef
Kordalewska M, Kalita J, Bakula Z, Brillowska-Dabrowska A, Jagielski T. PCR-RFLP assays for species-specific identification of fungi belonging to Scopulariopsis and related genera. Med Mycol, 2018; 57:643–8. CrossRef
Lau AF, Walchak RC, Miller HB, Slechta ES, Kamboj K, Riebe K, Robertson AE, Gilbreath JJ, Mitchell KF, Wallace MA, Bryson AL, Balada-Llasat JM, Bulman A, Buchan BW, Burnham CD, Butler-Wu S, Desai U, Doern CD, Hanson KE, Henderson CM, Kostrzewa M, Ledeboer NA, Maier T, Pancholi P, Schuetz AN, Shi G, Wengenack NL, Zhang SX, Zelazny AM, Frank KM. Multicenter study demonstrates standardization requirements for mold identification by MALDI-TOF MS. Front Microbiol, 2019; 10:2098.
Marcos-Zambrano LJ, Puig-Asensio M, Perez-Garcia F, Escribano, P, Sanchez-Carrillo C, Zaragoza O, Padilla B, Cuenca-Estrella M, Almirante B, Martin-Gomez MT, Munoz P, Bouza E, Guinea J. Candida guilliermondii complex is characterized by high antifungal resistance but low mortality in 22 cases of Candidemia. Antimicrob Agents Chemother, 2017; 61:e00099–17. CrossRef
Mirhendi H, Makimura K, Khoramizadeh M, Yamaguchi H. A one-enzyme PCR-RFLP assay for identification of six medically important Candida species. Nihon Ishinkin Gakkai Zasshi, 2006; 47:225–9. CrossRef
Normand AC, Cassagne C, Gautier M, Becker P, Ranque S, Hendrickx M, Piarroux R. Decision criteria for MALDI-TOF MS-based identification of filamentous fungi using commercial and in-house reference databases. BMC Microbiol, 2017; 17:25. CrossRef
Ojima-Kato T, Yamamoto N, Suzuki M, Fukunaga T, Tamura H. Discrimination of Escherichia coli O157, O26 and O111 from other serovars by MALDI-TOF MS based on the S10-GERMS method. PLoS One, 2014; 9:e113458. CrossRef
Otoupal PB, Ito M, Arkin AP, Magnuson JK, Gladden JM, Skerker JM. Multiplexed CRISPR-Cas9-Based genome editing of Rhodosporidium toruloides. mSphere, 2019; 4:e00099–19. CrossRef
Pacheco FLC, Pinto TJA. The bacterial diversity of pharmaceutical clean rooms analyzed by the fatty acid methyl ester technique. PDA J Pharm Sci Technol, 2010; 64:156–66.
Park HS, Jun SC, Han KH, Hong SB, Yu JH. Diversity, application, and synthetic biology of industrially important Aspergillus fungi. Adv Appl Microbiol, 2017; 100:161–202. CrossRef
Pasqualotto AC, Antunes AG, Severo LC. Candida guilliermondii as the aetiology of candidosis. Rev Inst Med Trop São Paulo, 2006; 48:123–7. CrossRef
Patel R. MALDI-TOF MS for the diagnosis of infectious diseases. Clin Chem, 2015; 61:100–11. CrossRef
Perrone G, Susca A. 2017. Penicillium species and their associated mycotoxins. Methods Mol Biol, 2017; 1542:107–19. CrossRef
Porte L, Garcia P, Braun S, Ulloa MT, Lafourcade M, Montana A, Miranda C, Acosta-Jamett G, Weitzel T. Head-to-head comparison of Microflex LT and Vitek MS systems for routine identification of microorganisms by MALDI-TOF mass spectrometry in Chile. PLoS One, 2017; 12:e0177929. CrossRef
Raja HA, Miller AN, Pearce CJ, Oberlies NH. Fungal identification using molecular tools: a primer for the natural products research community. J Nat Prod, 2017; 80:756–70. CrossRef
Robledo-Leal E, Rivera-Morales LG, Sangorrin MP, Gonzalez GM, Ramos-Alfano G, Adame-Rodriguez JM, Alcocer-Gonzalez JM, Arechiga-Carvajal ET, Rodriguez-Padilla C. Identification and susceptibility of clinical isolates of Candida spp. to killer toxins. Braz J Biol, 2018; 78:742–9. CrossRef
Sandle TA. Review of cleanroom microflora: types, trends, and patterns. PDA J Pharm Sci Technol, 2011; 65:392–403. CrossRef
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium, Fungal Barcoding Consortium Author List. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci USA, 2012; 109:6241–6. CrossRef
Schubert S, Kostrzewa M. MALDI-TOF MS in the microbiology laboratory: current trends. Curr Issues Mol Biol, 2017; 23:17–20. CrossRef
Singh G, Sinha S, Bandyopadhyay KK, Lawrence M, Prasad R, Paul D. Triauxic growth of an oleaginous red yeast Rhodosporidium toruloides on waste 'extract' for enhanced and concomitant lipid and beta-carotene production. Microb Cell Fact, 2018; 17:182. CrossRef
Spier FF, Tacuatiá LO, Agostini G, Eggers L, Chies TTS. Uso de marcadores PCR-RFLP como ferramenta na identificação de espécies da subfamília Iridoideae (Iridaceae) presentes no Parque Estadual de Itapuã, Viamão, RS, Brasil/ Use of PCR-RFLP markers as a tool to identify species of subfamily Iridoideae (Iridaceae) present in Itapuã State Park, Viamão, RS, Brazil. Brazilian J Biosci, 2008; 6:159–65.
USP—United States Pharmacopeia. General Chapter <1116> Microbiological control and monitoring of aseptic processing environments, 2012. [Online] Available via https://www.dcvmn.org/IMG/pdf/usp_1116_em_for_aseptic_processing.pdf (Accessed 2 March 2018).
Vilgalys R. Conserved primer sequences for PCR amplification and sequencing from nuclear ribosomal RNA, 2017. [Online] Available via https://sites.duke.edu/vilgalyslab/rdna_primers_for_fungi/ (Accessed 2 March 2018).
Xavier AREO, Cardoso L, Brito RVJ, Nobre SAM, Almeida AC, Oliveira AME, Xavier MAS. Detection and identification of medically important microorganisms isolated from pigeon excreta collected in a university in a newly industrialized country. Biotemas, 2019; 32:11–20. CrossRef
Xavier MP, Nogueira HS, Xavier MAS, Xavier AREO. Monitoramento microbiológico de áreas grau A e grau B de uma produção asséptica. Rev Unimontes Cient, 2017; 19:112–25.
Xavier MP, Vieira AARM, Silva ASS, Xavier MAS, Xavier AREO. Importância do monitoramento ambiental em áreas classificadas [Importance of environmental monitoring in controlled environments]. Rev Biol e Farmácia, 2013; 9:1–12.
Zhao Y, Tsang CC, Xiao M, Chan JFW, Lau SKP, Kong F, Xu Y, Woo PCY. Yeast identification by sequencing, biochemical kits, MALDI-TOF MS and rep-PCR DNA fingerprinting. Med Mycol, 2018; 56:816–27. CrossRef