INTRODUCTION
Actinobacteria are Gram-positive filamentous microorganisms that occur in diverse habitats both in terrestrial and aquatic environments. They are widely distributed in soils and some are strict saprophytes, but some are parasitic or live in mutualistic associations with plants and animals. These organisms have received more attention in recent years as producers of antibiotics, enzymes, and other proteins (Berdy, 2005; Goodfellow and Williams, 1983; Goodfellow et al., 1988; Inahashi et al., 2011). Recently, actinomycetes were found in other habitats, such as plant tissues (Taechowisan and Lumyong, 2003), root nodules (Trujillo et al., 2006), and the plant roots (Kuncharoen et al., 2019a; 2019b). The present study dealed with the isolation, identification, and antimicrobial activities of actinobacteria from plant rhizosphere soil of plants, including Barringtonia racemosa, Albizzia odoratissima, Spondia spinnata, and Azadirachta indica. at Kokkhumpoon Forest in the Sam Phrao campus, Udonthani Rajabhat University.
MATERIALS AND METHODS
Samples collection and isolation methods
Actinomycete strains were isolated from plant rhizosphere soils of four plants, including B. racemosa, A. odoratissima, S. spinnata, and A. indica. at Kokkhumpoon Forest in the Sam Phrao campus, Udonthani Rajabhat University, Udonthani province, Thailand. The pretreament of soil samples was done using an air dry at room temperature (37°C ± 2) for 2 days. The 10-fold dilution series to 10−3 were prepared using 1 g of soil sample suspended in 9 ml of sterile water. The suspension (0.1 ml) was spread on Humic acid-Vitamin (HV) agar plate (Hayakawa and Nonomura, 1978) and Arginine-Vitamin (AV) agar plate (Nonomura and Ohara, 1969) and then incubated at 30°C for 14 days. Nalidixic acid (25 mg/l) and cycloheximide (50 mg/l) were added into the medium to inhibit other bacteria and fungi, respectively. The colonies of actinomycete isolates were observed using a light microscope and they were purified and cultivated on ISP2 medium. The selected isolates were preserved by freezing at −80°C in 15% (v/v) glycerol solution and by lyophilization.
Identification methods
Phenotypic and genotypic characteristics
The isolates were identified based on morphological, cultural, physiological, and biochemical characteristics, including 16S rRNA gene sequence analyses. Morphological and cultural characteristics of the isolates were observed on the culture grown on ISP2 medium at 30°C for 14 days (Shirling and Gottlieb, 1966). The colour of upper colony, reverse colony, and soluble pigment were observed using the National Bureau of Standards/ the Inter-Society-Color-Council (NBS/ISCC) colour chart (Mundie, 1995). In addition, the spore morphology and aerial or substrate mycelium were observed using a light microscope and scanning electron microscope (JSM-5410LV, Japan).
The DNA was extracted from the cells as described by Tamaoka (1994). The 16S rRNA gene amplification was carried out using two primers 20F and 1500R as reported by Suriyachadkun et al. (2009). The polymerase chain reaction (PCR) product was purified and the nucleotides were sequenced using universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′), 518F (5′-CCAGCAGCCGCGGTAATACG-3′), 800R (5′-TACCAGGGTATCTAATCC-3′), 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) (Lane, 1991) (Macrogen; Seoul, Korea). The results of The Basic Local Alignment Search Tool (BLAST) analysis were assessed using the EzBioCloud server (Yoon et al., 2017).
Antimicrobial activities
The four different production media including 301 medium (2.4 g starch, 0.1 g glucose, 0.3 g peptone, 0.3 g meat extract, 0.5 g yeast extract, 0.4 g CaCO3, 100 ml distilled water, pH 7.0); 54 medium (2 g soluble starch, 0.5 g glycerol, 1 g defatted wheat germ, 0.3 g meat extract, 0.3 g yeast extract, 0.3 g CaCO3, 100 ml distilled water, pH 7.0–7.2); 51 medium (0.5 g glucose, 0.5 g corn steep powder, 1.0 g oatmeal, 1.0 g pharmamedia, 0.5 g K2HPO4, 0.5 g MgSO4.7H2O, 1 ml/l trace metal solution; 100 ml tap water), and Yeast extract-Dextrose (YD) broth (1.0 g yeast extract and 1.0 g dextrose, 100 ml distilled water) (Sripreechasak et al., 2013; 2014) were used. Each isolate was cultivated in YD broth as a seed culture for 4–7 days. The inoculum of seed (0.1 ml) was transferred to 10 ml of each the production medium incubated in a shaker (180 rpm) at 30°C for 7–14 days. The extract solution was collected by centrifugation (3,400 rpm) for 15 minutes after 10 ml of 95% ethanol was added into the culture broth and shaked (180 rpm) for 2 hours.
The antimicrobial activities were performed using agar-disc diffusion method (Qin et al., 2009). The paper disc (8 mm) was soaked into the extract solution of each strain and air-dried. The discs were put onto the surface of the agar plates containing the indicator strains. The antimicrobial inhibition zones (diameter, mm) were determined after incubation for 24 hours. The production medium without the culture was used as the negative control. Four bacterial strains used as the indicator strains are Bacillus subtilis ATCC 6633, Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, Pseudomonas aerogenosa ATCC 27853 and one yeast strain, Candida albicans ATCC 10231. The bacterial strains were cultivated and incubated at 37°C for 24 hours, while the yeast strain was incubated at 30°C for 48 hours.
RESULTS AND DISCUSSION
Isolation and identification of isolates
The isolates JA03, JP09, and JP15 from B. racemosa and KA03, KA26 and KP38 from A. odoratissima while strains MKA22, MKA49, MKA54, MKA56, MKP02, MKP14, MKP30, MKP31, and MKP33 from S. spinnata and SDP02 and SDA19 from A. indica collected in Udonthani province, Thailand were obtained as shown in Table 1. Seventeen isolates were belonged to the genus Streptomyces (10 isolates, Group 1), Micromonospora (5 isolates, Group 2), and 2 isolates were Kitazatospora (Group 3) based on their phenotypic characteristics and 16S rRNA gene sequence analysis as described below (Table 2).
.png) | Table 1. Isolate number, isolation source and cultural characteristics of isolates. [Click here to view] |
.png) | Table 2. Isolate number and nearest relatives of isolates based on 16S rRNA gene similarity (%). [Click here to view] |
Group 1 contained 10 isolates, JA03, JP09, KA03, MKA22, MKA54, MKA56, MKP02, MKP14, SDP02, and SDA19. They produced mature spore chains on ISP2 medium agar. Colonial appearance and scanning electron micrograph of isolates MKA56, MKA54, and SDP02 are shown in Figure 1. Variable characteristics of them were found in growth at pH 4–9 and on 0%–9% (w/v) NaCl, gelatin and starch hydrolysis, milk peptonization and coagulation, and nitrate reduction (Table 3). The isolates JA03, JP09, KA03, MKA22, MKA54, MKA56, MKP02, MKP14, SDA02, and SDA19 were belonged to the genus Streptomyces (Table 2) based on the phylogenetic analysis. Isolate JA03 was closely related to Streptomyces psammoticus NBRC 13971T with 98.95% similarity that might be the novel species. Isolates JP09 and MKP02 were closely related to each other and showed 100% and 99.8% similarity to Streptomyces sioyaensis NBRC 12820T. Isolates KA033 and MKA22 were closely related to each other and showed 99.98% and 99.8% similarity to Streptomyces vietnamensis CCTCCM 205143T. Isolates MKA54 and MKA56 were closely related to each other and showed 99% and 99.62% similarity to Streptomyces bungoensis NBRC 15711T. Isolate MKP14 were closely related to Streptomyces purpurascens NBRC 3389T (99.77%). Isolate SAP02 was closely related to Streptomyces hydrogenans DSM 40586 T (99.62%), while isolate SDA19 was closely related to Streptomyces lucensis DSM 40317T (99.23%). They were identified as S. sioyaensis, S. vietnamensis, S. bungoensis, S. purpurascens, S. hydrogenans, and S. lucensis, respectively (Zhu et al., 2007).
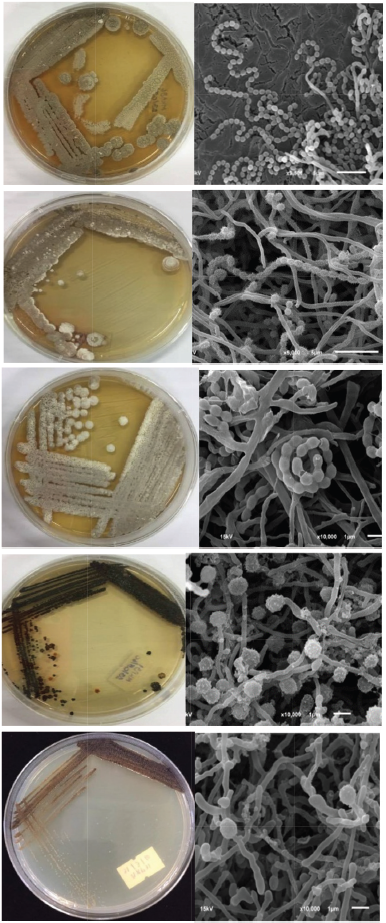 | Figure 1. Colonial appearance and scanning electron micrograph of isolates MKA56, MKA54, SDP02, MKP30, and MKP31. [Click here to view] |
Group 2 contained five isolates, JP15, KP38, MKP30, MKP31, and MKP33 (Table 1). They produced single spore on substrate mycelium on ISP2 medium agar at 30°C for 14 days (Fig. 1). Their colonies color on ISP2 medium were varied from dark grayish yellowish brown to blackish green as described in Table 1. Variable characteristics of them were growth at pH 5–9, on 0%–5% (w/v) NaCl, gelatin and starch hydrolysis, nitrate reduction, milk peptonization, and coagulation (Table 3).
The isolates JP15, KP38, MKP30, MKP31, and MKP33 revealed that they were belonged to the genus Micromonospora based on the phylogenetic analysis (Kawamoto, 1989) (Table 2). Isolate JP15 was closely related to Micromonospora schwarzwaldensis (99.55%), while isolate KP38 was closely related to Micromonospora chersina (99.70%). Isolate MKP30 was closely related to Micromonospora terminaliae (99.70%). Isolate MKP31 was closely related to Micromonospora chaiyaphumensis (99.26%), while isolate MKP33 was closely related to Micromonospora rhizosphaerae (99.05%). They were identified as M. schwarzwaldensis, M. chersina, M. terminaliae, M. chaiyaphumensis, and M. rhizosphaerae, respectively (Gurovic et al., 2013; Kaewkla et al., 2017; Wang et al., 2011).
Group 3 contained two isolates, KA26 and MKA49 (Table 1). They produced mature spore chains on ISP2 medium agar at 30°C for 14 days. Their colonies color on ISP2 medium were pale green and very dark bluish green, respectively (Table 1). The isolates grew at pH 4–9, on 3% (w/v) NaCl, and showed positive reaction for starch hydrolysis, peptonization, nitrate reduction, and milk coagulation (Table 3). Isolates KA26 and MKA49 were closely related to Kitasatospora putterlickiae with 99.26% and 99.85% similarity based on the phylogenetic analysis, respectively. Therefore, they were identified as K. putterlickiae (Groth et al., 2003).
Antimicrobial activities
Streptomyces isolates JA03 and MKA56 exhibited strong antimicrobial activity against C. albicans ATCC 10231, while isolates JP09, MKA54, MKP02, and MKP14 showed strong antimicrobial activity against S. aureus ATCC 25923 and B. subtilis ATCC 6633. Micromonospora sp. MKP30 exhibited strong antimicrobial activity against S. aureus ATCC 25923 and B. subtilis ATCC 6633 while Kitasatospora isolates KA26 and MKA49 exhibited against S. aureus ATCC 25923 and B. subtilis ATCC 663. Only isolates MKA22, MKA56, and KA26 exhibited strong antimicrobial activity against E. coli ATCC 25922. Fourteen strains did not showed antibacterial activity to P. aeruginosa ATCC 27853. The detailed antimicrobial activities are presented in Table 4.
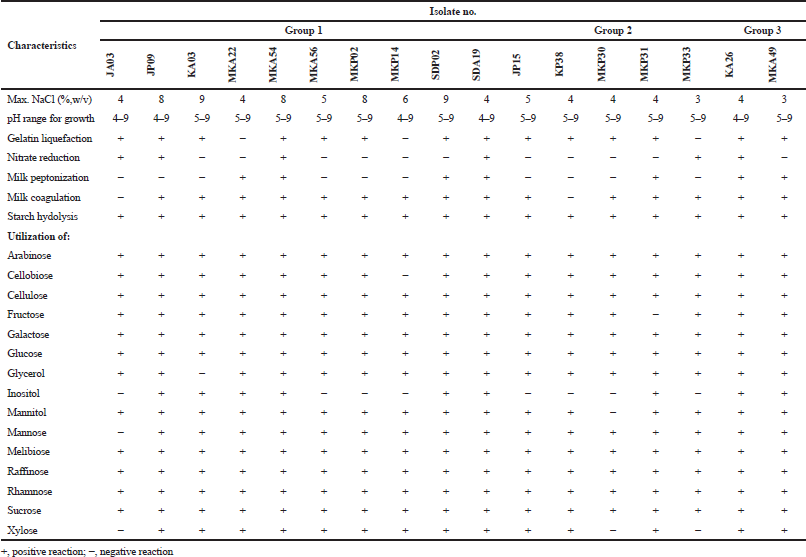 | Table 3. Phenotypic characteristics of isolates. [Click here to view] |
In Thailand, a large number of Streptomyces, Micromonospora, and Kitasatospora strains were isolated from soils (Anansiriwattana et al., 2006; Sripairoj et al., 2008; Sripreechasak et al., 2013; 2014). In addition, the strains of Streptomyces and Micromonospora, including the novel species of Micromonospora azadirachtae and Micromonospora globbae strains from plant roots are reported (Kuncharoen et al., 2018; 2019a; 2019b). In this study, we found diverse species of Streptomyces including S. psammoticus, S. sioyaensis, S. vietnamensis, S. bungoensis, S, purpurascens, S. hydrogenans S. lucensis; Micromonospora, M. schwarzwaldensis, M. chersina, M. chersina, Micromonospora terminalae, M. rhizosphaerae, and K. putterlickiae from rhizosphere soils that showed antimicrobial activities and their secondary metabolites are interesting for further study.
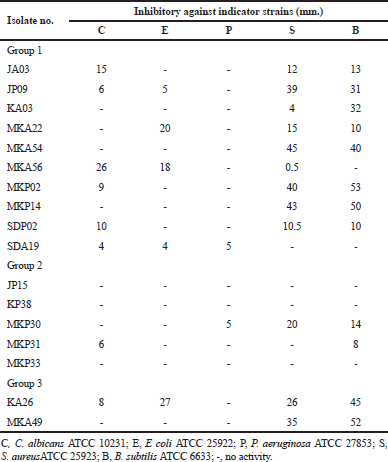 | Table 4. Antimicrobial activity of isolates. [Click here to view] |
CONCLUSION
Actinomycetes were isolated from rhizosphere soil samples collected from four plants including B. racemosa, A. odoratissima, S. spinnata, and A. indica. They were identified as S. psammoticus, S. sioyaensis, S. vietnamensis, S. bungoensis, S. purpurascens, S. lucensis, M. schwarzwaldensis, M. chersina, M. terminalae, M. chaiyaphumensis, M. rhizosphaerae, and Kitasatospora putterlickiae based on 16S rRNA gene sequence analysis. On the antimicrobial activities, the isolates exhibited the good activities against C. albicans ATCC 10231, E. coli ATCC 25922, S. aureus ATCC 25923, and B. subtilis ATCC 6633.
FINANCIAL SUPPORT AND SPONSORSHIP
This research was supported by the Grant of Program of Biology, Faculty of Science, Udonthani Rajabhat University and partially supported by Grant for International Research Integration: Research Pyramid, Ratchadaphiseksomphot Endowment Fund (GCURP_58_01_33_01), Chulalongkorn University.
CONFLICT OF INTERESTS
The authors declared that they have no conflicts of interest.
REFERENCES
Anansiriwattana W, Tanasupawat S, Amnuoypol S, Suwanborirux K. Identification and antimicrobial activities of actinomycetes from soils in Samed Island, and geldanamycin from strain PC4-3. Thai J Pharm Sci, 2006; 30:49–56.
Berdy J. Bioactive microbial metabolites. J Antibiot, 2005; 58:1–26. CrossRef
Goodfellow M, William ST. Ecology of actinomycetes. Ann Rev Microbiol, 1983; 37:189–216. CrossRef
Goodfellow M, Williams ST, Mordarski M. Actinomycetes in biotechnology. Academic Press Inc., London,UK, 1988.
Groth I, Schütze B, Boettcher T, Pullen CB, Rodriguez C, Leistner E, Goodfellow M. Kitasatospora putterlickiae sp. nov., isolated from rhizosphere soil, transfer of Streptomyces kifunensis to the genus Kitasatospora as Kitasatospora kifunensis comb. nov., and emended description of Streptomyces aureofaciens Duggar 1948. Int J Syst Evol Microbiol, 2003; 53:2033–40. CrossRef
Gurovic MS, Müller S, Domin N, Seccareccia I, Nietzsche S, Martin K, Nett M. Micromonospora schwarzwaldensis sp. nov., a producer of telomycin, isolated from soil. Int J Syst Evol Microbiol, 2013; 63:3812–7. CrossRef
Hayakawa M, Nonomura H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Ferment Technol, 1978; 65(5):501–9. CrossRef
Inahashi Y, Matsumoto A, Omura S, Takahashi Y. Streptosporangium oxazolinicum sp. nov., a novel endophytic actinomycete producing new antitrypanosomal antibiotics, spoxazomicins. J Antibiot (Tokyo), 2011; 64(4):297–302. CrossRef
Kaewkla O, Thamchaipenet A, Franco CM. Micromonospora terminaliae sp. nov., an endophytic actinobacterium isolated from the surface-sterilized stem of the medicinal plant Terminalia mucronata. Int J Syst Evol Microbiol, 2017; 67:225–30. CrossRef
Kawamoto I. Genus Micromonospora. In: Williams ST, Sharpe ME, Holt JG (eds.). Bergey’s manual of systematic bacteriology (vol. 4), Williams & Wilkins, Baltimore, MD, 1989.
Kuncharoen N, Fukasawa W, Mori M, Shiomi K, Tanasupawat S. Diversity and antimicrobial activity of endophytic actinomycetes isolated from plant roots in Thailand. Microbiology, 2019a; 88(4):479–88. CrossRef
Kuncharoen N, Kudo T, Ohkuma M, Tanasupawat S. Micromonospora azadirachtae sp. nov., isolated from roots of Azadirachta indica A. Juss. var. siamensis Valeton, Anton van Leeuw. Int J G, 2019b; 112:253–62. CrossRef
Kuncharoen N, Pittayakhajonwut P, Tanasupawat S. Micromonospora globbae sp nov., an endophytic actinomycete isolated from roots of Globba winitii C. H. Wright. Int J Sys Evol Microbiol, 2018; 68:1073–7. CrossRef
Lane DJ. 16S/23S rRNA sequencing. In Strackbrandt E, Goodfellow M (eds.). Nucleic acid techniques in bacterial systematics, Wiley, Chichester, pp 115–48, 1991.
Mundie DA. 1995. The NBS/ISCC Color System/David A. Mundie Pittsburgh, PA: Polymath Systems 535.6 dc-20. Available via: http://www.dodomagnifico.com/Colors/NBS.html (Accessed June 5, 2011).
Nonomura H, Ohara Y. Distribution of actinomycetes in soil. VI. A culture method effective for both preferential isolation and enumeration of Microbispora and Streptosporangium strains in soil. J Ferment Technol, 1969; 47:463–9.
Qin S, Li J, Chen HH, Zhao GZ, Zhu WY, Jiang CL, Xu LH, Li WJ. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl Environ Microbiol, 2009; 75(19):6176–86. CrossRef
Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol, 1966; 16:313–340. CrossRef
Sripairoj P, Tanasupawat S, Suwanborirux K. 16S rDNA sequence analyses and antimicrobial activities of Streptomyces strains from Thai soils. J Health Res, 2008; 22:1–8.
Sripreechasak P, Suwanborirux K, Tanasupawat T. Characterization and antimicrobial activity of Streptomyces strains from soils in southern Thailand. J Appl Pharm Sci, 2014; 4(10):24–31.
Sripreechasak P, Tanasupawat S, Matsumoto A, Inahashi Y, Suwanborirux K, Takahashi Y. Identification and antimicrobial activity of actinobacteria from soils in southern Thailand. Trop Biomed, 2013; 30(1):46–55.
Suriyachadkun C, Chunhametha S, Thawai C, Tamura T, Potacharoen W, Kirtikara K, Sanglier JJ. Planotetraspora thailandica sp. nov., isolated from soil in Thailand. Int J Syst Evol Microbiol, 2009; 59:1632–7. CrossRef
Taechowisan T, Lumyong S. Activity of endophytic actinomycetes from roots of Zingiber officinale and Alpinia galanga against phytopathogenic fungi. Ann Microbiol, 2003; 53(3):291–8.
Tamaoka J. Determination of DNA base composition. In Goodfellow M, O’Donnel AG (eds.). Chemical methods in prokaryotic systematics, Wiley, Chichester, pp 463–70, 1994.
Trujillo ME, Kroppenstedt RM, Schumann P, Carro L, Martinez-Molina E. Micromonospora coriariae sp nov., isolated from root nodules of Coriaria myrtifolia. Int J Syst Evol Microbiol, 2006; 56:2381–5. CrossRef
Wang C, Xu XX, Qu Z, Wang HL, Lin HP, Xie QY, Ruan JS, Hong K. Micromonospora rhizosphaerae sp. nov., isolated from mangrove rhizosphere soil. Int J Syst Evol Microbiol, 2011; 61:320–4. CrossRef
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol, 2017; 67:1613–7. CrossRef
Zhu HH, Guo J, Yao Q, Yang SZ, Deng MR, Phuong le TB, Hanh VT, Ryan MJ. Streptomyces vietnamensis sp. nov., a streptomycete with violet-blue diffusible pigment isolated from soil in Vietnam. Int J Syst Evol Microbiol, 2007; 57:1770–4. CrossRef