INTRODUCTION
Lactic acid-producing bacteria (LAB) are Gram-positive, non-spore forming, non-motile, non-respiring bacteria (Montet and Ray, 2016; Ray, 2020). The various antimicrobial and industrially important compounds produced by these LAB comprise lactic acid (Mayo et al., 2008), acetic acid (Ramsey et al., 2014), ethanol (Ray and Joshi, 2014), formic acid, fatty acids, hydrogen peroxide, and bacteriocin (Vanderbergh, 1993). Bacteriocins are ribosomal synthesized small antimicrobial proteins produced mainly by members of LAB and possess antimicrobial activity toward other bacteria, while synthesizing organisms are resistant to their own bacteriocins (Caulier et al., 2019; Chen and Hoover 2003; Perez et al., 2014). Bacteriocins are reputed as bio-preservatives due to their generally recognized as safe status (Singh, 2018). Bacteriocins are classified into different classes and turned out to be inactive as soon as they were treated with gastrointestinal enzyme in the stomach and were found to be harmless for human consumption (Khandelwal and Upendra, 2019; Khandelwal et al., 2015). Class I bacteriocins named Lantibiotics are bound to the type II lipid of the bacterial membrane which serves as a transporter of N-acetylmuramic acid, N-acetylglucosamine subunits of peptidoglycan layer from bacterial cytoplasm to its cell wall. This action prevents the synthesis of the bacterial cell wall and promotes cell death. In addition, bacteriocins apportioned in the class II type possess amphiphilic helical structures and insert themselves into to the bacterial membrane and promote depolarization, which in turn leads to the death of the bacterial cell. Class III bacteriocins catalyze the breakdown of the cell wall of Gram-positive bacteria, cause the lysis of bacteria, and promote its death (Tulini, 2014). The human gastrointestinal (GI) tract consists of layers such as mucosa, submucosa, epithelial cell lining, mucus layer, and serosa. Probiotic microorganisms are colonized in the gut of the human GI tract and produce bacteriocins to compete with the sensitive bacteria, hence reducing the load of bacteriocin-sensitive bacteria present at the GI tract. Due to the natural harsh conditions of the human gut, the colonized probiotic bacteria may produce bacteriocins lesser than the minimal inhibitory concentration levels, hence it inhibits the bacterial growth and are not harmful to humans (Dicks et al., 2018).
Bacteriocins, as a probiotic ingredient, exhibit different food applications, such as extend shelf life of food, preservation (Balciunas et al., 2013), control microbial spoilage of beer, wine, alcohol fermentation (Gabrielsen et al., 2014; Kjos et al., 2011), and are also used in antimicrobial packaging film to prevent microbial growth (Malhotra et al., 2015). A bacteriocin named Nisin was approved by the US-Food and Drug Administration as a food preservative and is widely used in canned foods, dairy products, meat products, and alcoholic beverages in more than 50 countries around the world (Barbour et al., 2020; Zhang and Jin, 2015).
Due to the wide use of conventional antibiotics in dealing with human diseases, multidrug resistance (MDR) strains appeared and are a major threat to mankind. To control MDR strains in food and feed products, bacteriocins can be used as antimicrobial substances instead of antibiotics. Bacteriocins are a viable alternative to traditional antibiotics in controlling infections caused by Gram-negative bacteria, i.e., Escherichia coli and Salmonella typhimurium, and Gram-positive bacteria, such as Listeria monocytogenes (Cotter et al., 2012; Helander et al., 1997; Khan et al., 2015). Bacteriocins are used for therapeutic purposes, i.e., atopic dermatitis, abdominal ulcers, and immune deficiency conditions (Perez et al., 2014). Nisin is used in the development of various healthcare products, such as toothpaste and skin care products, and in the treatment of cancer therapy (Mishra et al., 2020; Yang et al., 2014).
Several groups of LAB, i.e., Enterococcus, Oenococcus, Leuconostoc, Lactobacillus, Pediococcus, Lactococcus, and Streptococcus, were reported with bacteriocin-producing abilities (Lorca and de Valdez, 2009). Among these genera, nearly 415 species were reported to be LAB species (Euzéby, 1997; Parte, 2014). Bacteriocins such as pediocin AcH or pediocin PA-1 (Motlagh et al., 1992) isolated from the strains of Pediococcm acidilactici were used in meat and vegetable fermentations (Bhunia et al., 1998). In several food systems, bacteriocins (pediocins) were used successfully to inhibit foodborne pathogens such as L. monocytogenes (Pucci et al., 1988; Yousef et al., 1991). Pediococcus pentosaceus was isolated for the first time in the year 1953 from cucumber fermentation (Costilow et al., 1956). Several investigators proved the bacteriocin-producing abilities of the strain P. pentosaceus (Gutiérrez-Cortés et al., 2018; Svetoslav and Dicks, 2009; Wu et al., 2004; Zommiti et al., 2018).
The biggest challenge in the bioprocess was providing optimal fermentation conditions for the economically feasible bioprocesses (Upendra et al., 2013). Response surface methodology (RSM) is an effective and convenient method for designing experiments, building models, and screening key factors of process conditions (Kar et al., 2009; Upendra and Khandelwal, 2021; Upendra et al., 2015b). RSM employed with the hybrid artificial neural network-genetic algorithm (ANN-GA) will be able to address the nonlinear relationship between the actual and coded factors (Upendra et al., 2014a). The hybrid ANN-GA provides validated results and assesses the genetic fitness of organisms during the process.
In our earlier studies, bacteriocin-synthesizing LAB species, identified from unexplored food sources, were characterized as P. pentosaceus through 16S RNA typing. 16S RNA forward strand sequence was deposited in a nucleotide data bank, i.e., GenBank, of NCBI with issued accession number MF183113 (Upendra et al., 2016a). Scanty research is documented on the optimization of the submerged fermentation (SmF) process for higher bacteriocin yield applying RSM and hybrid ANN-GA. No study was found on the optimization of P. pentosaceus SmF culture for higher bacteriocin yield applying RSM and hybrid ANN-GA. With this lacuna, the aim of the present study is to optimize the conditions of the SmF process for the indigenous cultures of P. pentosaceus to achieve enhanced yield of bacteriocins by applying the RSM and hybrid ANN-GA design models. A full factorial central composite design (CCD) of RSM was used to evaluate the effect of four SmF process variables, such as pH, agitation, sucrose, and peptone, on the yield of bacteriocin. Furthermore, RSM results were validated by applying the hybrid ANN-GA methodology. The study reported a sixfold increase in bacteriocin yield (2.4 mg/l), with respect to the unoptimized process yield (0.4 g/l) for the SmF cultures of P. pentosaceus Sanna 14.
MATERIALS AND METHODS
The chemicals and all the reagents used in the preset study represent analytical grade quality (Merck and Qualigens).
Microorganism
Bacteriocin-producing strains employed in the study, such as P. pentosaceus Sanna 14 strain (GenBank MF183113), were isolated by the same research group (Khandelwal and Upendra, 2019; Khandelwal et al., 2017). Pediococcus pentosaceus LAB culture was grown on Mann Rogassa Sharpe (MRS) agar slants at 37°C with pH adjusted to 6.2 for 18–24 hours (Panda et al., 2009) and completely grown slants were preserved at 4°C for optimization studies (Thirumurugan et al., 2013). The inoculum was prepared on the MRS broth pH 6.2 by inoculating a loop full of microorganisms from a culture plate in aseptic conditions and incubated for 18–24 hours, 37°C at 120 rpm (Zamfir et al., 2000) in the orbital shaker incubator (Remi Pvt. Ltd, Bombay, India).
Response surface methodology (RSM)
Experimental design using CCD of RSM
RSM is a pool of mathematical and modeling tools applied in building an experimental model design to analyze the response impact of multivariable process parameters on the overall process yield (Kar et al., 2009; Upendra et al., 2014b, 2015b). Type of carbon source, type of nitrogen source, pH, temperatures, and agitation of the fermentation process were the most important process parameters influencing the bacteriocin yield (Gautam and Sharma, 2009; Upendra, 2017). The present study developed a four-factor experimental design applying a CCD of RSM with 30 experimental runs using the Design-Expert software version 9.0.0.7 to evaluate the optimum conditions of the four principle bacteriocin SmF process parameters selected from the literature survey (Biswas et al., 1991; Ray, 1995; Senbagam et al., 2013; Upendra et al., 2016b), i.e., pH (4.0–8.0), agitation (120–220 rpm), sucrose (20–40 g/l), and peptone (5–20 g/l). All were taken at a central-coded value considered as zero. It was observed from the literature review that sucrose was evidently the best source for the production of bacteriocin for the culture of P. pentosaceus (Suganthi and Mohanasrinivasan, 2015). The full experimental design layout is discussed in Table 1. Optimization experiments were carried out in batch phases considering the CCD of the RSM design, as shown in Table 1 in the conical flask (250 ml) with 100 ml volume as production media (MRS + optimized trail), along with MRS media alone conical flask as unoptimized process standard. 10% v/v (106 colony forming unit/ml) of culture of P. pentosaceus strain inoculum (Gutiérrez-Cortés et al., 2018) was transferred aseptically to 250 ml of production media (MRS + optimized trail) and unoptimized conical flask (MRS) and incubated at 37°C for the period of 72 hours.
Analysis of RSM optimization studies
The RSM optimized values of bacteriocin production were tested through the analysis of variance (ANOVA) study. A second-order polynomial response equation was applied to give the yield of bacteriocins (Eq. 1) as follows:
β0 + = 1nβiXi + =1nβiXi2 + i =1n j= nβij (1)
where Y is the bacteriocin yield, bo is the intercept, bi is the linear direct effect coefficient, and bij is the interaction effect coefficient. The coded equation is useful for predicting the combined influence of factors by comparing the factor coefficients (Myers and Montgomery, 1995; Upendra and Katta, 2021).
 | Table 1. Comparison of CCD-RSM and ANN results with bacteriocin yield. [Click here to view] |
Downstream processing of bacteriocin
After 72 hours of incubation, the bacteriocins produced were harvested from the spent broth by centrifuging at 10,000 g for 21 minutes at 4°C. Supernatant was treated with solid ammonium sulfate at 50% saturation and stirred at 4°C for 2 hours, centrifuged at 14,000 g for 1 hour at 4°C. The pellets thus obtained were suspended using 25 ml of 0.05 M potassium phosphate buffer (pH 7.0) and used in the estimation of bacteriocin with bovine serum albumin as standard by employing Lowry’s method (de Arauz et al., 2009; Upendra et al., 2016a).
Confirmation of bacteriocins by ATR FTIR
Qualitative determination of purified bacteriocin was achieved by employing the FTIR/Diamond ATR method. The FTIR model used in the present study was FTIR-8400S, Shimadzu brand. ATR was fixed to the FTIR instrument at 45° angle, with a sampling area of 1 mm diameter and a sampling depth of several microns. A salt disk was prepared compressing 10 mg sample and 100 mg of potassium bromide mixture and was placed on the ATR diamond disk. The sample was scanned at 4,000–400 wave numbers (cm−1) for absorbance measurements with 1 cm−1 as resolution (Halami et al., 2011).
Validation by hybrid ANN-GA
Artificial neural network
The CCD of the RSM design-optimized process parameters supporting a higher yield of bacteriocin was compared and validated by applying the multilevel feed forward model of ANN, designed using Neural Network MATLAB (version R 2014a software, USA) statistical software for simulation. The same experimental data of the CCD of RSM design were employed in designing the ANN analysis. The input variables taken were pH (4–8), agitation (120–220 rpm), sucrose (4.0–7.0), and fermentation time (8–14 days). The optimum yield of bacteriocin was used as a target. The data taken for the assessment were divided into three sets, such as training set with 70%, followed by validation (15%) and test (15%) datasets (Upendra et al., 2015a). The validation studies were carried out using the Levenberg–Marquardt algorithm consisting of trainlm training function. Assessed variables and response data were kept between 0 and 1 to reduce the network error. The normalization equation applied was as follows (Eq. 2):
Ya = Yi – YminYmax – Ymin (2)
where Yn, Ya, Ymin, and Ymax are normalized value, actual value, minimum value, and maximum value, respectively.
Genetic Algorithm (GA)
The genetic algorithm (GA) is a stochastic-based global optimizing evolutionary algorithm built on the principle of survival of the fittest theory proposed by Darwin.
The design follows five simple steps such as population, representation, variation, selection, and reproduction (Pasandideh and Niaki, 2006). GA was developed using MATLAB (version R 2014a software, USA). The ANN model employed was used to assess the fitness of GA design. At each step, the algorithm uses the individuals in the current generation to create the next population and screens the probable occurrence of variation on the population and accesses the genetic fitness of the organisms when exposed to the optimized conditions of the process (Peng et al., 2014), using Equation 3 as follows:
YWeight0 21 + e − 1 + hidden layer bias bH(3)
RESULTS AND DISCUSSION
Response surface methodology (RSM)
Experimental design using the CCD of RSM
UV spectrophotometric estimated values of extracted bacteriocin (mg/l) are discussed in Table 1. The results of the CCD of RSM experiments studied four independent variables of bacteriocin production, which are presented in Table 1. Based on these results, a quadratic polynomial equation was established to screen the correlation between bacteriocin yield and the studied process variables (Table 1). The final equation in terms of coded factors represents the yield of bacteriocin (Eq. 4) as follows:
Y (mg/l) = + 1.48 + 0.20* A + 0.25* B + 0.17* C + 0.27* D + 0.19* AB−0.063* AC + 0.13* AD−0.012* BC + 0.17* BD + 0.18* CD−0.21* A^2−0.31* B^2−0.16* C^2−0.23* D^2 (4)
where Y represents the bacteriocin yield (mg/l), A denotes pH, B represents agitation (rpm), C is the sucrose (g/l), and D specifies peptone (g/l). The specified equation is used in measuring the final bacteriocin yield. The values of coded factors were kept between high (+1) and low (−1) levels.
Statistical analysis of RSM optimization studies
The experimental values with respect to predicted values are compared in Table 1. The high F-value (157.89) denotes that the employed model was significant, with only 0.73% chance for the influence of noise in the model. The coefficient values and p-values discussed in Table 1 denote the mutual interaction between the coefficients. Lesser p-values suggest more impact of assessed factors on the final output (Senbagam et al., 2013). P-values in Table 2 specify that the coefficients of A, B, C, D, (A2), and (B2), all the quadratic coefficients (A2, B2, C2, D2), and five of interaction coefficients, i.e., AB, AD, BC, BD, and CD were found to be highly significant. Only AC was reported be non-significant. F-value of 2.66 indicates a insignificant impact of lack of fit relative to the pure error of the model (0.013).
The response surface graph studied explains the interactive effect of independent variables, i.e., pH, agitation, sucrose, and peptone, on the bacteriocin yield (Fig. 1). Figure 1A shows the response surface interaction between the variables pH and agitation (rpm), while keeping the other two variables (sucrose and peptone) at zero level. The results confirm that the increase in pH (7.0) and agitation (200 rpm) reportedly increased the bacteriocin yield to 1.8 mg/l. Figure 1B shows the effect of pH and peptone on bacteriocin yield, keeping agitation and sucrose at zero level. The graph shows that the maximum bacteriocin production (1.8 mg/l) occurred at pH (7.0) and peptone (20 g/l), which agrees with the model. Figure 1C shows the effect of agitation (rpm) and sucrose on bacteriocin production, keeping pH and peptone at zero level. The graph shows that the maximum bacteriocin production (1.9 mg/l) occurred at agitation (200 rpm) and sucrose (40 g/l) level. Figure 1D shows the outcome of agitation (rpm) and peptone on bacteriocin yield, with pH and sucrose at zero level. The graph explains that maximum bacteriocin yield (1.8 mg/l) measured at agitation (200 rpm) and peptone (20 g/l). Figure 1E shows the effect of agitation (rpm) and peptone on bacteriocin yield, considering pH and agitation at zero level. The graph shows that the maximum bacteriocin production (1.8 mg/l) occurred at sucrose (40 g/l) and peptone (20 g/l), which agrees with the model.
 | Table 2. ANOVA table for the response surface quadratic model. [Click here to view] |
The predicted RSM design R2 value (0.9658) was in close agreement with the measured R2 of 0.9933. This implies that more than 99.00% of the variation values for bacteriocin yield were address by the independent variables and the model does not explain only about less than 1.00% of variations. The adequate precision value is used to quantify the ratio of signal to background noise, which is usually greater than 4. The present ratio of 45.389 indicates that a polynomial-based quadratic model exhibits adequate signal; hence, the model directs the design space. The goodness of fit values of the RSM design employed indicates that the experimental output values lie on the 45°, indicating that the RSM design-predicted values are highly similar and express close agreement with the experimental data (Fig. 2). Maximum bacteriocin production was found in the experimental trial 26, whereas minimum in trial 01. RSM-optimized experimental results of pH (7.0), agitation (200), sucrose (40 g/l), and peptone (20 g/l) supported a higher yield (2.4 g/l) of bacteriocin in the SmF process for the culture of P. pentosaceus Sanna 14 (Table 1).
Confirmation of bacteriocins by ATR FTIR
The FTIR chromatogram of bacteriocin denotes peaks observed at 1,514.04 and 1,649.10 cm−1 confirms the presence of amide I and II functional groups, respectively; at 3,567.07, it indicates the occurrence of the free hydroxyl functional group, confirming the presence of peptides hence bacteriocin (Fig. 3). Upendra et al. (2016a) carried out the screening of indigenous strains of LAB species for their ability to produce bacteriocin and the produced bacteriocin in the fermentation broth was extracted as crude and was further purified using ammonium sulfate precipitation method. Purified bacteriocin was analyzed UV spectrophotometrically. The samples and the standard exhibited a peak at 225 nm in the UV spectrophotometer scanning spectra (200–240 nm) and was further confirmed by SDS-PAGE for the presence of low molecular weight proteins [SDS, molecular weight approximately less that 14 kDa (Upendra et al., 2016a)].
Validation by hybrid ANN-GA
Artificial neural network
The comparison of RSM- and ANN-predicted values is discussed in Table 1; the error of 0.005 indicates that the design applied was significant. The simulated value of the bacteriocin yield, predicted by the feed forward model (3.064 mg/g dry matter) of ANN, was in close agreement with the experimental values (3.065 mg/g dry matter) and higher than the predicted value of CCD of RSM (Table 1).
The study used the optimal architecture feed forward neural networks of ANN model topology (Fig. 4A), which possesses three layers of ANN, i.e., input layer consisting of the RSM design suggested optimized trail value; the hidden layer (tansig) has 11 neurons; and the output layer (purelin) has a linearized transfer function. 30 data points (n = 30) were taken to develop the ANN model, in that 70% data were used for training, 15% for testing, and 15% for validation.
In the present model, the training was completed after six iterations (epochs), and the study calculated the mean square error value (0.000466888) of the design (Fig. 4B). Furthermore, a regression-based assessment between ANN design outputs and the experimental received data was carried out and the results indicate the accurate prediction. The experimental data used in the prediction show the correlation coefficient (rr) value of 0.99416 for all data (Fig. 4C) and demonstrate that the established ANN model is significant and can be utilized to predict the optimal topology. The quality of input data was assessed through error histograms. For the present study, the error reported to be between 0.033 and 0.004 indicates that the employed design model is highly significant (Fig. 4D).
 | Figure 1. Three-dimensional graphs representing the linear relationship between the two parameters with respect to bacteriocin yield. [Click here to view] |
Genetic algorithm
The hybrid ANN-GA method was employed to optimize the input values of four variables studied and validated applying CCD of RSM and ANN models, respectively, with the aim of enhancing the final yield of bacteriocin for the SmF cultures of P. pentosaceus Sanna 14. The GA program was implemented in MATLAB (version R 2014a software, USA). The following expression was utilized to analyze the fitness assessment of an individual (solution) in a population:
εj = 1-1 J = 1,2...N
In this equation, εj represents the fitness score of the jth solution and yjpred defines lovastatin yield predicted by design model employed in response to the given candidate solution.
The optimum solution for the screened process was achieved by recapitulating the optimized process conditions for different GA input variable conditions. GA inputs of the previous literature reported that the solution must be a global optimal solution (Verma et al., 2014). The best fitness plot accessed during the analysis after 50 generations explains the steady progression of the results with respect to the optimal solution. The sum of mutations declines along with the average distance measures between individuals, which is nearly 0 for the final generation (Fig. 5A). The working model of the GA is shown in Figure 5B. The GA design assessment stops once the maximum generation value is attained (50). The maximum time limit measured in seconds and the results shown in the Figure 5B explain that 100% criteria were met. The selection function of GA is shown in Figure 5C. Fitness values at each generation is shown in Figure 5D; the vertical line at individual generations was smallest to the largest fitness value range; fitness measures indicate that the quantity of mutations declines. These plots represent that the dipping mutation values reduce the diversity rate of successive generations. GA reported that the optimal set of factors studied, i.e., pH (7.0), agitation (200 rpm), sucrose (40 g/l), and peptone (20 g/l), were found to influence the enhanced yield (2.4 g/l) of bacteriocin. The yield of bacteriocin achieved during the SmF process conditions was found to exactly match with the hybrid ANN-GA prediction.
 | Figure 2. Predicted versus actual values for the RSM design. The graph shows a linear relationship between predicted and actual values. The optimum bacteriocin yield is 2.4 mg/l. [Click here to view] |
 | Figure 3. Attenuated total reflectance-Fourier transform infrared spectroscopy spectra of extracted bacteriocin (Trail 26). [Click here to view] |
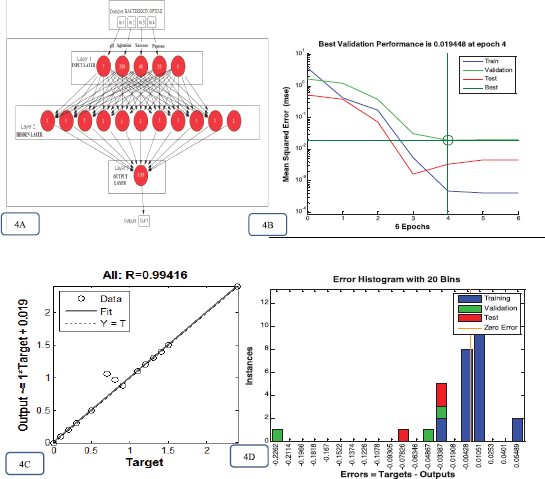 | Figure 4. Bacteriocin optimization process for ANN graphs. (A) Topology of the feed forward design of ANN with input, hidden, and output layer. The optimum yield obtained using Neuroph was 2.33~2.4. (B) MSE of the ANN model design measured during the phase of training. (C) ANN regression plots for the SmF bacteriocin yield and testing network. (D) ANN model Error histogram plots for the bacteriocin yield of the SmF process. [Click here to view] |
In the present study, the bacteriocin yield was optimized using biostatistical tools, namely RSM and ANN-GA. The optimized yield was found to be 2.4 mg/l, which showed a sixfold increase from the unoptimized bacteriocin yield (0.4 mg/l). The validation was carried out by artificial neural network (MATLAB). The ANN-predicted values and RSM-predicted values were compared, which showed an error of 0.005, and the fitness criteria of P. pentosaceus Sanna 14 were carried out using the GA and it was found that the organism is stable for 50 generations. Thirumurugan et al. (2015) optimized Lactobacillus plantarum using a statistical design, which was reported to be 5.75-fold lesser than the present study. Zhou et al. (2008) optimized the media composition for Nisin fermentation and reported a fourfold decrease than the present studies. Zommiti et al. (2018) extensively investigated the genus Pediococci. Research group isolated P. pentosaceus MZF16 strain from dried Ossban a meat products popular in Tunisia and experimented the growth pattern in different conditions such as pH and bile salts. Further probiotic inhibition activity of the P. pentosaceus MZF16 on the selected food spoilage and pathogenic bacteria, i.e., L. monocytogenes, was carried out. Bacteriocin-like compound which is 100% like coagulin was reported and it was concluded that the isolated strains of P. pentosaceus MZF16 proved that pediocins can act as a promising probiotic candidate (Zommiti et al., 2018).
 | Figure 5. Bacteriocin optimization process for GA graphs. (A) GA best fitness graph representing the performance progression of P. pentosaceus Sanna 14. (B) GA graphs representing the stopping criteria. (C) GA graphs representing the selection functions. (D) GA graphs representing the best, worst, and mean scores. [Click here to view] |
Gutiérrez-Cortés et al. (2018) investigated the bacteriocins yield abilities in coculture conditions of selected two LAB species, i.e., P. pentosaceus 147 and L. plantarum LE27. The study tested the coculture of the selected LAB strains on the cheese whey-based liquid media. The study reported 51,200 AU/ml of bacteriocin yield in coculture condition and concluded the potential of using cocultures of strains of the genera Pediococcus and Lactobacillus and using alternative substrates such as cheese whey for the enhanced production of bacteriocins (Gutiérrez-Cortés et al., 2018).
From the present study, it was concluded that the optimized condition of the SmF process of the present investigation, i.e., pH (7.0), agitation (200 rpm), sucrose (40 g/l), and peptone (20 g/l), using P. pentosaceus had shown maximum yield (2.4 g/l) of bacteriocin. Optimized values of these parameters were validated by the feed forward model of ANN and genetic fitness of the process was accessed through GA. The present investigation explored the applications of biostatical tools in the optimization studies and the optimized conditions of the present study raised the bacteriocin yield (2.4 g/l) approximately by a 6.0-fold compared with the yield (0.4 mg/l) of unoptimized SmF process conditions.
ACKNOWLEDGMENTS
The authors wish to express their gratitude to the officials and other staff members who rendered their help during the period of this research work.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
FUNDING
The authors declare that the manuscript was not funded by any external agency.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Balciunas EM, Castillo Martinez FA, Todorov SD, Franco DG de M, Converti A, Oliveira RP de S. Novel biotechnological applications of bacteriocins: a review. Food Control, 2013; 32:134–42. CrossRef
Barbour A, Wescombe P, Smith L. Evolution of lantibiotic salivaricins: new weapons to fight infectious diseases. Trends Microbiol, 2020; 28(7):578–93. CrossRef
Bhunia AK, Johnson MC, Ray B. Purification, characterization, and antimicrobial spectrum of a bacteriocin produced by Pediococctls acidilactici. J Appl Bacteriol, 1988; 65:261–8. CrossRef
Biswas SR, Ray P, Johnson MC, Ray B. Influence of growth conditions on the production of a bacteriocin, pediocin AcH, by Pediococcus acidilactici H. Appl Environ Microbiol, 1991; 57:1265–7. CrossRef
Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon, J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol, 2019; 10:302. CrossRef
Chen H, Hoover DG. Bacteriocins and their food applications. Compr Rev Food Sci Food Saf, 2003; 2:82–100. CrossRef
Costilow RN, Coughlin FM, Robach DL, Ragheb HS. A study of the acid-forming bacteria from cucumber fermentations in Michigan. Food Res Int, 1956; 21:27–33. CrossRef
Cotter PD, Ross RP, Hill C. Bacteriocins – a viable alternative to antibiotics? Nat Rev Microbiol, 2012; 11:95–105. CrossRef
de Arauz LJ, Jozala AF, Mazzola PG, Penna TCV. Nisin biotechnological production and application: a review. Trends Food Sci Technol, 2009; 20(3–4):146–54. CrossRef
Dicks L, Dreyer L, Smith C, & van Staden AD. A review: the fate of bacteriocins in the human gastro-Intestinal tract: do they cross the gut-blood barrier? Front Microbiol, 2018; 9:2297. CrossRef
Euzéby JP. List of bacterial names withstanding in nomenclature: a folder available on the internet. Int J Syst Bacteriol, 1997; 47:590–2. CrossRef
Gabrielsen C, Brede DA, Nes IF, Diep DB. Circular bacteriocins: biosynthesis and mode of action. Appl Environ Microbiol, 2014; 80:6854–62. CrossRef
Gautam N, Sharma N. Bacteriocin: safest approach to preserve food products. Indian J Microbiol, 2009; 49(3):204–11. CrossRef
Gutiérrez-Cortés C, Suarez H, Buitrago G, Nero LA, Todorov SD. Enhanced bacteriocin production by Pediococcus pentosaceus 147 in co-culture with Lactobacillus plantarum LE27 on cheese whey broth. Front Microbiol, 2018; 9:2952. CrossRef
Halami PM, Badrinath V, Manjulata devi S, Vijayendra SVN. Partial charecterization of heat stable, antilisterial and cell lytic bacteriocin of Pediococcus pentosaceus CFR SIII isolated from a vegetable source. Ann Microbiol, 2011; 6:323–30. CrossRef
Helander IM, von Wright A, Mattila-Sandholm TM. Potential of lactic acid bacteria and novel antimicrobials against Gram-negative bacteria. Trends Food Sci Technol, 1997; 8:146–50. CrossRef
Kar S, Swain MR, Ray RC. Statistical optimization of alpha-amylase production with immobilized cells of Streptomyces erumpens MTCC 7317 in Luffa cylindrica L. sponge discs. Appl Biochem Biotechnol, 2009; 152:177–88. CrossRef
Khan A, Vu KD, Riedl B, Lacroix M. Optimization of the antimicrobial activity of nisin, Na-EDTA and pH against gram-negative and gram-positive bacteria. LWT – Food Sci Technol, 2015; 61(1):124–9. CrossRef
Khandelwal P, Gaspar FB, Crespo MTB, Upendra RS. Lactic acid bacteria general characteristics, food preservation and health benefits. In: D Montet, RC Ray (eds.). Lactic acid bacteria. Fermented foods, part I: biochemistry and biotechnology, CRC Press, Boca Raton, FL, pp 127–47, 2015. CrossRef
Khandelwal P, Upendra RS. Nanotechnology and bacteriocins: perspectives and opportunities. In: HAR Suleria, LK Dasarahally-Huligowda, MR Goyal (eds.). Nanotechnology applications in dairy Science. Apple Academic Press, Waretown, NJ, pp 187–224, 2019. CrossRef
Khandelwal P, Upendra RS, Aishwarya Rani BR, Shivani R, Kruthika Prasanna B, Sai Rakshita M. Studies on Bacteriocin producing abilities of indigenously isolated lactic acid bacteria strains. Int J Fermented Foods, 2017; 6(1):1–10. CrossRef
Kjos M, Borrero J, Opsata M, Birri DJ, Holo H, Cintas LM, Snipen L, Hernandez PE, Nes IF, Diep DB. Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology, 2011; 157(12):3256–67. CrossRef
Lorca GL, de Valdez GF. Lactobacillus stress responses. In: Å Ljungh, T Wadström (eds.). Lactobacillus molecular biology: from genomics to probiotics. Caister Academic Press, Norfolk, UK, pp 115–38, 2009. CrossRef
Malhotra B, Keshwani A, Kharkwal H. Antimicrobial food packaging: potential and pitfalls. Front Microbiol, 2015; 6:611. CrossRef
Mayo B, Van Sinderen D, Ventura M. Genome analysis of food grade lactic acid- producing bacteria: from basics to applications. Curr Genomics, 2008; 9(3):169–83. CrossRef
Mishra S, Rath S, Mohanty N. Probiotics-A complete oral healthcare package. J Integr Med, 2020; 18(6):462–9. CrossRef
Montet D, Ray RC. Fermented foods, Part I: biochemistry and biotechnology. CRC Press, Boca Raton, FL, 2016. CrossRef
Motlagh AM, Bhunia AK, Szostek F, Hansen TR, Johnson MC, Ray B. Nucleotide, and amino acid sequence of PAP gene pediocin AcH production in Pediococctls acidilactici H. Lett Appl Microbiol, 1992; 15(2):45–8. CrossRef
Myers RH, Montgomery DC. Response surface methodology: process and product optimization using designed experiments. 1st edition, John Wiley & Sons, Inc, New York, NY, 1995. CrossRef
Panda SH, Naskar SK, Sivakumar PS, Ray RC. Lactic acid fermentation of anthocyanin-rich sweet potato (Ipomoea batatas L.) into lacto-juice. Int J Food Sci Technol, 2009; 44(2):288–96. CrossRef
Parte AC. LPSN–list of prokaryotic names withstanding in nomenclature. Nucleic Acids Res, 2014; 42:613–9. CrossRef
Pasandideh SHR, Niaki STA. Multi-response simulation optimization using genetic algorithm within desirability function framework, Appl Math Comput, 2006; 175:366–82. CrossRef
Peng W, Zhong J, Yang J, Ren Y, Xu T, Xiao S, Zhou J, Tan H. The artificial neural network approach based on uniform design to optimize the fed-batch fermentation condition: application to the production of iturin A. Microb Cell Fact, 2014; 13(1):1–0.
Perez RH, Zendo T, Sonomoto K. Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb Cell Fact, 2014; 13(1):1–3.
Pucci MJ, Vedarnuthu ER, Kunka B, Vandenbergh PA. Inhibition of Listeria monocytogenes by using bacteriocin PA-1 produced by Pediococcus acidilactici PAC 1.O. Appl Environ Microbiol, 1988; 54:2349–53.
Ramsey M, Hartke A, Huycke M. The physiology and metabolism of Enterococci. In: Gilmore MS, Clewell DB, ike Y, Shankar N, (eds.), Enterococci: from commensals to leading causes of drug resistant infection [Internet]. Massachusetts Eye and Ear Infirmary, Boston, MA, p 106, 2014.
Ray B. Pediococcus in fermented foods. In: Hui YH, Khachatourians GG (eds.). Food biotechnology. Wiley-VCH, New York, NY, pp 745–95, 1995.
Ray RC. Microbial biotechnology in food and health. Academic Press, Cambridge, MA, 2020.
Ray RC, Joshi VK. Fermented foods: past, present and future scenario. In: Ray RC, Montet D (eds.). Microorganisms and fermentation of traditional foods. CRC Press, Boca raton, FL, pp 1–36, 2014.
Senbagam D, Gurusamy R, Senthilkumar B. Physical chemical, and biological characterization of a new bacteriocin produced by Bacillus cereus NS02. Asian Pac J Trop Med, 2013; 6(12):934–41. CrossRef
Singh VP. Recent approaches in food bio-preservation-a review. Open Vet J, 2018; 8(1):104–11. CrossRef
Suganthi V, Mohanasrinivasan V. Optimization studies for enhanced bacteriocin production by Pediococcus pentosaceus KC692718 using response surface methodology. J Food Sci Technol, 2015; 52(6):3773–83. CrossRef
Svetoslav DT, Dicks LMT. Bacteriocin production by Pediococcus pentosaceus isolated from marula (Scerocarya birrea). Int J Food Microbiol, 2009; 132(2):117–26. CrossRef
Thirumurugan A, Ramachandran S, Gobikrishnan S. Optimization of medium components for maximizing the bacteriocin production by Lactobacillus plantarum ATM11 using statistical design. Int Food Res J, 2015; 22(3):1272.
Thirumurugan A, Ramachandran S, Shiamala Gowri A. Combined effect of bacteriocin with gold nanoparticles against food spoiling bacteria-an approach for food packaging material preparation. Int Food Res J, 2013; 20(4):1909–12.
Tulini FL. Isolation of lactic acid bacteria from milk and cheese with potential for food biopreservation and utilization for increasing whey digestibility. Diss. Doctoral thesis, Universidade de São Paulo (USP). Faculdade de Ciências Farmacêuticas de Ribeirão Preto Ribeirão Preto, Ribeirão Preto, Brazil, 2014.
Upendra RS. Biotechnological interventions for production optimization and downstream processing (DSP) of medicinally important secondary metabolites from natural & standard fungal cultures. Doctoral dissertation, Ph. D Thesis. Visvesvaraya Technological University, Belgaum, India, 2017. CrossRef
Upendra RS, Katta SS. Biosimulation studies-based optimization of fungal derived lovastatin. Int J Pharm Res, 2021; 13(1);400–7.
Upendra RS, Khandelwal P. Recent advancements in fermentation studies for lovastatin biosynthesis. In: Ray RC (ed.). Microbial biotechnology in food and health. Academic Press, Cambridge, MA, pp 251–88, 2021.
Upendra RS, Khandelwal P, Akshitha N, Chakraborty I, Monis SA, Shruthi NA. Optimization of submerged state fermentation process for terreic acid production applying biostatistical tools. Int J Pharm Sci Res, 2016b;7(3):144–51.
Upendra RS, Khandelwal P, Amiri ZR, Achar A, Kumari BG, Sowmaya M, Tejaswini J. Optimization of fluoride removal system using Ocimum sp. leaves and ragi seed husk by applying bio-statistical tools. J Environ Res Dev, 2015b; 9(4):109–116.
Upendra RS, Khandelwal P, Amiri ZR, Banu R, Barade A, Veena K, Gayathri V, Yamini DE. Artificial neural network: a novel method for optimization of bioproducts and bioprocesses: a critical review. MSR J Sci, 2014a; 1(1):21–34.
Upendra RS, Khandelwal P, Amiri ZR, Nair BM. Invited session key lecture on “Optimization of process for producing lovastatin from novel fungal isolates”. In Proceedings of the IV International Conference on SUTPBM. New Delhi Publishers (ND Publishers), New Delhi, India, pp 28–34, 2014b
Upendra RS, Khandelwal P, Ausim MS. A novel approach for enhancement of lovastatin production using Aspergillus species. Int J Agric Environ Biotech, 2013; 6(4S):779.
Upendra RS, Khandelwal P, Jana K, Ajay Kumar N, Gayathri Devi M, Stephaney ML. Bacteriocin production from indigenous strains of lactic acid bacteria isolated from selected fermented food sources. Int J Pharm Res Health Sci, 2016a; 4(1):982–90.
Upendra RS, Khandelwal P, Veeresh AV. Application of artificial neural network statistical design (ann) in enhanced production of biopharmaceuticals. Int J Comput Eng Technol, 2015a; 6(3):46–52.
Vanderbergh PA. Lactic acid bacteria, their metabolic products and interference with microbial growth. FEMS Microbiol Rev, 1993; 12:221–38. CrossRef
Verma DK, Hasan SH, Singh DK, Singh S, Singh Y: Enhanced biosorptive remediation of hexavalent chromium using chemotailored biomass of a novel soil isolate Bacillus aryabhattai ITBHU02: process variables optimization through artificial neural network linked genetic algorithm. Ind Eng Chem Res, 2014; 53(9):3669–81. CrossRef
Wu CW, Yin LJ, Jiang ST. Purification, and characterization of bacteriocin from Pediococcus pentosaceus ACCEL. J Agric Food Chem, 2004; 52(5):1146–51. CrossRef
Yang SC, Lin CH, Sung CT, Fang JY. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol, 2014; 5:241. CrossRef
Yousef AE, Luchansky JB, Degnan A, Doyle MP. Behaviour of Listeria monoytogenes in weiner exudates in the presence of Pediococcus acidilactici H or pediocin AcH during storage at 4 or 25oC. Appl Environ Microbiol, 1991; 57:1461–7. CrossRef
Zamfir M, Callewaert R, Cornea PC, De Vuyst L. Production kinetics of acidophilin 801, a bacteriocin produced by Lactobacillus acidophilus IBB 801. FEMS Microbiol Lett, 2000; 190(2):305–8. CrossRef
Zhang J, Zhong J. The journey of nisin development in China, a natural-green food preservative. Protein Cell, 2015; 6(10):709–11. CrossRef
Zhou XX, Pan YJ, Wang YB, Li WF. Optimization of medium composition for nisin fermentation with response surface methodology. J Food Sci, 2008; 73(6):245–9. CrossRef
Zommiti M, Bouffartigues E, Maillot O, Barreau M, Szunerits S, Sebei K, Feuilloley M, Connil N, Ferchichi M. In vitro assessment of the probiotic properties and bacteriocinogenic potential of Pediococcus pentosaceus MZF16 isolated from artisanal Tunisian meat “Dried Ossban”. Front Microbiol, 2018; 9(9):2607. CrossRef